
Research Article
Austin Immunol. 2016; 1(2): 1010.
Effect of 1,25(OH)2D3 and 25(OH)D3 on FOXP3, IgE Receptors and Vitamin D Regulating Enzymes Expression in Lymphocyte Cell Lines
Vijayendra Chary A and Hemalatha R*
Department of Clinical Division, Microbiology and Immunology, National Institute of Nutrition (NIN), India
*Corresponding author: R. Hemalatha, Scientist F & HOD, Dept of Clinical Division, Microbiology and Immunology, National Institute of Nutrition (NIN), Hyderabad-07, Telangana, India
Received: October 03, 2016; Accepted: November 07, 2016; Published: November 10, 2016
Abstract
Impact of 25(OH)D3 and 1,25(OH)2D3 was studied on transcription regulation of FOXP3, regulatory cytokines (TGF-β, & IL-10) and IgE receptors (CD23 and CD21) in human immortalized JURKAT and 8E5 cell lines. In addition, vitamin D regulating enzymes (CYP27B1, CYP2R1 & CYP24A1), VDR, RXR and VDBP mRNA expressions were quantified by RT-PCR. The mRNA expression of FOXP3 was significantly (p < 0.05) upregulated, while IgE receptors were downregulated (P < 0.05) in both the cell lines upon treatment with 25(OH)D3 or 1,25(OH)2D3. As for the regulatory cytokines, IL-10 was not affected, whereas TGF-β increased significantly (P < 0.05) with 1,25(OH)2D3 treatment in both the cell lines. Vitamin D metabolizing enzyme (CYP24A1) was upregulated (p < 0.05) with 1,25(OH)2D3 treatment; in contrast, CYP27B1 and CYP2R1 expression remained unaltered. This study shows that 1,25(OH)2D3 and 25(OH)D3 impact transcription regulation of FOXP3, TGF-β and IgE receptors expression in lymphocyte cell lines.
Keywords: 25(OH)D3; CYP27B1; FOXP3; CD23; CD21
Abbreviations
FOXP3: Forkhead Box P3; 25-hydroxyvitamin D3 1α-hydroxylase; CYP2R1: Vitamin D3 25-hydroxylase; CYP24A1: Cytochrome P24 A1 or 1,25-dihydroxyvitamin D3 24-hydroxylase; VDR: Vitamin D Receptor; RXR: Retinoic acid Receptor; VDBP: Vitamin D Binding Protein; 1,25(OH)2D3, 1,25-dihydroxyvitamin D3; IgE: Immunoglobulin E; CD: Cluster of Differentiation; TGF β: Transforming Growth Factor Beta; IL-10: Interleukin-10; CYP27B1: RQ: Relative Quantification
Introduction
Several in vitro and in vivo studies have shown that vitamin D plays an important role in shaping of the innate and adaptive immune system [1]. It is supported by the fact that almost all immune cells, including T lymphocytes, B lymphocytes, neutrophils, and antigen presenting cells express vitamin D receptor and vitamin D activating enzyme, 1α-hydroxylase [1-5]. Immune cells possess the ability to convert 25(OH)D3 to 1,25(OH)2D3, which in turn is regulated by circulating levels of 25(OH)D3 [1-5].
The main targets for vitamin D regulation appear to be T Helper cells (TH cells), especially regulatory T cells (CD4+/CD25+/CD127-/ FOXP3 or Treg cells) [6]. Treg cells are subsets of T lymphocytes described by the expression of a specific transcription factor, Forkhead box P3 (FOXP3) [7]. There are reports implicating reduction in the numbers and functionality of Treg cells in both human and mouse, during allergic conditions [7,8]. Furthermore, Treg cells produce regulatory cytokines such as Interleukin 10 (IL-10) and transforming growth factor beta (TGF-β), both of which have anti-inflammatory activity relevant to the inhibition of asthma pathology [9,10]. IL-10 acts on antigen-presenting cells and suppresses T cell activation, including Th2 cells [9]. On the other hand, TGF-β induces peripheral expression of the transcription factor FOXP3. Interaction between IL-10 and TGF-β is likely to be vital in the regulation of IgE and its substrates, which play an essential role in all allergic conditions [11].
CD23 and CD21 are substrates on the B lymphocytes that act as receptors for IgE antibody and are further linked to asthma/ allergy in children as well as in adults [12-14]. B cells can be identified by their characteristic presence of CD19 receptor, which is expressed during all stages of B-cell differentiation, maturation and proliferation [12,15,16]. Recently, we found impaired immune regulation and increased CD23 and CD21 expression in pregnant women with vitamin D deficiency [17].
Several enzymes are involved in the homeostasis of 25(OH) D3. Upon intake, vitamin D3 (vitamin D) is metabolized to 25-hydroxycholecalciferol (calcidiol or 25-hydroxyvitamin D3 or 25(OH)D3) by the hepatic 25-hydroxylases (CYP2R1) in the liver. Subsequently, 25(OH)D3 is converted to 1,25 dihydroxycholecalciferol (calcitriol or 1,25(OH)2D3) by 1α-hydroxylase (CYP27B1) in the kidney [18]. The active form of vitamin D (1,25(OH)2D3) form complexes with Vitamin D Receptor (VDR) and forms a heterodimer with 9-cis-retinoic acid receptor (RXR) and interacts with genes in the immune cells [19]. 1,25(OH)2D3 formation also takes place in several other cell types, including dermal, intestinal epithelial cells, lymph nodes, monocytes and placenta, which express CYP27B1 enzyme, that aids in extra renal activation of 25(OH)D3 to the active hormonal form1,25(OH)2D3 [20,21]. Altered expression of VDR, RXR and vitamin D regulating enzymes are associated with vitamin D deficiency [17].
