
Review Article
Austin Alzheimers J Parkinsons Dis. 2014;1(3): 10.
The Role of MicroRNA in the Pathogenesis and Diagnosis of Neurodegenerative Diseases
Michal Prendecki and Jolanta Dorszewska*
Department of Neurology, Poznan University of Medical Sciences, Poland
*Corresponding author: Jolanta Dorszewska, Laboratory of Neurobiology, Department of Neurology, Poznan University of Medical Sciences, 49 Przybyszewskiego St, PL 60-355 Poznan, Poland.
Received: October 16, 2014; Accepted: November 09, 2014; Published: November 10, 2014
Abstract
Today, Neurodegenerative Diseases (NDs) constitute one of the most significant issues in public healthcare. One of these NDs, Alzheimer's disease (AD), affects more than 24 million people worldwide. Scientists all over the world are searching for biomarkers that are vital for ND pathogenesis and diagnosis. It seems that one of these promising biomarkers might be microRNA (miRNA), whose biosynthesis is understood quite well.
Currently known human miRNAs (~2600) are involved in numerous physiological and pathological processes. Recent studies have sought specific miRNAs that are significant for the pathogenesis and diagnosis of NDs. Most miRNAs are common for many NDs, however, few seem to be specific to individual diseases: AD (e.g. let-7f, miR-125b, -193b), Parkinson's disease (e.g. miR-19b, -34b/c, -133b), and frontotemporal dementia (e.g. miR-132, -212).
It seems that finding specific miRNAs for individual NDs may contribute to early and certain diagnosis and to introducing effective therapy.
Keywords: miRNA; Neurodegeneration; Alzheimer's disease; AD; Parkinson's disease; PD
Abbreviations
3'-UTR: 3'-Untranslated Region of mRNA; AD: Alzheimer's disease; APP: Amyloid Precursor Protein; ASN: α-synuclein; Aβ: Beta-Amyloid; BACE1: Beta-Site APP Cleaving Enzyme 1; CNS: Central Nervous System; DNs: Dopaminergic Neurons; FTD: Frontotemporal Dementia; LRRK2: Leucine-Rich Repeat Kinase 2; MCI: Mild Cognitive Impairment; miRISC: miRNA-Induced Silencing Complex; miRNA: microRNA; MMSE: Mini-Mental State Examination; ncRNA: Noncoding RNA; NDs: Neurodegenerative Diseases; nt: Nucleotides; PD: Parkinson's disease; pre-miRNA: Premature microRNA; pri-miRNA: Primary microRNA; ROS: Reactive Oxygen Species; RT-qPCR: Real Time Quantitative PCR
Introduction
Neurodegenerative Diseases (NDs), including Alzheimer's disease (AD) and Parkinson's disease (PD), are a non-homogeneous group of various disorders affecting the Central Nervous System (CNS), mostly by excessive apoptosis of neurons in diverse locations of the human brain. Both the region and neuron types that are affected determine the nature of the cognitive, behavioral and motor deficits, which are fairly specific to each disease. Moreover, the significant heterogeneity of clinical symptoms of NDs makes a definitive diagnosis feasible only upon a postmortem histopathological examination of brain tissue.
NDs have become one of the most significant public health issues in recent years, since 24 million people are suffering from AD worldwide and the number of patients is expected to double in the next 15 years [1]. Yet current knowledge is still incomplete regarding the genetic basis underlying changes taking place on both a molecular and cellular level. As a consequence, there are no available therapies that would effectively modify the disease, and contemporary diagnostic tools are not able to detect early changes that take place for years prior to the actual symptoms, which manifest themselves when most of the brain damage has already been done [2].
Attempts to explain the pathogenesis of neurodegenerative diseases via genetic mutations were not entirely fruitful and researchers realized that there must be another level of neuronal homeostasis regulation [2]. Another piece of the puzzle was uncovered along with sequencing of the human genome, when it turned out that more than 95% of human cellular RNAs are noncoding RNAs (ncRNAs) [3]. These small molecules seem to be plentiful in the human brain and in other parts of the nervous system; later on it was shown that they control its proper function and development [4]. Although previously underappreciated, ncRNAs have proved to be pivotal in degenerative processes, hence neurodegeneration may be regarded as an "RNA disorder" where one class of ncRNAs, namely microRNAs (miRNAs), seems to play the leading role [2].
miRNAs are a conserved group of short (about 22 nucleotides (nt)), single-stranded RNA molecules [5] that play a significant role in "fine-tuning" gene expression by semi-complementary hybridizing to mRNA and suppressing its effective translation [6]. Since their discovery in 1993 by Lee et al. [7], miRNAs have become one of the mostly researched groups of small noncoding nucleic acids. There were over 35 thousand records on PubMed as of September 2014 (www.pubmed.org), and information on miRNAs is being gathered in specialized databases, e.g. at www.mirbase.org, whose most recent version (21st edition; June 2014) contains information on nearly 2600 sequences of mature human miRNAs.
To select relevant studies for this review, the authors conducted multiple searches through public databases, including PubMed, Scopus and Google Scholar, by using the following search strategy: ("Alzheimer's disease" or "AD", or "Parkinson's disease" or PD, or "Frontotemporal dementia" or "FTD") and ("microRNA" or "miRNA") and ("biomarker" or "SNP", or "genetic polymorphism" or "mutation"). The last search was performed in September of 2014. A subsequent search through review articles and references facilitated finding additional eligible studies.
Role of miRNA in physiological conditions
Biosynthesis of miRNA
The first step of miRNA biosynthesis is RNA polymerase IImediated transcription of primary miRNA (pri-miRNA) either from independent miRNA genes or the introns of protein-coding mRNAs [8]. Similarly to coding mRNAs, pri-miRNAs are polyadenylated and their expression is regulated by transcription factors [9].
pri-miRNAs tend to fold into secondary structures containing imperfectly base-paired stem loops which are subsequently cleaved into about 70-nucleotide hairpins of premature-miRNAs (premiRNAs) by the nuclear complex RNase III type endonuclease Drosha and the DGCR8 protein [8]. Alternative pathways have also been described, e.g. pre-miRNAs may bypass the Drosha/DGCR8 step and be synthesized from very short introns (mirtrons) as a result of debranching or splicing [8,10].
pre-miRNAs, after processing by the Drosha complex, are transferred through the nuclear membrane to the cytoplasm via a Ran-GTP-dependent mechanism by Exportin-5 [8]. Outside the nucleus, pre-miRNAs are cleaved close to the terminal loop by the second RNase III-type enzyme, a complex of Dicer and its cofactor TAR RNA-binding protein 2, thus giving RNA duplexes of roughly 22 nt [8]. The newly created short RNA duplexes bind to a glycinetryptophan repeat-containing protein of 182 kDa and an argonaute protein forming the miRNA-Induced Silencing Complex (miRISC) [11]. Next, one of the two strands, the so-called "passenger miRNA" (also known as "complementary star-form miRNA", "miRNA*" or "miRNA-3p"), is released, while the other strand, designated as the "guide strand", "mature miRNA", or "miRNA-5p", remains within miRISC [12,13]. Current studies have implied that both arms (3' and 5' for -3p and -5p, respectively [14]), of the pre-miRNA hairpin can give rise to mature miRNAs [15].
