
Research Article
J Bacteriol Mycol. 2023; 10(1): 1202.
Neonatal Bacteraemia and Antibiotic Resistance at the Angre University Hospital, Abidjan, 2020
Yapi IAD1*, Coulibaly-Diallo YM1, Bahan GAJ1, Tué Bi CG2, Koné IT2 and Kacou-N’Douba A1,3
1Bacteriology Unit, Medical Biology Department, Angre University Hospital, Côte d’Ivoire
2Neonatalogy Unit, Paediatric and Specialties Department, Angre University Hospital, Côte d’Ivoire
3Department of Microbiology, Faculty of Medical Sciences, University of Felix Houphouët Boigny, Côte d’Ivoire
*Corresponding author: Yapi IADBacteriology Unit, Medical Biology Department, Angre University Hospital, Côte d’Ivoire, 28 BP 1530 Abidjan 28, Cote d’Ivoire
Received: January 17, 2023; Accepted: March 10, 2023; Published: March 17, 2023
Abstract
Background: Multi-drug resistant bacteria are an increasingly important cause of neonatal sepsis. Nowadays, they are a great concern in neonates because few therapeutic options are available.
Aim: To determine the bacteria responsible of neonatal sepsis and their antibiotic susceptibility pattern in order to improve the quality of antibiotic prescription.
Methods: A retrospective data review on positive blood cultures from the neonatal department at the university hospital of Angre between January to December 2020 was conducted. All neonates with clinical suspicion of sepsis with positive blood cultures were identified. Patient demographics, clinical details, and laboratory data were recorded and analyzed by Epi info software version 7.2.5.0
Results: Out of 221 blood cultures samples, 82 were positive (37%). The predominant age group was that between day 0 and day 7. A preponderance of bacteria of the genus Staphylococcus (48.78%) compared to enterobacteria (43.90%) was observed. The main isolated bacteria were coagulase-negative Staphylococcus (29.27%), Klebsiella pneumoniae (26.83%) and Staphylococcus aureus (19.51%). Streptococcus agalactiae was isolated in 2 cases (2.44%). Among the Enterobacteriaceae strains, 80.56% produced an Extended Spectrum Beta-Lactamase (ESBL). The rate of methicillin-resistant was observed in coagulase-negative Staphylococcus and Staphylococcus aureus in 37.50% and 31.25% of cases respectively. Out of ESBL strains, 89.65% were multi-resistant. Of the 14 strains of methicillin-resistant staphylococci, 13 (92.86%) were multidrug resistant.
Conclusion: Coagulase-negative Staphylococcus and Klebsiella pneumoniae were the common causes of neonatal sepsis. The high rate of multi-drug resistant bacteria resistant represents a great threat to neonatal survival and warrants modification of existing empirical therapy.
Keywords: Neonatal bacteraemia; Antibiotic resistance; Hospital; Abidjan
Abbreviations: ATCC: American Type Culture Collection; AMC: Amoxicillin-Clavulanic Acid; AMK: Amikacin; BLSE: Extended Spectrum Beta Lactamases; BMR: Multi-drug Resistant Bacteria; CBC: Blood Count; CIP: Ciprofloxacin; CMN: Clindamycin; CN: Gentamycin; CHU: Hospital and University Center; CO2: Carbon Dioxide; CoNS: Coagulase-Negative Staphylococcus; CRO: Ceftriaxone; CRP: C-Reactive Protein; CTZ: Ceftazidime; ESBLs: Extended Spectrum Beta Lactamases; E. Coli: Escherichia Coli; ERY: Erythromycin; EUCAST: European Committee on Antimicrobial Susceptibility Testing; FOX: Cefoxitin; K. Pneumoniae: Klebsiella Pneumonia; LEV: Levofloxacin; MDR: Multi-Drug Resistant; MRSA: Methicillin-Resistant Staphylococcus Aureus; MR-CoNS: Methicillin-Resistant Coagulase-Negative Staphylococcus; NS: Neonatal Sepsis; PTN: Pristinamycine; SA: Staphylococcus Aureus; SXT: Sulfamethoxazole-Trimetroprime; TIC: Ticarcillin-Clavulanic Acid; TOB: Tobramycin; VAN: Vancomycin; WHO: World Health Organization
Introduction