To demonstrate the effect of vitamin D on Treg cells, IgE receptors and vitamin D regulating enzymes, we used different concentrations of vitamin D and studied the mRNA expression of FOXP3, IgE receptors, vitamin D regulating enzymes (CYP2R1, CYP27B1 and CYP24A1) and Vitamin D Receptors (VDR & RXR) in T lymphocyte (JURKAT) and B lymphocyte (8E5) cell lines.
Materials and Methods
Cell culture
Human immortalized T lymphocytes (JURKAT) and B lymphocytes (8E5) were obtained from National Centre for Cell Science (NCCS), Pune, India. These cells were cultured in RPMI 1640 medium containing heat inactivated 10% Fetal Bovine Serum (FBS) (Thermo Fischer scientific, USA), 2 mM L-glutamine (Thermo Fischer scientific, USA), MEM nonessential amino acids (Thermo Fischer scientific, USA), 1% anti mycotic antibiotic (Thermo Fischer scientific, USA) and was incubated in a humified (95 %) atmosphere with 5 % CO2 at 37°C. For expression assays the cells were treated with Phorbol 12-Myristate 13 Acetate (PMA) (Sigma Aldrich, St. Louis, MO, USA) for 12 and 24 h. Following the treatment with PMA, the cells were treated either with 25(OH)D3 or 1,25(OH)2D3 in different concentrations such as 25 nM, 50nM and 100nM solvent (Ethanol, 0.1 % final concentration). All the assays were run in triplicates and were repeated in three independent experiments. PMA stimulated cells were taken as control; PMA stimulated cells with 0.1% ethanol were taken as vehicle control and PMA stimulated cells with 0.1% ethanol and treatment with either 25(OH)D3 or 1,25(OH)2D3 were taken as treated cells.
FOXP3, CD23, CD21, VDR, CYP2R1, CYP27B1, CYP24A1, VDBP and RXR mRNA expression by Real-time PCR
Total RNA extraction from the cells: The total RNA was extracted using the method described by Chomzinsky and Sacchi [22]. The Total RNA was extracted using TRIZOL reagent from the cells that were treated with PMA for 24 h, followed by exposure to the 25(OH)D3 or 1,25(OH)2D3 in different concentrations (100nM, 50nM and 25nM) after 12 and 24 h incubation. Total RNA, thus obtained was treated with DNase I (Ambion) according to manufacturer’s protocol. The quality and the yield of total RNA were checked using Agilent Bioanalyzer 2100 (Agilent Technologies) and Nanodrop 1000 (Thermo Scientific). Electrophoresis was employed to check the integrity and purity of the RNA for cDNA synthesis by Masek et al. procedure [23].
cDNA synthesis and gene expression from mRNA: Five micrograms of total RNA were transcribed into cDNA using Revert Aid first strand cDNA synthesis kit (Thermo scientific).The cDNA thus obtained was aliquoted and stored at –20°C. For the RT-PCR assay, primers were designed by Geneiou’s pro software (Version; 5.4.6, New Zealand) and synthesized by Imperial life sciences, New Delhi, India (Table 1). Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) was used as internal control. In order to quantify gene expression of target genes like VDR, CD23, CD21 and FOXP3, the ABI step one plus system was used to programme, which consisted of 95°C for 10 min followed by 40 cycles of 95°C for 15 sec and 60°C for one minute. The difference in cycle threshold [24] value between the control gene (GAPDH) and the target genes (CD23, CD21, FOXP3, VDR, CYP2R1, CYP27B1, CYP24A1, VDBP and RXR) was obtained for each sample. The critical threshold cycle [24] values were determined using the ABI system and Step one plus software. The difference in CT values (ΔCT) between the target genes and GAPDH was normalized to the corresponding ΔCT of the calibrator (ΔΔCT) and was expressed in fold expression (2-(ΔΔCT)) or Relative Quantification (RQ) to assess the relative difference in mRNA for each gene.
S.
No
Gene
Primer sequence
Base pairs
1
GAPDH
Forward: 51- TCGACAGTCAGCCGCATCTTCTTT -31
Reverse: 51- ACCAAATCCGTTGACTCCGACCTT - 31
94
2
VDR
Forward:51- TGTAATCCCAGCAGTTTGGGAGGT - 31
Reverse: 51- AGGGTTTCTCCATGTTGGTCAGGT -31
88
3
FOXP3
Forward: 51AGATCTACCACTGGTTCACACGCA-31
Reverse: 51-GCACAAAGCACTTGTGCAGACTCA-31
105
4
CD23
Forward: 51-GGAATTGAACGAGAGGAACGAAG-31
Reverse: 51-AAAGCCGCTGGACACCTG-31
97
5
CD21
Forward: 51-CCCATAGTACCAGGAGGATACA-31
Reverse: 51-CCGTTCATGGAGAAGTTGGT- 31
100
6
CYP27B1
Forward: 51-CCATGTGGCAGAAGGGATAA-31
Reverse: 51-AAACCGTAAACCAGGCTAGG-31
100
7
CYP2R1
Forward: 51-GACAGACCATGCCTTCCTTTA-31
Reverse: 51-ATCGTCTGTGATCAACCCATC-31
94
8
CYP24A1
Forward: 51-CGCCTCAGATGGTGGTATTT-31
Reverse: 51-AGCAGTGAACCCTGTAGAATG-31
92
9
RXR
Forward: 51-GGACCCTCCTTTGGTGAAAT-31
Reverse: 51-AGGATTGGGAACGGCTAAAG-31
105
10
VDBP
Forward: 51-GGTACTTGAGCCAACCCTAAA-31
Reverse: 51-GTAGAGGGCCCTTAGCATTAAA-31
86
Table 1: Primers used in RT-PCR.