Full miRISCs recognize target mRNAs by hybridizing the seed region of miRNA (between the 2nd and 8th nt of the miRNA) to the complementary region in the 3' untranslated region of mRNAs (3'- UTR) [16-18]. Bounding miRISCs to mRNAs inflicts translational repression [11].
Current studies have highlighted that the down-regulation effects of miRNAs/miRISCs occur mostly through mRNA degradation rather than translational repression [19]. Moreover, recent data show that efficient repression requires the presence of typically > 100 copies of miRNA per cell [20]. Hence, poorly expressed miRNAs may play little or no part in adjusting gene expression [21]. Additionally, molecular sponges may bind free miRNA and prevent hybridization to mRNA targets [22,23].
Various functions of miRNA
miRNA research on the mammalian brain started in 2003, when Krichevsky et al. conducted microarray studies and showed significant changes in miRNA levels during brain development [24]. Next-generation sequencing as used by Landgraf et al. in 2007 constituted another leap which allowed identifying differences inmiRNA expression in various cell types and parts of the brain [25].
Today, miRNAs are believed to take part in both neuronal and brain development as well as in many physiological processes. The role of miRNA in developing neurons was proven by Yoo et al. in 2011. They discovered that miR-9* and miR-124 induce compositional changes of SWI/SNF-like BAF chromatin-remodeling complexes and control multiple genes regulating neuronal differentiation and function. They showed that expression of miR-9/9* and miR-124 in human fibroblasts induced (further augmented by Neurogenic differentiation factor 2) conversion into neurons. In their experiment the addition of ASCL1 and MYT1L transcription factors enhanced the speed of conversion and differentiation but was not alone-sufficient to trigger the conversion [26].
In physiological conditions, miRNAs act as expression controllers and are responsible for maintaining proper levels of various proteins in cells [27]. miRNA genes are not translated into proteins, instead they usually bind with the 6-nucleotide long semi-complementary seed sequence to the 3'-UTR and sporadically to the 5'-UTR or coding regions of target mRNAs [19], thus inducing gene silencing or, rarely, over-expression [23]. Bioinformatic analyses show that miRNAs may regulate the expression of over 60% of all human proteincoding genes [28,29]. MiRNAs are involved in countless biological processes, such as development, differentiation, and growth [6,15,30]. What has been shown lately is that a single miRNA molecule interacts with numerous mRNAs, and mRNA expression may be regulated by various miRNAs, thus starting a huge net of co-interactions [31-33]. miRNAs are mostly considered to "fine-tune" gene expression and to regulate development and tissue homeostasis [34].
It has been found that miRNAs are also involved in synaptic plasticity, as shown by Gao et al. They investigated SIRT1, sirtuin 1, a gene having systemic roles in cardiac function, DNA repair and genomic stability. Recent studies suggest that SIRT1 plays a role in normal brain physiology as well as in neurological disorders. Gao et al. also found that proper activity of SIRT1 increases, whereas its lossof- function impairs synaptic plasticity via a microRNA-mediated mechanism involving CREB and miR-134. They also showed that SIRT1 limits the expression of miR-134 via a repressor complex containing transcription factor YY1. Overexpression of miR-134 has been shown to lower the levels of CREB and the brain-derived neurotrophic factor, thus impairing synaptic plasticity, which is the key mechanism controlling memory [35].
Other examples of miRNA involved in the function and development of neurons and the CNS are: miR-9 - responsible for neuronal differentiation, formation of the cortex, neurogenesis and brain development; miR-124 - controlling serotonin synaptic facilitation, neuritis development, neuron differentiation; miR-125b - adjusting spine width, dendritic branching and weakening synaptic transmission; miR-132 - inducing neural outgrowth, regulating dendritic complexity, spine width, stimulating synaptic transmission; miR-137 - inhibiting spine development and maturation as well as inducing proliferation of neuronal progenitor cells; miR-138 - regulating the size of the spine; miR-375 - repressing the density of dendrites; and miR-379/410 cluster - inhibiting dendritic outgrowth [36]. Further miRNAs, such as let-7 and miR-9, have been described as stimulating differentiation, while miR-25 has been shown to induce the proliferation of neural stem cells [37].
Various miRNA-mediated mechanisms of neuronal damage in neurodegenerative diseases
miRNAs are probably associated with numerous pathological processes, such as response to oxidative stress, cell cycle disorders, neuroinflammation, clearance of pathological proteins and cholesterol trafficking. All of these contribute to the development of NDs (see Figure 1).
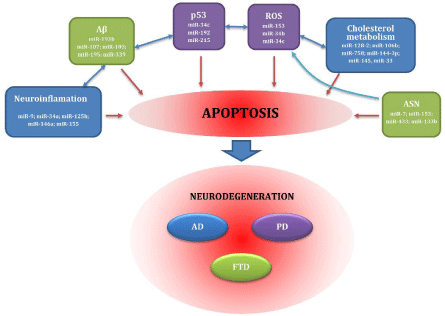
Figure 1: Factors important for pathogenesis of neurodegenerative diseases. Abbreviations: Aβ: β-amyloid; AD: Alzheimer's disease: ASN: α-synuclein; FTD: Frontotemporal Dementia; miR: MicroRNA; p53: Tumor Protein 53; PD: Parkinson's disease; ROS: Reactive Oxygen Species
Oxidative stress responses have been observed in postmortem ND brains and in many rodent mouse models of NDs [38]. Oxidative stress in NDs is the center of a complex interaction network affecting DNA repair, proper protein folding, aggregation and clearance of damaged proteins, membrane lipid peroxidation and calcium homeostasis [39]. Reactive Oxygen Species (ROS) are principally generated by mitochondrial respiration and inflammation, thus creating an association between oxidative stress, mitochondrial dysfunction and neuroinflammatory responses [38]. Recent papers indicate that the role of miRNA is particularly visible in response to physiological as well as pathological stresses [40-42].
