Introduction
SMG-1 (Suppressor of Morphogenesis in Genitalia-1) is an evolutionally conserved serine/threonine-protein kinase and belongs Phosphatidylinositol-3-Kinase (PI3K) related kinases (PIKKs) that include Ataxia Telangiectasia Mutated (ATM), ATM and Rad3 Related (ATR), Mammalian Target of Rapamycin (mTOR), DNADependent Protein Kinase Catalytic Subunit (DNA-PKcs) and Transformation/Transcription Domain-Associated Protein (TRRAP) [1]. The deduced 3,521-amino acid protein has a calculated molecular mass of 410kD [2]. PIKKs have diverse functions. For example, ATM, ATR and DNA-PKcs are involved in the response to DNA damage. ATM and DNA-PKcs respond to DNA Double Strand Breaks (DSBs) and ATR to DNA replication blockers or formation of long stretches of single strand DNA [3]. mTOR is a nutrient-regulated kinase that controls a wide variety of pathways involved in metabolism and cell growth [4]. TRRAP functions as part of a multiprotein co-activator complex which is involved with the transcriptional activity of c-Myc and other transcriptional factors [5]. SMG-1 is a part of the mRNA surveillance complex that regulates Nonsense-Mediated mRNA Decay (NMD) [6]. SMG-1 was firstly reported as a member of the informational suppression in Caenorhabditis elegans (C. elegans) which affected several mRNA processes in 1989 [7]. Pulak et al. reported that loss of function mutations affecting seven C. elegans smg genes eliminates NMD [8] and later demonstrated that smg- 1 kinase activity was essential for NMD [9]. In 2001, Deming et al. reported a partial sequence of human SMG-1 as C. elegans SMG-1 related protein [10]. Yamashita et al. also reported the full-length sequence for human smg-1 which encoded a 3657 aa protein that was a novel PIKK and showed the involvement of SMG1 in mammalian NMD [11]. In 2004, Brumbaugh et al. reported the activation of SMG-1 by DNA damage and involvement of SMG-1 in genotoxic stress-induced phosphorylation of P53 [2]. Besides NMD, SMG- 1 roles as a protective agency in genotoxic stress such as radiation, tumor proliferation, and apoptosis. I will briefly summarize the roles of SMG-1 related with NMD and others in this paper.
SMG-1 as a Player in NMD
NMD is a cellular defense mechanism eliminating mRNA that harbors Premature Translation Termination Codons (PTCs) and encode nonfunctional or potentially harmful polypeptides [6,12,13]. NMD pathway involves a cascade of fine-coordinated events. A SURF complex (SMG-1, SMG-8, SMG-9, UPF1 and the eukaryotic release factors 1 and 3 (eRF1 and 3) is formed on a ribosome which encounters a PTC [14,15]. Interaction of UPF1 with UPF2 and UPF3B bound to the downstream EJC triggers UPF1 phosphorylation by the SMG-1 and remodels the SURF complex to form the decayinducing complex [16-19]. The primary structure of PIKKs contains a conserved C terminus preceded by a long stretch of helical, mostly HEAT (huntington, elongation factor 3, a subunit of PP2A and TOR1) repeats which are units of two anti-parallel α-helices connected by flexible loops and large superhelical frequently twisted structures [20]. A bent arm comprising a long region of HEAT repeats at the N-terminus of SMG-1 functions as a scaffold for SMG- 8 and SMG-9, and projects from C-terminal core containing PI3K domain, which modulates NMD [21,22]. Resultant NMD processes decrease the production of potentially harmful polypeptides and enhances the accuracy of gene expression. SMG-1 is one of the essential players triggering an NMD response, because it is the kinase that phosphorylates the UPF1 protein. SMG-1 functions in NMD by selective degrading PTCs mRNAs which can be generated by gene mutations, splicing or transcription errors and by phosphorylating specific serine residues in UPF1 helicase which is a crucial regulator of NMD [23]. Phosphorylated UPF1 leads the releases of eRF1 and eRF3 and recruits NMD factors such as SMG-5~7 [24]. They recognize its phosphorylation by inducing the remodeling of the mRNA surveillance complex which discriminates the PTC-containing mRNA from the normal mRNA as an essential step in NMD [11,14]. Recently, González-Huici et al. declared that SMG-1 kinase activity could be activated following DNA damage to phosphorylate specific DNA repair proteins and/or NMD inactivation may lead to aberrant mRNAs leading to synthesis of malfunctioning DNA repair proteins [25]. Consequently, SMG-1 play viral roles in the regulation of cell growth, proliferation, survival, and cellular responses to stresses as a member of PIKKs.