Regulatory cytokines (TGF-β and IL-10) secretion in cell supernatant
Cytokines such as TGF-β and IL-10 were quantified by Sandwich ELISA. The cell lysates were used for this assay. JURKAT and 8E5 cell lines were stimulated with PMA for 24 hours and were treated with different concentrations (25nM, 50nM and 100nM) of 25(OH)D3 or 1,25(OH)2D3 and were incubated for 12 and 24 hrs. Briefly, the micro plate was coated with capture antibody. Then, samples containing an unknown amount of the target analyte of interest were added. After washing, to get rid of the micro plate of unbound substances, an HRP conjugate was added and absorbance was determined according to the manufacturer’s instructions (E biosciences, USA). The lower limit of detection for total IgE was 7.8 ng/mL and the standard curve range given with the kit was 7.8 to 1000 ng/mL. The lower limit of detection for IL-10 was 0.8 ng/mL and the standard curve range given with kit was 0.8 to 50 ng/mL. The lower limit of detection for TGF-β was 80 pg/mL and the standard curve range given with kit was 80 to 5000 ng/mL.
Data analysis and statistics: Data are presented as Mean ± Standard Error (SE), derived from a minimum of three independent experiments. Statistical analysis was conducted using the SPSS software, version 22.0 (IBM software, USA). Graphical analysis was performed in SigmaPlot™12.3 (Systat Software, USA). Significance in differences between treated versus control was analyzed using Analysis of variance (ANOVA) with post hoc multiple comparison test by Dunnet.
Results
The mRNA expressions of FOXP3, CD23, CD21, TGF-β, IL-10, CYP27B1, CYP2R1, CYP24A1, VDR, RXR and VDBP were expressed in relative quantification (RQ) values and were compared between the treated and the control. Control cells (JURKAT or 8E5) with PMA and without 25(OH)D3 or 1,25(OH)2D3 was taken as calibrator and GAPDH was used as internal control. The mRNA expressions of FOXP3, CD23, CD21, TGF-β, IL-10, CYP27B1, CYP2R1, CYP24A1, VDR, RXR and VDBP in the control cells were always comparable with vehicle control (cells with PMA and ethanol but without 25(OH) D3 or 1,25(OH)2D3).
Effect of 1,25(OH)2D3 on immune parameters in JURKAT cells
The mRNA expression of Treg cell transcription factor, FOXP3 and regulatory cytokine, TGF-β increased significantly (p<0.05) at 100nM and 50nM of 1,25(OH)2D3 when compared to control cells (Figures 1A & 1B). However, mRNA expression of IL10 was not affected at different concentrations (100nM, 50nM and 25nM) of 1,25(OH)2D3. In contrast, the mRNA expression of CD23 and CD21 was significantly (p < 0.05) downregulated upon treatment with 1,25(OH)2D3 at 100nM and 50nM concentrations (Figures 1A & 1B). 25nM of 1,25(OH)2D3 had no effect on any of the immune parameters tested (25nM concentration data not shown).
Effect of 25(OH)D3 on immune parameters in JURKAT cells
The mRNA expression of Treg cell transcription factor, FOXP3 and regulatory cytokine, TGF-β increased significantly (p < 0.05) at 100nM of 25(OH)D3 when compared to control cells, but 50nM & 25nM had no effect (Figures 1C & 1D). In contrast, the mRNA expression of CD23 and CD21 was significantly (p < 0.05) downregulated upon treatment with 25(OH)D3 at 100nM concentrations, but 50nM & 25nM had no effect. However, mRNA expression of IL-10 was not affected at different concentrations (100nM, 50nM and 25nM) of 25(OH)D3 (Figures 1C & 1D).
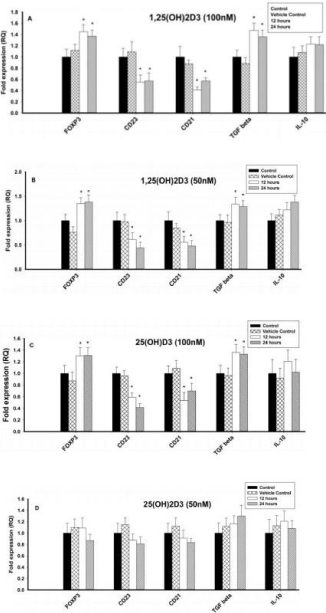
Figure 1: Effect of 1,25(OH)2D3 and 25(OH)D3 on the mRNA expression of
immune markers (FOXP3, CD23, CD21, TGF-β and IL-10) in T lymphocytes
(JURKAT cells) for 12 and 24 hours. The mRNA expression has been
analyzed by qRT-PCR and is expressed as the fold expression (RQ) in
related to the expression in control and vehicle control (Ethanol treated)
cells. GABDH was used as an internal control and the expression in control
cells was set to one and data are presented as the mean ± SE. *p < 0.05,
statistically different from controls. A) Effect of 1,25(OH)2D3 (100 nM) on the
mRNA expression of immune markers. B) Effect of 1,25(OH)2D3 (50 nM) on
the mRNA expression of immune markers. C) Effect of 25(OH)D3 (100 nM)
on the mRNA expression of immune markers. D) Effect of 25(OH)D3 (50 nM)
on the mRNA expression of immune markers.