A study by Narasimhan et al. showed a connection between ROSdependent miR-153 and NDs. They investigated an environmental toxin, paraquat, which may increase the risk of developing PD by damaging Dopaminergic Neurons (DNs). They performed Real Time Quantitative PCR (RT-qPCR) analysis which revealed that paraquat significantly increased the expression of brain-enriched miR-153 with an associated decrease in nuclear factor Nrf2. The transcription initiator ARE involved in redox balance is activated via binding to Nrf2, which suggests a critical role for ROS-mediated miR-153-Nrf2/ ARE pathway interaction in paraquat neurotoxicity on DNs [43].
The pivotal gene controlling apoptosis, TP53, has been found to be mutated in several cases of AD [44]. Moreover, it has recently been shown that the p53 protein, a transcript of the TP53 gene, regulates the expression of several miRNAs, e.g. the miR-34 family. Ectopic expression of miR-34s inhibits proliferation, migration, invasion and metastasis of various cancer cells. On the other hand, miR-34 plays an important role in aging, stem cell differentiation and neuronal development [45]. The work of Zovoilis et al. identified miR-34c as a negative limit of memory consolidation and showed that miR- 34c levels are elevated in the hippocampus of AD patients and corresponding mouse models. Moreover, blocking miR-34 activity restored learning ability in the studied animals [46]. Braun et al. used array hybridization to show that p53 induces other microRNAs, namely miR-192 and miR-215, thus leading to their up-regulation. Alternatively, miR-192 and miR-215 can each contribute to enhanced cyclin-dependent kinase A1 and cyclin-dependent kinase inhibitor p21 levels, thus promoting neuronal cell colony suppression, cell cycle arrest and increased cell movability. These effects seem to depend on the presence of wild-type p53. miR-192 and miR-215 seem to induce p53-dependent apoptosis and cell cycle arrest through p21 buildup [47].
Zhao et al. have shown that TREM2 is down-regulated in AD, and its 3'-UTR may also be targeted by miR-34a, which was shown to be regulated by NF-κB [48]. TREM2 is thought to outline native immune and phagocytic responses that contribute to inflammatory neurodegeneration [49].
Other NF-κB-sensitive miRNAs have been studied by Alexandrov et al. using a highly sensitive LED-Northern dot-blot assay. They showed that several miRNAs, namely miRNA-9, miRNA-125b, miRNA-146a, and miRNA-155, were present in the cerebrospinal fluid and extracellular fluid of AD patients. They suggested involvement of the studied miRNAs in the modulation of neuronal proliferation in the CNS as well as an association with progressive spreading of inflammatory neurodegeneration [50].
These data suggest that in AD, memory impairment may be mediated by the miR-34/TP53 pathway through excessive neuron apoptosis, but also through miRNA/NF-κB-mediated neuroinflammation.
Microglia, the immune cells of the CNS that protect it against pathogens, play an important role in neuroinflammation. Dysregulation of microglial activity may result in a prolonged proinflammatory state and in subsequent neurodegeneration [51]. In 2013, Jayadev et al. found that miR-146, a negative regulator of the monocyte pro-inflammatory response, is constitutively downregulated in mice microglia with dysfunctional presenilin 2 [52], whose mutations were shown to condition autosomal dominant AD [53,54].
Microglia may also be activated by ROS, initiating the above-mentioned transcription factor p53 which stimulates glial proinflammatory functions [55]. Recently, the Jayadev group identified a new miRNA/p53-dependent pathway that modulates functional differentiation of microglia both in cell culture and in vivo. They showed that miR-34a -145 and -155 are regulated by p53and negatively regulate the c-Maf transcription factor and its transcription activator Twist2. They suggested that p53 activated by ROS and oxidative DNA damage may induce microglial-mediated neuroinflammation [56].
The main component of AD associated with senile plaques is amyloid β (Aβ), a 40 (Aβ40) or 42 (Aβ42) amino acid peptide [57]. It is produced by proteolytic cleavage of the Amyloid Precursor Protein (APP) performed by two proteolytic enzymes: β- and γ-secretase.
According to Liu et al., the expression of APP is controlled by miRNA. They used bioinformatic analyses to predict that APP 3'-UTR is a potential target of miR-193b. The results indicated that overexpression of miR-193b could reduce the efficiency of mRNA translation, thus diminishing the expression of APP. By using luciferase assay they identified the miR-193b binding sites in APP 3'-UTR. Furthermore, they measured the concentration of Aβ42 in the cerebrospinal fluid and the expression of exosomal miR-193b, and confirmed a negative correlation between these biomarkers [58].
Another interesting study was performed by Augustin et al., who used in silico analysis of ADAM10, a gene coding an active unit of α-secretase [59], to search for miRNAs involved in its regulation. They found two promising candidates, namely, miR-107 and miR- 103. It seemed that both miRNAs reduced the expression of ADAM10 as shown by reporter assay [60]. These findings were confirmed by Leidinger et al., who used the RT-qPCR approach to show that miR- 107 and miR-103 were down-regulated in the peripheral blood of both AD and PD and schizophrenia patients [61].
Beta-site APP cleaving enzyme 1 (BACE1) is an active unit of β-secretase involved in Aβ formation. Increased BACE1 expression is a significant risk factor for sporadic AD [62]. The study of Wang et al. showed that according to bioinformatic predictions, BACE1 3'-UTR possesses several binding sites of miR-107. Cell culture reporter assays confirmed this suspicion. The study was validated by a correlation of miRNA profiling, in situ hybridization, and Affymetrix microarrays, and showed that BACE1 mRNA levels tended to rise as miR-107 levels decreased with the advancement of AD [63]. This negative correlation was convergent with the results of Nelson et al. [64].
Zhu et al. found that BACE1 3'-UTR is also targeted by miR-195. They validated the study by luciferase assay on HEK293 cells. Their study also demonstrated that miR-195 is capable of decreasing Aβ levels, therefore this may be an opportunity for future AD treatment [65]. Another miRNA associated with BACE1, namely miR-339, was found by Long et al. They identified two distinct miR-339-5p target sites in BACE1 3'-UTR by in silico analyses. Through a series of experiments they demonstrated a negative correlation between miR- 339 and BACE1 mRNA. This association was abolished by mutation in target sites. The synthetic mimic of miR-339 significantly inhibited expression of the BACE1 protein in human primary brain cultures and human glioblastoma cells. They also found that miR-339 levels were significantly lowered in AD patients' brains as compared to controls [66].