Neonatal Sepsis (NS) is a major cause of morbidity and mortality and is the third leading cause of death worldwide [1]. The rapid evolution of these infections is the source of diagnostic concern for clinicians, which contributes to prescribe a significant and sometimes inappropriate prescription of broad-spectrum antibiotics. This exposition to antibiotics contributes to the emergence of resistant bacteria [2], which complicates the therapeutic treatment. In 2016, the first estimate of neonatal deaths attributable to antimicrobial resistance was published [3]. Multidrug-resistant pathogens accounted for approximately 30% of all neonatal sepsis mortality worldwide [3]. In Africa, the situation is more worrisome [4] due to insufficient access to last-resort antibiotics, a higher burden of infectious diseases, weak health systems, and limited resources. However, in a study conducted in Côte d'Ivoire in 2010, most bacteria causing neonatal sepsis were susceptible [5]. Moreover, in recent years, it is clear that few studies concerning antibiotic resistance in neonatology have been conducted. Therapeutic management may therefore be ineffective. The objective of this study was therefore to determine the bacteria responsible of neonatal sepsis and to describe their antibiotic susceptibility patterns.
Materials and Methods
Conception and Study Design
This was a retrospective cross-sectional study that took place at the medical biology department of the CHU of Angre. Data from the neonatal unit from January 1 to December 31, 2020 were examined. On the one hand, clinical data and biological parameters (CBC and CRP) were collected from the patients' medical files. On the other hand, microbiological data that include dates of blood cultures, blood culture positivity, identified organisms, and antibiotic susceptibility were extracted from the laboratory computerized database.
Study Population
Culture-proven NS was defined as one or more blood culture obtained at 0–28 days of life growing a recognized pathogenic bacterium or fungus and a clinical diagnosis of sepsis [6]. After analysis of the medical files, all clinically suspected cases of neonatal sepsis admitted to the neonatal department of the CHU of Angre were listed. Out of them, only neonates with a positive blood culture were included in the study. Neonates were defined as patients whose age ranged from 0 to 28 days of life.
Culture and Identification
Microorganism identification and culture were conducted according to the routine diagnostic standard operation procedure used by the clinical laboratory in the study hospital: For any new-born, 0.5-3 mL of blood sample was inoculated into a commercial culture bottle exclusively for new-borns and analyzed using an automated monitoring system for bacterial detection (BioMérieux Bact/ALERT). Incubation was continued until a positive result was observed or up to a maximum of 7 days. Subcultures are typically made on to blood and chocolate agar. They are incubated aerobically at 37°C with 5 to 10% CO2 for 24 to 48 hours. The microorganisms were identified to species level by conventional biochemical techniques or automated methods with the VITEK system (BioMérieux). The bacterial isolates were tested for susceptibility to antibiotics using VITEK MD 2 Compact® (BioMérieux) or the manual Kirby–Bauer disc diffusion method, as per interpretive standards established by the CA-SFM/ EUCAST 2019. Quality control strains were used: Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603 and Staphylococcus aureus ATCC 29213. Routinely at the laboratory, positive blood cultures are evaluated to determine whether they represent true bacteraemia or contamination. They are each reviewed by the clinical microbiologist for decisions on the significance of the organism and the appropriate antibiotic susceptibility data to report. The clinical microbiologists have access to patients’ clinical information. Thus, Coagulase-Negative Staphylococci (CoNS), micrococci, propionibacteria, and corynebacteria grown alone in a single sample were considered contaminants and excluded. CoNS were included only when cultivated in two or more blood samples. Bacteria like Staphylococcus Aureus (SA), Enterobacteriaceae, Pseudomonas, and Acinetobacter which grown alone in a single sample without clinical signs were considered contaminants and excluded too. Repeatedly positive samples were considered to represent the same episode of infection.