Effect of 1,25(OH)2D3 on vitamin D regulating enzymes and vitamin D receptors in JURKAT cells
Different concentrations (100nM, 50nM and 25nM) of 1,25(OH)2D3 had no effect on the mRNA expression of CYP27B1, CYP2R1, RXR and VDBP and were comparable with control cells (Figures 2A & 2B). However, the vitamin D metabolizing enzyme, CYP24A1, was upregulated (p < 0.05) at 100nM and 50nM of 1,25(OH)2D3 when compared to control, but 25nM had no effect. VDR was also upregulated (p < 0.05) with different concentrations of 1,25(OH)2D3 when compared to control cells (Figures 2A & 2B).
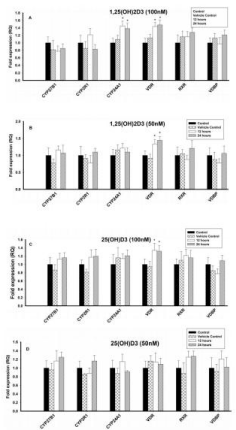
Figure 2: Effect of 1,25(OH)2D3 and 25(OH)D3 on the mRNA expression of
vitamin D regulating enzymes (CYP27B1, CYP2R1, CYP24A1 & VDBP) and
Vitamin D Receptors (VDR & RXR) in T lymphocytes (JURKAT cell lines) for
12 and 24 hours. The mRNA expression has been analyzed by qRT-PCR
and is expressed as the fold expression (RQ) in related to the expression in
control and vehicle control (Ethanol treated) cells. GABDH was used as an
internal control and the expression in control cells was set to one and data are
presented as the mean ± SE. *p < 0.05, statistically different from controls. A)
Effect of 1,25(OH)2D3 (100 nM) on mRNA expression of vitamin D regulating
enzymes and vitamin D receptors. B) Effect of 1,25(OH)2D3 (50 nM) on
mRNA expression of vitamin D regulating enzymes and vitamin D receptors.
C) Effect of 25(OH)D3 (100 nM) on mRNA expression of vitamin D regulating
enzymes and vitamin D receptors. D) Effect of 25(OH)D3 (50 nM) on mRNA
expression of vitamin D regulating enzymes and vitamin D receptors.
Effect of 25(OH)D3 on vitamin D regulating enzymes and vitamin D receptors in JURKAT cells
Both the vitamin D activating enzymes (CYP27B1 & CYP2R1) and the RXR and the VDBP mRNA expression were not affected at different concentrations (100nM, 50nM and 25nM) of 25(OH)D3 (Figures 2C & 2D). However, the vitamin D metabolizing enzyme, CYP24A1, was significantly upregulated (p < 0.05) with 25(OH)D3 at 100nM, but 50nM and 25nM had no effect. Similarly, vitamin D receptor (VDR) was upregulated (p < 0.05) upon treatment with 25(OH)D3 at 100nM, but 50nM & 25nM had no effect (Figures 2C & 2D).
Cytokine secretion in cell supernatant in JURKAT cells
Regulatory cytokine TGF-β was significantly increased in JURKAT cells with 100nM and 50nM of 1,25(OH)2D3 and 100nM of 25(OH)D3 after 12 or 24 hours incubation. However, lower concentrations of 1,25(OH)2D3 (25nM) and 25(OH)D3 (50nM and 25nM) had no effect on TGF-β. IL-10 concentration, however, was not effected with both forms of vitamin D such as 1,25(OH)2D3 and 25(OH)D3 (Figures 3A & 3B).
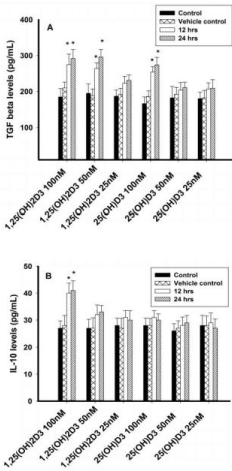
Figure 3: Regulatory cytokines analysis (TGF-β and IL-10) in T lymphocytes
(JURKAT cells) by ELISA. Values are expressed as mean ± SE. A) TGF β
concentrations (pg/mL) in T lymphocytes (JURKAT cell lines) after treatment
with 1,25(OH)2D3 (100 nM, 50 nM & 25 nM) and 25(OH)D3 (100 nM, 50
nM & 25 nM) at both 12 or 24 hours. B) IL-10 concentrations (pg/mL) in T
lymphocytes (JURKAT cells) after treatment with 1,25(OH)2D3 (100 nM, 50
nM & 25 nM) and 25(OH)D3 (100 nM, 50 nM & 25 nM) at 12 or 24 hours.
Effect of 1,25(OH)2D3 on immune parameters in 8E5 cells
The mRNA expression of Treg cell transcription factor, FOXP3 and regulatory cytokine, TGF-β increased significantly (p < 0.05) at 50 and 100nM of 1,25(OH)2D3 when compared to control cells (Figures 4A & 4B). However, mRNA expression of IL10 was not affected at different concentrations (100nM, 50nM and 25nM) of 1,25(OH)2D3. In contrast, the mRNA expression of CD23 and CD21 was significantly (p < 0.05) downregulated upon treatment with 1,25(OH)2D3 at 100nM and 50nM concentrations (Figures 4A & 4B). 25nM of 1,25(OH)2D3 had no effect on any of the immune parameters tested.