SMG-1 as a Player in Tumor Suppression
SMG-1 has been suggested as a tumor suppressor, though its role as an NMD effector has been precisely documented. In the planarians study, the planarian mTORC1 signaling compounds and SMG-1 performed as a key regulator of regeneration and growth antagonistically. Rapamycin as an inhibitor of mTORC1 increased the survival rate of smg-1 animals by decreasing cell proliferation. They showed the possibility of the tumor suppressor function of SMG-1 [26]. Gubanova et al. reported that in Human Papillomavirus (HPV)-positive head and neck squamous cell carcinoma, SMG- 1 was underexpressed and exhibited tumor suppressive activity [27]. They also reported that SMG-1 suppresses CKD2 and tumor growth by regulating both the p53 and Cdc25A signaling pathways in 2013. They showed that SMG-1 regulates the G 1/S checkpoint through both a p53-dependent, and a p53-independent pathway and depletion of SMG-1 increased tumor growth [28]. In 2014, Nam et al. declared that SMG-1 as a promising modulator of sorafenib resistance by using an unbiased genome-wide massive genetic screen method. They showed that the inhibition of SMG-1 reduced sorafenib sensitivity in the several hepatocellular carcinoma cell lines and could be an agent to reverse sorafenib resistance [29]. The results suggested the possibility of SMG-1 as a potent tumor suppressor (Figure 1). At a similar time, Han et al. demonstrated that expression of SMG-1 was significantly lower in the HCC tissue than that in the normal tissues and declared that SMG-1 expression was an independent prognostic marker for overall survival [30]. A novel AKT inhibitor inhibited phosphorylation of AKT downstream molecules and activated phosphorylation of mTOR and SMG-1 in the several liver cancer cell lines [31]. This report suggested the promising possibility of usage as anti-cancer regimen of SMG-1. Recently, Zhang et al. reported that SMG-1 was suppressed by miR-192 and -215 and functioned as a tumor suppressor in gastric cancer by inactive Wnt signaling and suppressing epithelial-mesenchymal transition [32].
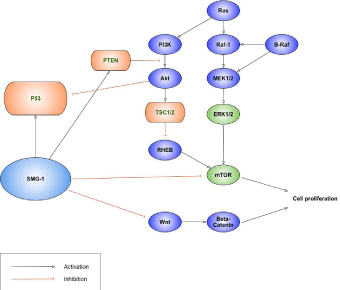
Figure 1: SMG-1 as a tumor suppressor.
Conclusion
SMG-1 is involved in NMD as an active effector and tumorigenesis as a tumor suppressor. Moreover, SMG-1 may be a promising interrupter during the cancer proliferation mechanisms for development of anti-cancer agent and represent a biomarker for predicting the prognosis of cancer. SMG-1 may warrant investigation to defeat cancer in the future. Therefore, more well-coordinated studies are needed to elucidate the overall precise mechanisms of SMG-1 involving NMD and cancer development processes.
References
- Baretic D and Williams RL. PIKKs-the solenoid nest where partners and kinases meet. Curr Opin Struct Biol. 2014; 29: 134-142.
- Brumbaugh KM, Otterness DM, Geisen C, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004; 14: 585-598.
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003; 3: 155-168.
- Wullschleger S, Loewith R and Hall MN. TOR signaling in growth and metabolism. Cell. 2006; 124: 471-484.
- McMahon SB, Wood MA and Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000; 20: 556-562.
- Yamashita A, Kashima I and Ohno S. The role of SMG-1 in nonsensemediated mRNA decay. Biochim Biophys Acta. 2005; 1754: 305-315.