Another interesting discovery was made by Shioya et al. They used RT-qPCR to find significant down-regulation of miR-29a in AD brains. Subsequently, they performed a database search and identified the potential gene target, namely NAV3. Further evaluation confirmed elevated levels of NAV3 mRNA in AD brains. NAV3 down-regulation via miR-29a was verified by luciferase reporter assay. By immunohistochemistry they also showed that NAV3 protein expression was visibly higher in the degenerating neurons of AD patients' brains [67]
The other mechanism involved in neuronal loss is dysregulation of cholesterol metabolism in the brain. This sequence of biochemical reactions has been associated with many neurodegenerative disorders, especially AD. Specifically, rare variants of genes involved in cholesterol efflux (ABCA1 and APOE) were suggested to play a role in sporadic AD [68,69].
Adlakha et al. showed that miR-128-2 is a pro-apoptotic regulator of ABCA1 and ABCG1. This complex interaction network is mediated by SREBP and RXR. miR-128-2 increased the expression of SREBP2 and decreased the expression of SREBP1 in various cell lines. Moreover, miR-128-2 lowered the protein and mRNA levels of ABCA1, ABCG1 and RXRa by direct hybridizing to 3'-UTRs of corresponding genes [70]. Other miRNAs, such as miR-144-3p, miR- 145-5p, miR-106b-5p, miR-33-5p, and miR-758-5p, have also been shown to regulate the expression of ABCA1 [71-77].
Frontotemporal Dementia (FTD) is rather seldom and a poorly understood cause of cognitive impairment. On the other hand, standard dementia treatment may worsen the condition, hence the need to develop new therapeutic approaches. A subtype of FTD with TDP-43 inclusions (FTLD-TDP) is associated with mutations in the GRN causing a decrease in progranulin levels. Additionally, variants in TMEM106B were linked to FTLD-TDP. A study by Chen-Plotkin et al. showed that TMEM106B expression levels were increased in the FTLD-TDP brain and were controlled by miR-132 and miR-212 [78]. These findings show new directions for the development of miRbased therapies in FTLD-TDP.
The loss of brain DNs responsible for the development of PD occurs in the presence of, for example, intracellular inclusions, namely Lewy bodies [79], whose main component is α-synuclein (ASN). In a healthy brain, active ASNs form tetramers which constitute the functional tertiary structure [80]. Even a slight misbalance in ASN metabolism may lead to its aggregation and to toxic effects to DNs. This process may be promoted by mutations in parkin and α-synuclein genes [81,82] as well as in leucine-rich repeat kinase 2 (LRRK2) [83]. Moreover, it has recently been shown that the metabolism of ASN may be "fine-tuned" by various miRNAs [84]. By a series of mouse cell culture experiments, Junn et al. demonstrated an inverse correlation of ASN expression and miR-7 levels [85]. These data were convergent with the results of Doxakis, who also found that miR-153 is another ASN repressor. He used luciferase-transfected HEK293 cells to validate the study [86].
Cho et al., in their work from 2013, showed that expression of another gene involved in PD pangenesis, namely LRRK2, may be regulated via the miRNA network [83]. It was demonstrated that over-expression of LRRK2 may accelerate ASN-mediated neurodegeneration, while LRRK2 silencing may lead to ASN-induced neuropathology [87]. Several studies have associated LRRK2 with familial PD [88,89], yet there are others which tie this kinase with sporadic PD. Cho et al. showed that miR-205 was significantly underexpressed in the frontal cortex of sporadic PD patients. They also demonstrated that miR-205 binds to LRRK2-3'UTR, thus leading to its down-regulation [83].
Other evidence of miRNA involvement in ASN regulation was presented by Wang et al., who found that miR-433 may be linked to the accumulation of ASN by increasing expression of FGF20 both in vitro and in vivo. They showed, by luciferase assay, that downregulation of miR-433 and polymorphism in FGF20-3'UTR may raise the FGF20 protein level, followed by increased expression of ASN [90].
Kim et al. discovered that miR-133b is expressed mainly in midbrain DNs and is deficient in PD patients. miR-133b regulates the maturation and function of midbrain DNs within a negative feedback loop that includes the paired-like homeodomain transcription factor Pitx3 [91].
A study by Miñones-Moyano et al. identified two more miRNAs to be down-regulated in the postmortem brains of PD patients, namely miR-34b and miR-34c. Their work also demonstrated the negative correlation between these two miRNAs and parkin as well as DJ1 proteins. This may induce oxidative stress and dysfunction of mitochondrial metabolism in affected brain areas [92].
A recent study by Alvarez-Erviti et al. identified six miRNAs that were over-expressed in the brains of postmortem PD patients, namely, miR-21-3p, -26b, -106a*, -224, -301b, and -373-3p. The authors performed a bioinformatic prediction which showed putative targets in hsc70 and LAMP-2A, previously linked to PD. Luciferase assay confirmed that miR-26b, -106a*, -301b, and miR-21-3p, 224, 373-3p may down-regulate hsc70 and LAMP-2A, respectively [93].
Cardo et al. characterized the expression of miRNA in the substantia nigra of 8 postmortem PD and 4 healthy subjects using TaqMan low-density arrays for 733 human miRNAs. They found 10 down-regulated and one over-expressed miRNA (check Table 1). Subsequently, they performed a bioinformatic analysis predicting that miR-135b could bind to genes implicated in several neurodegenerative pathways [94].