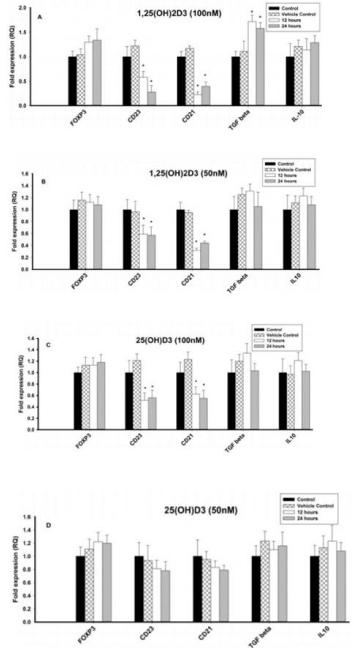
Figure 4: Effect of 1,25(OH)2D3 and 25(OH)D3 on the mRNA expression of
immune markers (FOXP3, CD23, CD21, TGF-β and IL-10) in B lymphocytes
(8E5 cell lines) for 12 and 24 hours. The mRNA expression has been
analyzed by qRT-PCR and is expressed as the fold expression (RQ) in
related to the expression in control and vehicle control (Ethanol treated) cells.
GABDH was used as an internal control and the expression in control cells
was set to 1 and data are presented as the mean ± SE. *p < 0.05, statistically
different from controls. A) Effect of 1,25(OH)2D3 (100 nM) on the mRNA
expression of immune markers. B) Effect of 1,25(OH)2D3 (50 nM) on the
mRNA expression of immune markers. C) Effect of 25(OH)D3 (100 nM) on
the mRNA expression of immune markers. D) Effect of 25(OH)D3 (50 nM) on
the mRNA expression of immune markers.
Effect of 25(OH)D3 on immune parameters in 8E5 cells
The mRNA expression of Treg cell transcription factor, FOXP3 and regulatory cytokine, TGF-β increased significantly (p<0.05) at 100nM of 25(OH)D3 when compared to control cells, but 50nM & 25nM had no effect (Figures 4C & 4D). In contrast, the mRNA expression of CD23 and CD21 was significantly (p < 0.05) downregulated upon treatment with 25(OH)D3 at 100nM concentrations, but 50nM & 25nM had no effect. However, mRNA expression of IL-10 was not affected at different concentrations (100nM, 50nM and 25nM) of 25(OH)D3 (Figures 4C & 4D).
Effect of 1,25(OH)2D3 on vitamin D regulating enzymes and vitamin D receptors in 8E5 cells
Different concentrations (100nM, 50nM and 25nM) of 1,25(OH)2D3 had no effect on CYP27B1, CYP2R1, RXR and VDBP. However, Vitamin D metabolizing enzyme, CYP24A1 was significantly (P < 0.05) upregulated with 1,25(OH)2D3 at 100nM when compared to control cells, but had no effect was observed with 50nM and 25nM. However, VDR was significantly (P < 0.05) upregulated with different concentrations of 1,25(OH)2D3 when compared to control cells (Figures 5A & 5B).
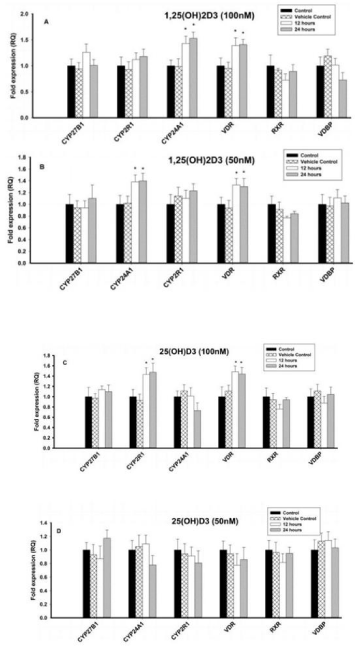
Figure 5: Effect of 1,25(OH)2D3 and 25(OH)D3 on the mRNA expression
of vitamin D regulating enzymes (CYP27B1, CYP2R1, CYP24A1 & VDBP)
and vitamin D receptors (VDR & RXR) in B lymphocytes (8E5 cell lines) for
12 and 24 hours. The mRNA expression has been analyzed by qRT-PCR
and is expressed as the fold expression (RQ) in related to the expression in
control and vehicle control (Ethanol treated) cells. GABDH was used as an
internal control and the expression in control cells was set to 1 and data are
presented as the mean ± SE. *p < 0.05, statistically different from controls. A)
Effect of 1,25(OH)2D3 (100 nM) on mRNA expression of vitamin D regulating
enzymes and vitamin D receptors. B) Effect of 1,25(OH)2D3 (50 nM) on
mRNA expression of vitamin D regulating enzymes and vitamin D receptors.
C) Effect of 25(OH)D3 (100 nM) on mRNA expression of vitamin D regulating
enzymes and vitamin D receptors. D) Effect of 25(OH)D3 (50 nM) on mRNA
expression of vitamin D regulating enzymes and vitamin D receptors.
Effect of 25(OH)D3 on vitamin D regulating enzymes and vitamin D receptors in 8E5 cells
Different concentrations (100nM, 50nM and 25nM) of 1,25(OH)2D3 had no effect on of CYP27B1, CYP2R1, RXR, VDBP and vitamin D metabolizing enzyme (CYP24A1). In contrast, VDR mRNA expression was significantly (P < 0.05) upregulated with 25(OH)D3 at 100nM when compared to control cells. But 50nM and 25nM of 1,25(OH)2D3 had no effect on VDR mRNA (Figures 5C & 5D).
Cytokine secretion in cell supernatant in 8E5 cells
Different concentrations (100nM, 50nM and 25nM) of 1,25(OH)2D3 and 25(OH)D3 had no effect on TGF-β or IL-10 concentrations (Figures 6A & 6B).