- Hodgkin J, Papp A, Pulak R, Ambros V and Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989; 123: 301-313.
- Pulak R and Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993; 7: 1885-1897.
- Grimson A, O’Connor S, Newman CL and Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsensemediated mRNA Decay in Caenorhabditis elegans. Mol Cell Biol. 2004; 24: 7483-7490.
- Denning G, Jamieson L, Maquat LE, Thompson EA and Fields AP. Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J Biol Chem. 2001; 276: 22709- 22714.
- Yamashita A, Ohnishi T, Kashima I, Taya Y and Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001; 15: 2215- 2228.
- Holbrook JA, Neu-Yilik G, Hentze MW and Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004; 36: 801-808.
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004; 5: 89-99.
- Kashima I, Yamashita A, Izumi N, et al. Binding of a novel SMG-1-Upf1- eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006; 20: 355-367.
- Yamashita A, Izumi N, Kashima I, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009; 23: 1091-1105.
- Lykke-Andersen J, Shu MD and Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000; 103: 1121-1131.
- Chamieh H, Ballut L, Bonneau F and Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008; 15: 85-93.
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW and Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008; 27: 736- 747.
- Melero R, Buchwald G, Castano R, et al. The cryo-EM structure of the UPFEJC complex shows UPF1 poised toward the RNA 3’ end. Nat Struct Mol Biol. 2012; 19: 498-505, S1-2.
- Lempiainen H and Halazonetis TD. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 2009; 28: 3067-3073.
- Arias-Palomo E, Yamashita A, Fernandez IS, et al. The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev. 2011; 25: 153-164.
- Deniaud A, Karuppasamy M, Bock T, et al. A network of SMG-8, SMG-9 and SMG-1 C-terminal insertion domain regulates UPF1 substrate recruitment and phosphorylation. Nucleic Acids Res. 2015; 43: 7600-7611.
- Schweingruber C, Rufener SC, Zund D, Yamashita A and Muhlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013; 1829: 612-623.
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F and Ohno S. N- and C- terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5: SMG-7 during NMD. Nucleic Acids Res. 2012; 40: 1251-1266.
- Gonzalez-Huici V, Wang B and Gartner A. A Role for the Nonsense-Mediated mRNA Decay Pathway in Maintaining Genome Stability in Caenorhabditis elegans. Genetics. 2017; 206: 1853-1864.
- Gonzalez-Estevez C, Felix DA, Smith MD, et al. SMG-1 and mTORC1 act antagonistically to regulate response to injury and growth in planarians. PLoS Genet. 2012; 8: e1002619.
- Gubanova E, Brown B, Ivanov SV, et al. Downregulation of SMG-1 in HPV-positive head and neck squamous cell carcinoma due to promoter hypermethylation correlates with improved survival. Clin Cancer Res. 2012; 18: 1257-1267.
- Gubanova E, Issaeva N, Gokturk C, Djureinovic T and Helleday T. SMG-1 suppresses CDK2 and tumor growth by regulating both the p53 and Cdc25A signaling pathways. Cell Cycle. 2013; 12: 3770-3780.
- Nam SW, Park KC, Yang KJ, Lee B and Kim SW. Genetic screen identifies suppressor of morphogenesis in genitalia-1 (SMG-1) as a modulator of sorafenib resistance in hepatocellular carcinoma cell lines. Int J Oncol. 2014; 45: 1450-1456.
- Han LL, Nan HC, Tian T, et al. Expression and significance of the novel tumor-suppressor gene SMG-1 in hepatocellular carcinoma. Oncol Rep. 2014; 31: 2569-2578.
- Zhang Y, Zheng Y, Faheem A, et al. A novel AKT inhibitor, AZD5363, inhibits phosphorylation of AKT downstream molecules, and activates phosphorylation of mTOR and SMG-1 dependent on the liver cancer cell type. Oncol Lett. 2016; 11: 1685-1692.
- Zhang X, Peng Y, Huang Y, et al. SMG-1 inhibition by miR-192/-215 causes epithelial-mesenchymal transition in gastric carcinogenesis via activation of Wnt signaling. Cancer Med. 2018; 7: 146-156.