miRNA
Source
Change
Size of group
References
Alzheimer's disease
miR-9, -125b, -146a, -155
CSF, BD-ECF
Over-expression
3P, 3C
Lukiw, [121]
miR-34a, -181b
PBMC
Over-expression
16P, 16C
Schipper et al., [110]
miR-26a, -27b, -30e-5p, -34a, -92, -125, -145, -200c,
-381, -422a, -423Hippocampus,
cerebellum, medial
frontalgyrus
Over-expression
15P, 12C
Cogswell et al., 2008 [109]
miR-9, -132, -146b, -212
Down-regulation
let-7f, miR-105, -125a, -135a, -138, -141, -151,
-186, -191, -197, -204, -205, -216, -302b, -30a-5p,
-30a-3p, -30b, -30c, -30d, -32, -345, -362, -371,
-374, -375, -380-3p, -429, -448, -449, -494, -501,
-517, -517b, -518b, -518f, -520a*, -526a
CSF
Over-expression
10P, 10C
miR-10a, -10b, -125, -126*, -127, 142-5p, -143,
-146b, -154, -15b, -181a, -181c, -194, -195,
-199a*, -214, -221, -328, -422b, -451, -455, -497,
-99a
Down-regulation
miR-29a
Frontal cortex
Down-regulation
7P, 4C
Shioya et al., [67]
miR-9, -125b, -146a, -155
CSF, BD-ECF
Over-expression
6P, 6C
Alexandrov et al., [50]
miR-26b
SN
Over-expression
10P, 8C
Absalon et al., [122]
let-7d-3p, miR-112, -151a-3p, -161, -5010-3p
Peripheral blood
Over-expression
106P, 22C
Leidinger et al., [61]
let-7f, miR-26a, -26b, -103a, -107, -532, -1285
Down-regulation
miR-9
Serum
Over-expression
105P, 150C
Tan et al., [111]
miR-125b, -181c
Down-regulation
Parkinson's disease
miR-133b
SNC
Down-regulation
3P, 5C
Kim et al., [91]
miR-34b, -34c
SNC
Down-regulation
11P, 6C
Minones-Moyano et al., [92]
miR-1, -22*, -29a
Peripheral blood
Down-regulation
8P, 8C
Margis et al., [123]
miR-181c, -331-5p, -193a-p, -196b, -454, -125a-3p,
-137Plasma
Over-expression
31P, 25C
Cardo et al., [124]
miR-21-3p, -224, -373-3p, -26b, -106a, -301b
SN
Over-expression
?
Alvarez-Erviti et al. [93]
miR-205
Frontal cortex
Down-regulation
16P,
7C
Cho et al., [83]
miR-19b, -29a -29c
Serum
Down-regulation
65P
65C
Botta-Orfila et al., [120]
Abbreviations: BD-ECF: Brain Derived Extracellular Fluid; CSF: Cerebrospinal Fluid; PBMC: Peripheral Blood Mononuclear Cells; SN: Substantia Nigra; SNC: Substantia Nigra Compacta; P: Patients; C: Controls
Table 1: The changes in expression of chosen miRNAs in AD and PD.
Single nucleotide polymorphisms and miRNA dysregulation in neurodegenerative diseases
The dysregulation of miRNA-mediated pathways may be associated with numerous variants in miRNA-coding genes as well as miRNA targets. Cui et al. studied two polymorphisms in miR146a, namely, rs2910464 and rs57095329, in 292 AD cases and 300 healthy controls. They found that the AA genotype of rs57095329 led to significantly higher neuroinflammation associated with miR-146a expression than the GG and GA genotypes of rs2910164 in peripheral blood mononuclear cells [95]. Delay et al. investigated the relationship between several miRNAs (miR-24, -186, and -455) and nicastrin - a subunit γ-secretase involved in Aβ generation. Using luciferasebased assays, they demonstrated that rs113810300 and rs141849450 SNPs in nicastrin 3'-UTR affected its miRNA-mediated repression. Notably, rs141849450 completely eliminated the miR-455-mediated repression of nicastrin [96].
Genomic changes in miRNA target sites were also observed in FTD. According to Rademakers et al., rs5848SNP associated with FTD-TDP is located in the miR-659 target site of the GRN and affects the miRNA-mediated repression of progranulin [97].
Similarly, variants in 3'-UTRs have been noticed in PD. In 2008 Wang et al. described a rs12720208 polymorphism in 3'UTR of FGF20 which affected the target site of miR-433. This led to higher levels of the FGF20 protein, thus promoting aggregation of ASN [90]. Cardo et al. described the SNP in 3'-UTR in the LRKK2 rs66737902 C allele which was more frequent in PD patients. They also found a significantly lower level of the LRRK2 transcript in the substantia nigra of PD postmortem patients who possessed the rs66737902 C genotype. In silico analyses predicted that the polymorphism affects the miR-138-2-3p target site [98].
In the future, knowledge on the genetic variants of miRNA genes and target sites could lead to the development of individualized therapy targeting and fixing misbalanced pathways responsible for developing ND in a given patient [99].
miRNAs as biomarkers important for the diagnosis of neurodegenerative diseases
As very movable, independent, small genetic entities that are plentiful in brain cell cytoplasm, cerebrospinal fluid and in peripheral circulation, miRNAs may serve as diagnostic biomarkers for AD and other human CNS diseases [50,61,100-105].
As was stated before, AD is becoming a valid public health issue since the number of 24 million AD patients worldwide is expected to double in the next 15 years [1]. Even though various approaches to developing minimally invasive tests for early detection of AD have been tested [106-108], there are no reliable molecular tests for diagnosing AD at the pre-symptomatic stage yet.
Cogswell et al. conducted a vast analysis of AD-associated miRNAs extracted from various parts of the brain and cerebrospinal fluid and described potential AD-specific miRNA biomarkers, which are shown in Table 1 [109].
Schipper et al. used the microRNA microarray (MMChip) for 462 human miRNAs in order to assess miRNA levels in the peripheral blood mononuclear cells of 16 AD patients and 16 controls, and validated the results by RT-qPCR. They demonstrated that miR-34a and 181b were significantly over-expressed (see Table 1). Subsequently, they performed bioinformatic target sequence prediction that yielded genes involved in synaptic activity [110].
Tan et al. conducted a serum study on 105 probable AD patients and 150 age- and gender-matched normal controls. The concentrations of 6 miRNAs were measured using RT-qPCR. They found that both miR-125b and miR-181c were down-regulated, while miR-9 was up-regulated in the serum of AD patients as compared with that of normal controls. The calculated specificity and sensitivity of miR-125b were up to 68.3% and 80.8%, respectively. Moreover, miR-125b correlated with the Mini-Mental State Examination (MMSE) of AD patients [111].
Leidinger et al. investigated the blood samples of 44 AD patients, finding 140 unique mature miRNAs whose expression levels were significantly changed. Subsequently, they verified the results by RTqPCR on 202 patients and developed an assay of 12 blood-based biomarkers able to discriminate AD cases from controls with an accuracy of 93%, a specificity of 95% and a sensitivity of 92% (see Table 1). They also demonstrated the possibility of being able to distinguish between AD and other neurological conditions with accuracies between 74% and 78%. They also showed that miR-107 decreases most significantly in AD, but also in PD and schizophrenia and depression. On the other hand, it was over-expressed in Mild Cognitive Impairment (MCI) [61]. A study by Wang et al. showed that miR-107 levels decrease significantly in the brains of AD patients at a very early stage of the disease [63]. These findings suggest that testing for miR-107 might provide useful information about a patient's condition in the pre-symptomatic stage of dementia.