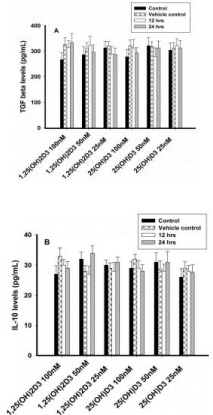
Figure 6: Regulatory cytokines analysis (TGF-β and IL-10) in T lymphocytes
(8E5 cell lines) by ELISA. Values are expressed as mean ± SE. A) TGF β
concentrations (pg/mL) in T lymphocytes (JURKAT cell lines) after treatment
with 1,25(OH)2D3 (100 nM, 50 nM & 25 nM) and 25(OH)D3 (100 nM, 50
nM & 25 nM) at both 12 or 24 hours. B) IL-10 concentrations (pg/mL) in T
lymphocytes (JURKAT cells) after treatment with 1,25(OH)2D3 (100 nM, 50
nM & 25 nM) and 25(OH)D3 (100 nM, 50 nM & 25 nM) at 12 or 24 hours.
Discussion
The current study demonstrates effect of 1,25(OH)2D3 and 25(OH)D3 on transcription regulation of FOXP3, IgE receptors (CD23 & CD21) and regulatory cytokine expressions in the T and B lymphocyte cell lines. In our earlier study, vitamin D deficiency leads to impaired regulatory T cell population and its transcription factor (FOXP3) in pregnant women and in children with asthma [17]. In the present study, 1,25(OH)2D3 and 25(OH)D3 augmented Treg cell transcription factor (FOXP3) expression in both T and B lymphocytes, similar to that observed by other investigators in PBMC [25-28]. Moreover, increasing expression of FOXP3 was associated with TGF-β mRNA expression and protein production from both the cell lines, suggesting a direct role of 25(OH)D3 and 1,25(OH)2D3 on TGF-β induction and FOXP3 expression. In contrast, we did not register increase in IL-10 mRNA expression or protein production with either 25(OH)D3 or 1,25(OH)2D3 even after 24 hours incubation. The effect of 1,25(OH)2D3 treatment on IL-10 production has been suggested to be bi-directional, with experiments showing suppression during the first 8 hours and upregulation after 48 hours of incubation. Since our experiments were completed by 24 hours, predictably the IL-10 production was not increased with 1,25(OH)2D3 treatment.
TGF-β is a key regulator of the signaling pathways that initiate and maintain FOXP3 expression via smad proteins (Small dependent transcription factors) or MAPK (Mitogens activated protein kinase) pathway [29]. TGF-β is capable of inducing FOXP3 expression in naïve peripheral CD4+CD25-FOXP3- T cells into FOXP3+ Treg cells [30]. The TGF-β induced FOXP3+ Treg cells are phenotypically and functionally indistinguishable from the natural FOXP3+ Treg cells generated in the thymus [31]. However, FOXP3 expression in the present study was not associated with IL-10 expression, suggesting that the induced FOXP3 may not be IL-10 producing phenotype [32]. Together, these results suggest that 1,25(OH)2D3 and 25(OH) D3 may induce FOXP3 expression, perhaps via TGF-β induction in naïve peripheral lymphocytes, but may have little role on IL-10 production [32,33]. Also, these data suggest that IL-10 production is not a prerequisite for FOXP3 expression in vitro, and suggest that TGF-β and FOXP3 may have distinct origins as compared to IL-10 [32].
CD23 and CD21 on B cells may hold a central role as receptors of IgE antibody and possibly are linked with increased respiratory morbidity amongst vitamin D deficient subjects [34,35]. It has been hypothesized that vitamin D insufficiency may increase early life respiratory morbidity by increasing B cells bearing IgE receptors [36]. In the present study, IgE receptors such as CD23 and CD21 were down regulated in both JURKAT and 8E5 cells with the active form of vitamin D i.e. 1,25(OH)2D3. These results may confirm that vitamin D might affect allergy related IgE receptors such as CD23 and CD21 and thus provide additional evidence of important immunomodulatory properties of 1,25(OH)2D3 that may help to control immune mediated diseases.
Vitamin D shows its action on immune regulation due to the presence of VDR in immune mediated cells. Through VDR receptors, 1,25(OH)2D3 acts directly on immune system cells and more specifically on T lymphocytes in autoimmune disease [37,38]. 1,25(OH)2D3 protects VDR protein from proteasomal degradation, and as such VDR protein is stabilized with 1,25(OH)2D3 as reported in the current study and elsewhere [38,39]. The active forms of 1,25(OH)2D2 or 1,25(OH)2D3 act via the VDR, although some have effects unrelated to VDR binding such as CYP24A1 enzyme inhibition [40]. VDR moves constantly between the nucleus and cytoplasm, is a negative regulator of NF-κB signaling and thus mediates various cellular effects [41].
The activity of vitamin D regulating enzymes is poorly understood and it is unknown if they are directly regulated by 25(OH)D3 or 1,25(OH)2D3 [42]. Additionally, the regulation of CYP27B1 or CYP2R1 may not be critical in determining the cell’s ability to generate the active form of vitamin D [43]. mRNA expression was important to know the role of 1,25(OH)2D3 and 25(OH)D3 on CYP27B1 and CYP2R1’s true activity, especially since CYP2R1 has been reported to be the most biologically active enzyme in humans [43]. In our earlier studies, we have shown a strong relationship between 25(OH)D3 and vitamin D regulating enzymes in pregnant women as well as in children with asthma [17]. Earlier studies have shown increased expression of CYP27B1 with 1,25(OH)2D3 in human Peripheral Blood Mononuclear Cells (PBMC) [44,45]. In contrast, in our study, no change in the expression of vitamin D activating enzymes CYP27B1 and CYP2R1 were observed in both T and B lymphocyte cell lines, this may not be wholly unexpected, since, CYP27B1 enzyme converts 7-dehydrocholesterol to 25(OH)D3 and CYP2R1 converts 25(OH)2D3 to active form of 1,25(OH)2D3 [18]. Thus, the availability of both the forms of vitamin D (25(OH)D3 & 1,25(OH)2D3) in our experiment might have prevented upregulation of CYP27B1 and CYP2R1 by a negative feedback mechanism [46]. Conversely, CYP2R1 was expressed in higher concentration with 25(OH)D3, but not with 1,25(OH)2D3, indicating the requirement of activating enzyme (CYP2R1) to convert 25(OH)D3 to 1,25(OH)2D3. CYP24A1 is the vitamin D metabolizing enzymes and as expected was highly expressed in both the cell lines after treatment with 100nM of 1,25(OH)2D3, suggesting that the CYP24A1 enzyme is driven by high concentration of 1,25(OH)2D3.
Conclusion
It should be noted, however, that most of the earlier studies were done on human peripheral blood mononuclear cells or in the mouse. The T cells come from different tissues and the functions of the T cells depend large on where they are located. Few investigators have studied the effect of vitamin D and immune regulation on the human T and B lymphocytes. This study demonstrates role of 1,25(OH)2D3 and 25(OH)D3 on transcription regulation of FOXP3, TGF-β and IgE receptors.
Acknowledgment
Authors greatly acknowledged the National Institute of Nutrition (ICMR) for providing financial support and facilities to conduct the study. The Authors sincerely acknowledge the CSIR-HRDG for the SRF fellowship to Mr. Vijayendra Chary. RH conceived and designed the study, interpreted the data and approved the final manuscript. AV collected and analyzed the data, performed laboratory work and prepared the manuscript.
References
- Prietl B, Treiber G, Pieber TR, Amrein K. Vitamin D and immune function. Nutrients. 2013; 5: 2502-2521.
- Wu S, Ren S, Nguyen L, Adams J, Hewison M. Splice variants of the CYP27b1 gene and the regulation of 1, 25-dihydroxyvitamin D3 production. J Endocrinol. 2007; 148: 3410-3418.
- Hewison M. Vitamin D and the immune system. J Endocrinol. 1992; 132: 173-175.
- Aranow C. Vitamin D and the immune system. J Investig Med. 2011; 59: 881-886.
- Borges MC, Martini LA, Rogero MM. Current perspectives on vitamin D, immune system, and chronic diseases. Nutrition. 2011; 27: 399-404.
- Van Etten E, Mathieu C. Immunoregulation by 1, 25-dihydroxyvitamin D 3: basic concepts. J Steroid Biochem Mol Biol. 2005; 97: 93-101.
- Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010; 10: 490-500.
- Corthay A. How do regulatory T cells work? Scand J Immunol. 2009; 70: 326-336.
- O’Garra A, Barrat FJ, Castro AG, Vicari A, Hawrylowicz C. Strategies for use of IL-10 or its antagonists in human disease. Immunol Rev. 2008; 223: 114-131.
- Li MO, Wan YY, Sanjabi S, Robertson A-KL, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006; 24: 99-146.
- Taylor A, Verhagen J, Blaser K, Akdis M, Akdis CA. Mechanisms of immune suppression by interleukin-10 and transforming growth factor-β: the role of T regulatory cells. Immunology. 2006; 117: 433-442.
- Rabatic S, Gagro A, Medar-Lasic M. CD21-CD23 ligand pair expression in children with allergic asthma. Clin Exp Immunol. 1993; 94: 337-340.
- Vijayendrachary A, Hemalatha R, Narendrababu K, Rameshkumar R, Sivaramaprasad K, Dineshkumar B. CD23, Th1/Th2 cytokines in bronchial asthma, bronchiolitis and bronchial pneumonia in pediatric patients. Asian J Pharm Clin Res. 2012; 5: 111-114.
- Vijayendra C, Hemalatha R, Narendra B, Ramesh K, Sathavahana C, Dinesh K. CD23, Total IgE and Th1/Th2 Cytokines in Asthma Patients. Int J Pharm Pharm Sci. 2012; 4: 175-177.
- Otero DC, Rickert RC. CD19 function in early and late B cell development. II. CD19 facilitates the pro-B/pre-B transition. J Immunol. 2003; 171: 5921-5930.
- Bowles SL, Jaeger C, Ferrara C, Fingeroth J, Van De Venter M, Oosthuizen V. Comparative binding of soluble fragments (derCD23, sCD23, and exCD23) of recombinant human CD23 to CD21 (SCR 1-2) and native IgE, and their effect on IgE regulation. Cellular Immunology. 2011; 271: 371-378.
- Vijayendra Chary A, Hemalatha R, Seshacharyulu M, Vasudeva Murali M, Jayaprakash D, Dinesh Kumar B. Vitamin D deficiency in pregnant women impairs regulatory T cell function. J Steroid Biochem Mol Biol. 2015; 147: 48-55.
- Hewison M, Burke F, Evans KN, Lammas DA, Sansom DM, Liu P, et al. Extra-renal 25-hydroxyvitamin D 3-1alpha-hydroxylase in human health and disease. J Steroid Biochem Mol Biol. 2007; 103: 316-321.
- Pike JW, Meyer MB, Bishop KA. Regulation of target gene expression by the vitamin D receptor-an update on mechanisms. Rev Endocr Metab Disord. 2012; 13: 45-55.
- Shinkyo R, Sakaki T, Kamakura M, Ohta M, Inouye K. Metabolism of vitamin D by human microsomal CYP2R1. Biochem Biophys Res. 2004; 324: 451-457.