Many studies have shown that AD dementia is preceded by 10-20 years of disease progress, primarily without any significant clinical signs (pre-symptomatic AD) before it shows the first symptoms of MCI [112,113]. Also, early stages of many neurodegenerative diseases, such as FTD and PD, might be misdiagnosed as MCI [114]. Current methods of psychological evaluation, e.g. MMSE, are not able to predict whether the patient is suffering from stable MCI or will develop fully symptomatic dementia [114,115]. This creates the need to develop a minimally invasive screening test which would help to monitor disease progression and response to currently experimental treatment [112]. It is also worth realizing that some unsuccessful clinical trials may deliver an effective disease modifier for a fraction of patients with early dementia [116]. Despite the obvious advancements of neuroimaging techniques, the high costs associated with them make them impractical as a tool for screening for early changes in the AD brain [117]. Moreover, a desperate need to develop better methods for early AD detection has also been highlighted in recent FDA reports [118].
Sheinerman et al. recently proposed a method for early detection of MCI based on an analysis of cell-free circulating miRNAs in the plasma by using RT-qPCR [119]. They identified two sets of circulating brain-abundant miRNAs, i.e. the miR-132 family (miR- 128, miR-132, miR-874) normalized per miR-491-5p, and the miR-134 family (miR-134, miR-323-3p, miR-382) normalized per miR-370, both of which are able to discriminate MCI from an agematched control with high accuracy. The miR-132 family biomarkers demonstrated 84%-94% sensitivity and 96%-98% specificity, and the miR-134 family biomarkers showed 74%-88% sensitivity and 80-92% specificity. Combining two miRNAs from the same family increases these values even more [119].
In 2014, Botta-Orfila et al. proposed a panel of 3 serum-derived miRNAs as potential biomarkers of PD. They conducted three sets of experiments, each using more patients, in order to validate their findings. Finally, they showed that miR-19b, -29a, and -29c were significantly under-expressed as compared to controls in the serum of idiopathic PD cases. These miRNAs were not found to be significantly changed in other NDs, thus they seem to be promising candidates for diagnostic purposes [120].
Conclusion
The role of miRNA in the pathogenesis of NDs is not fully understood. As of today (i.e. October 2014), the most reliable miRNA database (www.mirbase.org) contains ca. 2600 sequences of mature human miRNAs, and the function of most of them still remains unknown. The significant number of described miRNAs has been associated with AD, PD and other neurodegenerative diseases. However, currently there are no known miRNAs that are specifically responsible for the development of individual neurological diseases and that could be useful for diagnosis of NDs. Also, currently intensive effort is being made to develop miRNA-based assays which could combine data on the expression of several cerebrospinal fluid- or peripheral blood-enriched miRNAs. This could provide a means for introducing sensitive and specific diagnostic tests for early detection of neurodegeneration.
Scientists' efforts are also directed at discovering the genetic origin of miRNA-mediated disorders.
References
- Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, et al. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005; 366: 2112-2117.
- Johnson R, Noble W, Tartaglia GG, Buckley NJ. Neurodegeneration as an RNA disorder. Prog Neurobiol. 2012; 99: 293-315.
- Pearson H. Genetics: what is a gene? Nature. 2006; 441: 398-401.
- Saba R, Schratt GM. MicroRNAs in neuronal development, function and dysfunction. Brain Res. 2010; 1338: 3-13.
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001; 107: 823-826.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116: 281-297.
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75: 843-854.
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010; 11: 597-610.
- Rüegger S, GroΒhans H. MicroRNA turnover: when, how, and why. Trends Biochem Sci. 2012; 37: 436-446.
- Westholm JO, Lai EC. Mirtrons: microRNA biogenesis via splicing. Biochimie. 2011; 93: 1897-1904.
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012; 19: 586-593.
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005; 123: 631-640.
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003; 115: 199-208.
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004; 32: D109-111.
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008; 9: 102-114.
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005; 3: e85.
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004; 18: 504-511.
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007; 27: 91-105.
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010; 466: 835-840.
- Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007; 25: 1457-1467.
- Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, et al. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods. 2012; 9: 840-846.
- Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010; 20: R858-861.
- Lee HJ. Exceptional stories of microRNAs. Exp Biol Med (Maywood). 2013; 238: 339-343.
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003; 9: 1274-1281.
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007; 129: 1401-1414.
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011; 476: 228-231.
- Ouellet DL, Perron MP, Gobeil LA, Plante P, Provost P. MicroRNAs in gene regulation: when the smallest governs it all. J Biomed Biotechnol. 2006; 2006: 69616.
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19: 92-105.
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120: 15-20.
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008; 9: 831-842.
- Lin CC, Mitra R, Zhao Z. A tri-component conservation strategy reveals highly confident microRNA-mRNA interactions and evolution of microRNA regulatory networks. PLoS One. 2014; 9: e103142.
- Forrest ARR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 2010; 24: 460-466.
- Hon LS, Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 2007; 8: R166.
- Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010; 18: 510-525.
- Gao J, Wang WY, Mao YW, Gräff J, Guan JS, Pan L, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010; 466: 1105-1109.
- Olde Loohuis NF, Kos A, Martens GJ, Van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci. 2012; 69: 89-102.
- Meza-Sosa KF, Pedraza-Alva G, Pérez-Martínez L. microRNAs: key triggers of neuronal cell fate. Front Cell Neurosci. 2014; 8: 175.
- Coppedè F, Migliore L. DNA repair in premature aging disorders and neurodegeneration. Curr Aging Sci. 2010; 3: 3-19.
- Jellinger KA. Recent advances in our understanding of neurodegeneration. J Neural Transm. 2009; 116: 1111-1162.
- Elton TS, Selemon H, Elton SM, Parinandi NL. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. 2013; 532: 1-12.
- Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010; 40: 205-215.
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012; 148: 1172-1187.
- Narasimhan M, Riar AK, Rathinam ML, Vedpathak D, Henderson G, Mahimainathan L. Hydrogen peroxide responsive miR153 targets Nrf2/ARE cytoprotection in paraquat induced dopaminergic neurotoxicity. Toxicol Lett. 2014; 228: 179-191.
- Dorszewska J, Oczkowska A, Suwalska M, Rozycka A, Florczak-Wyspianska J, Dezor M, et al. Mutations in the exon 7 of Trp53 gene and the level of p53protein in double transgenic mouse model of Alzheimer's disease. Folia Neuropathol. 2014; 52: 30-40.
- Rokavec M, Li H, Jiang L, Hermeking H. The p53/miR-34 axis in development and disease. J Mol Cell Biol. 2014; 6: 214-230.
- Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, et al. microRNA-34c is a novel target to treat dementias. EMBO J. 2011; 30: 4299-4308.
- Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, et al. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008; 68: 10094-10104.
- Zhao Y, Bhattacharjee S, Jones BM, Dua P, Alexandrov PN, Hill JM, et al. Regulation of TREM2 expression by an NF-?B-sensitive miRNA-34a. Neuroreport. 2013; 24: 318-323.
- Bhattacharjee S, Zhao Y, Lukiw WJ. Deficits in the miRNA-34a-regulated endogenous TREM2 phagocytosis sensor-receptor in Alzheimer's disease (AD); an update. Front Aging Neurosci. 2014; 6: 116.
- Alexandrov PN, Dua P, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. microRNA (miRNA) speciation in Alzheimer's disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int J Biochem Mol Biol. 2012; 3: 365-373.
- Li Y, Tan MS, Jiang T, Tan L. Microglia in Alzheimer's disease. Biomed Res Int. 2014; 2014: 437483.
- Jayadev S, Case A, Alajajian B, Eastman AJ, Möller T, Garden GA. Presenilin 2 influences miR146 level and activity in microglia. J Neurochem. 2013; 127: 592-599.
- Contino M. Biomarkers for the early diagnosis of Alzheimer's disease: The challenge of XXI century. Adv. Alzheimer's Dis. 2013; 02: 13-30.
- Cruchaga C, Haller G, Chakraverty S, Mayo K, Vallania FL, Mitra RD,et al. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer's disease families. PLoS One. 2012; 7: e31039.
- Jayadev S, Nesser NK, Hopkins S, Myers SJ, Case A, Lee RJ, et al. Transcription factor p53influences microglial activation phenotype. Glia. 2011; 59: 1402-1413.
- Su W, Hopkins S, Nesser NK, Sopher B, Silvestroni A, Ammanuel S, et al. The p53transcription factor modulates microglia behavior through microRNA-dependent regulation of c-Maf. J Immunol. 2014; 192: 358-366.
- Pung L, Wang X, Li M, Xue L. The role of APP in Alzheimer's disease. Adv. Alzheimer's Dis. 2013; 02: 60-65.
- Liu CG, Song J, Zhang YQ, Wang PC. MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer's disease. Mol Med Rep. 2014; 10: 2395-2400.
- Postina R. A closer look at alpha-secretase. Curr Alzheimer Res. 2008; 5: 179-186.
- Augustin R, Endres K, Reinhardt S, Kuhn PH, Lichtenthaler SF, Hansen J, et al. Computational identification and experimental validation of microRNAs binding to the Alzheimer-related gene ADAM10. BMC Med Genet. 2012; 13: 35.
- Leidinger P, Backes C, Deutscher S, Schmitt K, Mueller SC, Frese K, et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013; 14: R78.
- Hébert SS, Horré K, NicolaÏ L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008; 105: 6415-6420.
- Wang W-X, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, et al. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008; 28: 1213-1223.
- Nelson PT, Wang WX. MiR-107 is reduced in Alzheimer's disease brain neocortex: validation study. J Alzheimers Dis. 2010; 21: 75-79.
- Zhu HC, Wang LM, Wang M, Song B, Tan S, Teng JF, et al. MicroRNA-195 downregulates Alzheimer's disease amyloid-β production by targeting BACE1. Brain Res Bull. 2012; 88: 596-601.
- Long JM, Ray B, Lahiri DK. MicroRNA-339-5p down-regulates protein expression of Β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J Biol Chem. 2014; 289: 5184-5198.
- Shioya M, Obayashi S, Tabunoki H, Arima K, Saito Y, Ishida T, et al. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol Appl Neurobiol. 2010; 36: 320-330.
- Goedeke L, Fernández-Hernando C. microRNAs: A connection between cholesterol metabolism and neurodegeneration. Neurobiol Dis. 2014.
- Koldamova R, Staufenbiel M, Lefterov I. Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. J Biol Chem. 2005; 280: 43224-43235.
- Adlakha YK, Khanna S, Singh R, Singh VP, Agrawal A, Saini N. Pro-apoptotic miRNA-128-2 modulates ABCA, ABCG1 and RXRa expression and cholesterol homeostasis. Cell Death Dis. 2013; 4: e780.
- de Aguiar Vallim TQ, Tarling EJ, Kim T, Civelek M, Baldán Á, Esau C, et al. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ Res. 2013; 112: 1602-1612.
- Kang MH, Zhang LH, Wijesekara N, de Haan W, Butland S, Bhattacharjee A, et al. Regulation of ABCA1 protein expression and function in hepatic and pancreatic islet cells by miR-145. Arterioscler Thromb Vasc Biol. 2013; 33: 2724-2732.
- Kim J, Yoon H, Ramírez CM, Lee SM, Hoe HS, Fernández-Hernando C, et al. MiR-106b impairs cholesterol efflux and increases Aβ levels by repressing ABCA1 expression. Exp Neurol. 2012; 235: 476-483.
- Marquart TJ, Allen RM, Ory DS, Baldán A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010; 107: 12228-12232.
- Ramirez CM, Dávalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol. 2011; 31: 2707-2714.
- Ramírez CM, Rotllan N, Vlassov AV, Dávalos A, Li M, Goedeke L, et al. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ Res. 2013; 112: 1592-1601.
- Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010; 328: 1570-1573.
- Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L, et al. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci. 2012; 32: 11213-11227.
- Schapira AH. Pathogenesis of Parkinson's disease. Baillieres Clin Neurol. 1997; 6: 15-36.
- Oczkowska A, Kozubski W, Dorszewska J. [Alpha-synuclein in Parkinson's disease]. Przegl Lek. 2014; 71: 26-32.
- Oczkowska A, Kozubski W, Lianeri M, Dorszewska J. Mutations in PRKN and SNCA Genes Important for the Progress of Parkinson's Disease. Curr Genomics. 2013; 14: 502-517.
- Hardy J, Cai H, Cookson MR, Gwinn-Hardy K, Singleton A. Genetics of Parkinson's disease and parkinsonism. Ann Neurol. 2006; 60: 389-398.
- Cho HJ, Liu G, Jin SM, Parisiadou L, Xie C, Yu J, et al. MicroRNA-205 regulates the expression of Parkinson's disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet. 2013; 22: 608-620.
- Mouradian MM. MicroRNAs in Parkinson's disease. Neurobiol Dis. 2012; 46: 279-284.
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009; 106: 13052-13057.
- Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010; 285: 12726-12734.
- Lin X, Parisiadou L, Gu XL, Wang L, Shim H, Sun L, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant alpha-synuclein. Neuron. 2009; 64: 807-827.
- Paisán-Ruíz C, Jain S, Evans EW, Gilks WP, Simón J, van der Brug M, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004; 44: 595-600.
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004; 44: 601-607.
- Wang G, van der Walt JM, Mayhew G, Li YJ, Züchner S, Scott WK, et al. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008; 82: 283-289.
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007; 317: 1220-1224.
- Miñones-Moyano E, Porta S, Escaramís G, Rabionet R, Iraola S, Kagerbauer B, et al. MicroRNA profiling of Parkinson's disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 2011; 20: 3067-3078.
- Alvarez-Erviti L, Seow Y, Schapira AH, Rodriguez-Oroz MC, Obeso JA, Cooper JM. Influence of microRNA deregulation on chaperone-mediated autophagy and a-synuclein pathology in Parkinson's disease. Cell Death Dis. 2013; 4: e545.
- Cardo LF, Coto E, Ribacoba R, Menéndez M, Moris G, Suárez E, et al. MiRNA Profile in the Substantia Nigra of Parkinson's Disease and Healthy Subjects. J Mol Neurosci. 2014.
- Cui L, Li Y, Ma G, Wang Y, Cai Y, Liu S, et al. A functional polymorphism in the promoter region of microRNA-146a is associated with the risk of Alzheimer disease and the rate of cognitive decline in patients. PLoS One. 2014; 9: e89019.
- Delay C, Dorval V, Fok A, Grenier-Boley B, Lambert JC, Hsiung GY, et al. MicroRNAs targeting Nicastrin regulate Aβ production and are affected by target site polymorphisms. Front Mol Neurosci. 2014; 7: 67.
- Rademakers R, Eriksen JL, Baker M, Robinson T, Ahmed Z, Lincoln SJ, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008; 17: 3631-3642.
- Cardo LF, Coto E, Ribacoba R, Mata IF, Moris G, Menéndez M, et al. The screening of the 3'UTR sequence of LRRK2 identified an association between the rs66737902 polymorphism and Parkinson's disease. J Hum Genet. 2014; 59: 346-348.
- Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009; 10: 399-416.
- Dorval V, Nelson PT, Hébert SS. Circulating microRNAs in Alzheimer's disease: the search for novel biomarkers. Front Mol Neurosci. 2013; 6: 24.
- Geekiyanage H, Jicha GA, Nelson PT, Chan C. Blood serum miRNA: non-invasive biomarkers for Alzheimer's disease. Exp Neurol. 2012; 235: 491-496.
- Provost P. Interpretation and applicability of microRNA data to the context of Alzheimer's and age-related diseases. Aging (Albany NY). 2010; 2: 166-169.
- Kumar P, Dezso Z, MacKenzie C, Oestreicher J, Agoulnik S, Byrne M, e al. Circulating miRNA biomarkers for Alzheimer's disease. PLoS One. 2013; 8: e69807.
- Bekris LM, Lutz F, Montine TJ, Yu CE, Tsuang D, Peskind ER, et al. MicroRNA in Alzheimer's disease: an exploratory study in brain, cerebrospinal fluid and plasma. Biomarkers. 2013; 18: 455-466.
- Henriksen K, O'Bryant SE, Hampel H, Trojanowski JQ, Montine TJ, Jeromin A, et al. The future of blood-based biomarkers for Alzheimer's disease. Alzheimers Dement. 2014; 10: 115-131.
- Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K, et al. Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med. 2007; 13: 1359-1362.
- Reddy MM, Wilson R, Wilson J, Connell S, Gocke A, Hynan L, et al. Identification of candidate IgG biomarkers for Alzheimer's disease via combinatorial library screening. Cell. 2011; 144: 132-142.
- Nagele E, Han M, Demarshall C, Belinka B, Nagele R. Diagnosis of Alzheimer's disease based on disease-specific autoantibody profiles in human sera. PLoS One. 2011; 6: e23112.
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008; 14: 27-41.
- Schipper HM, Maes OC, Chertkow HM, Wang E. MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul Syst Bio. 2007; 1: 263-274.
- Tan L, Yu JT, Liu QY, Tan MS, Zhang W, Hu N, et al. Circulating miR-125b as a biomarker of Alzheimer's disease. J Neurol Sci. 2014; 336: 52-56.
- Pillai JA, Cummings JL. Clinical trials in predementia stages of Alzheimer disease. Med Clin North Am. 2013; 97: 439-457.
- Sperling RA, Jack CR Jr, Aisen PS. Testing the right target and right drug at the right stage. Sci Transl Med. 2011; 3: 111cm33.
- Karakaya T, Fuβer F, Schröder J, Pantel J. Pharmacological Treatment of Mild Cognitive Impairment as a Prodromal Syndrome of Alzheimer’s Disease. Curr Neuropharmacol. 2013; 11: 102-108.
- Snyder EM, Olin J, David FS. Maximizing the value of diagnostics in Alzheimer's disease drug development. Nat Rev Drug Discov. 2012; 11: 183-184.
- Cunningham EL, Passmore AP. Drug development in dementia. Maturitas. 2013; 76: 260-266.
- Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2013; 9: e111-194.
- FDA AD Drug Development Guidance. J Nucl Med. 2013; 54: 16N.
- Sheinerman KS, Tsivinsky VG, Abdullah L, Crawford F, Umansky SR. Plasma microRNA biomarkers for detection of mild cognitive impairment: biomarker validation study. Aging (Albany NY). 2013; 5: 925-938.
- Botta-Orfila T, Morat X, Compta Y, Lozano JJ, Falgàs N, Valldeoriola F, et al. Identification of blood serum micro-RNAs associated with idiopathic and LRRK2 Parkinson’s disease. J Neurosci Res. 2014; 92: 1071-1077.
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007; 18: 297-300.
- Absalon S, Kochanek DM, Raghavan V, Krichevsky AM. MiR-26b, upregulated in Alzheimer's disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J Neurosci. 2013; 33: 14645-14659.
- Margis R, Margis R, Rieder CR. Identification of blood microRNAs associated to ParkinsonÄ–s disease. J Biotechnol. 2011; 152: 96-101.
- Cardo LF, Coto E, de Mena L, Ribacoba R, Moris G, Menéndez M, et al. Profile of microRNAs in the plasma of Parkinson's disease patients and healthy controls. J Neurol. 2013; 260: 1420-1422.