- Zehnder D, Evans KN, Kilby MD, Bulmer JN, Innes BA, Stewart PM, et al. The ontogeny of 25-hydroxyvitamin D3 1α-hydroxylase expression in human placenta and decidua. Am J Pathol. 2002; 161: 105-114.
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987; 162: 156-159.
- Masek T, Vopalensky V, Suchomelova P, Pospisek M. Denaturing RNA electrophoresis in TAE agarose gels. Anal Biochem. 2005; 336: 46-50.
- Sim JJ, Lac PT, Liu ILA, Meguerditchian SO, Kumar VA, Kujubu DA, et al. Vitamin D deficiency and anemia: a cross-sectional study. Annals of Hematology. 2010; 89: 447-452.
- Daniel C, Sartory NA, Zahn N, Radeke HH, Stein JM. Immune modulatory treatment of trinitrobenzene sulfonic acid colitis with calcitriol is associated with a change of a T helper (Th) 1/Th17 to a Th2 and regulatory T cell profile. J Pharmacol Exp Ther. 2008; 324: 23-33.
- Penna G, Roncari A, Amuchastegui S, Daniel KC, Berti E, Colonna M, Adorini L. Expression of the inhibitory receptor ILT3 on dendritic cells is dispensable for induction of CD4+Foxp3+ regulatory T cells by 1,25-dihydroxyvitamin D3. Blood. 2005; 106: 3490-3497.
- Gorman S, Kuritzky LA, Judge MA, Dixon KM, McGlade JP, Mason RS, et al. Topically applied 1,25-dihydroxyvitamin D3 enhances the suppressive activity of CD4+CD25+ cells in the draining lymph nodes. J Immunol. 2007; 179: 6273-6283.
- Jeffery LE, Burke F, Mura M, Zheng Y, Qureshi OS, Hewison M, et al. 1, 25-Dihydroxyvitamin D3 and IL-2 combine to inhibit T cell production of inflammatory cytokines and promote development of regulatory T cells expressing CTLA-4 and FoxP3. J Immunol. 2009; 183: 5458-5467.
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006; 24: 179-189.
- Johnston CJ, Smyth DJ, Dresser DW, Maizels RM. TGF-β in tolerance, development and regulation of immunity. Cellular Immunology. 2016; 299: 14-22.
- Chen L, Cencioni MT, Angelini DF, Borsellino G, Battistini L, Brosnan CF. Transcriptional profiling of γδ T cells identifies a role for vitamin D in the immunoregulation of the Vγ9Vδ2 response to phosphate-containing ligands. J Immunol. 2005; 174: 6144-6152.
- Matilainen JM, Husso T, Toropainen S, Seuter S, Turunen MP, Gynther P, et al. Primary effect of 1α, 25(OH)2D3 on IL-10 expression in monocytes is short-term down-regulation. BBA-Mol Cell Res. 2010; 1803: 1276-1286.
- Koizumi T, Nakao Y, Matsui T, Nakagawa T, Katakami Y, Fujita T. Effect of 1, 25-dihydroxyvitamin D3 on cytokine-induced thymocyte proliferation. Cellular immunology. 1985; 96: 455-461.
- Bowles SL, Jaeger C, Ferrara C, Fingeroth J, Van De Venter M, Oosthuizen V. Comparative binding of soluble fragments (derCD23, sCD23, and exCD23) of recombinant human CD23 to CD21 (SCR 1-2) and native IgE, and their effect on IgE regulation. Cell Immunol. 2011; 271: 371-378.
- Hibbert RG, Teriete P, Grundy GJ, Beavil RL, Reljic R, Holers VM, et al. The structure of human CD23 and its interactions with IgE and CD21. J Exp Med. 2005; 202: 751-760.
- Morales E, Romieu I, Guerra S, Ballester F, Rebagliato M, Vioque J, et al. Maternal vitamin D status in pregnancy and risk of lower respiratory tract infections, wheezing, and asthma in offspring. Epidemiology. 2012; 23: 64-71.
- Di Rosa M, Malaguarnera G, De Gregorio C, Palumbo M, Nunnari G, Malaguarnera L. Immuno-modulatory effects of vitamin D3 in human monocyte and macrophages. Cellular Immunology. 2012; 280: 36-43.
- Kongsbak M, Levring TB, Geisler C, Von Essen MR. The vitamin d receptor and T cell function. Front Immunol. 2013; 4: 148.
- Masuyama H, MacDonald PN. Proteasome-mediated degradation of the Vitamin D Receptor (VDR) and a putative role for SUG1 interaction with the AF-2 domain of VDR. J Cell Biochem. 1998; 71: 429-440.
- Bikle DD. Vitamin D metabolism, mechanism of action, and clinical applications. Chem Biol. 2014; 21: 319-329.
- Chen Y-H, Yu Z, Fu L, Wang H, Chen X, Zhang C, et al. Vitamin D3 inhibits lipopolysaccharide-induced placental inflammation through reinforcing interaction between vitamin D receptor and nuclear factor kappa B p65 subunit. Sci Rep. 2015; 5.
- Prosser DE, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci. 2004; 29: 664-673.
- Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc Natl Acad Sci USA. 2004; 101: 7711-7715.
- Tsoukas CD, Provvedini DM, Manolagas SC. 1,25-dihydroxyvitamin D3: a novel immunoregulatory hormone. Science. 1984; 224: 1438-1440.
- Rigby WF, Yirinec B, Oldershaw RL, Fanger MW. Comparison of the effects of 1,25-dihydroxyvitamin D3 on T lymphocyte subpopulations. Eur J Immunol. 1987; 17: 563-566.
- Wacker M, Holick MF. Sunlight and Vitamin D: A global perspective for health. Dermatoendocrinol. 2013; 5: 51-108.