Statistical Analyses
Data were entered into an Excel spreadsheet (Microsoft excel version 2019). Epi info version 7.2.5.0 was used for data analyses. Mean and standard deviation were used to describe the central tendency of continuous variables based on the distribution of the data. Categorical variables were described using frequency and percentages.
Ethical Approval and Consent to Participate
The study was based entirely on routine clinical and laboratory data. The consent to participate wasn’t so obligatory. Ethics approval was obtained from the Biomedical Research Ethics Committee of the University of Felix Houphouet Boigny and the medical and scientific management of the CHU of Angre. All data were de-identified from the routine database used and shared with the investigators in a password-protected file, to which only the principal investigator and the laboratory manager had access.
Results
General Characteristics
Between the study period, the microbiology laboratory received 221 blood cultures of new-borns and a total of 98 (44.34%) organisms were isolated. Out of the isolates, 16 were considered contamination by commensals and were excluded in further analyses: Staphylococcus aureus n=7, CoNS n=4, Klebsiella pneumoniae n=4, andEscherichia coli n=1. The rate of clinically relevant bacteraemia amongst the neonates was 37.10% (82/221). Of the 82 neonates included, 44 (53.65%) were male with a sex ratio of 1.15. The median age was 3.76 ±7.53 days. According to the time of sepsis, 70/82 (85.36%) cases were in the age range of day 0 to day 7. 24.39% (20/82) were born at term and 13.41% (11/82) were preterm. At birth, 22 (26.83%) were eutrophic, 7 (8.54%) hypotrophic, and 1 (1.22%) had macrosomia. Age and birth weight were not found in 62.20% (51/82) and 63.41% (52/82) of the cases respectively. It should be noted that these clinical data were not recorded by the prescribers in the patients' files.
Isolated Bacteria from Neonatal Sepsis
A total of 82 isolates were identified (Table 1). Sepsis was primarily caused by Gram-positive bacteria (n=45, 54.88%), compared with Gram-negative bacteria (n=37, 45.12%). The most frequent pathogens were CoNS (n=24, 29.27%), K. pneumoniae (n=22, 26.83%), and S. aureus (n=16, 19.51%). Others pathogens included are shown in the (Table 1).
GRAM
Species
Frequency n (%)
Gram-positive bacteria
N=45Coagulase negative Staphylococcus
24 (29,27)
Staphylococcus aureus
16 (19,51)
Enterococcus spp
2 (2,44)
Streptococcus agalactiae
2 (2,44)
Streptococcus pneumoniae
1 (1,22)
Gram-negative bacteria
N=37Klebsiella pneumoniae
22 (26,83)
Enterobacter cloacae
7 (8,54)
Escherichia coli
5 (6,10)
Citrobacter koseri
1 (1,22)
Citrobacter freundii
1 (1,22)
Acinetobacter baumannii
1 (1,22)
Total
82 (100)
Table 1: Distribution of isolated bacteria neonatal sepsis.
Antibiotic Susceptibility Pattern
For Gram-positive cocci, the (Table 2) showed the distribution of resistance pattern of CoNS and SA. Out of CoNS, the resistance to methicillin was observed in 37.50% (9/24) of cases. CoNS was resistant to gentamycin (n=5; 20.83%), ciprofloxacin (n=7, 29.16%), levofloxacin (n=7, 29.16%), sulfamethoxazole-trimetroprime (n=9, 37.50%), and erythromycin (n=6, 25%). All strains of CoNS were sensitives to vancomycin.
Species
Resistance to antibiotics tested n (%)
FOX
KMN
TOB
CN
CIP
LEV
ERY
CMN
PTN
SXT
VAN
CoNS
(N=24)9 (37.50)
5
(20.83)7
(29.16)7
(29.16)6
(25%)1
(4.17)1
(4.17)9
(37.50%)0
SA
(N=16)5
(31.25)
2
(12.50)3
(18.75)2
(12.50)5
(31.25)4
(25%)2
(12.50)5
(31.25)2
(12.50)CoNS: Coagulase Negative Staphylococcus; SA: Staphylococcus aureus; FOX: Cefoxitin; KMN: Kanamycine; TOB: Tobramycin; CN: Gentamycin; CIP: Ciprofloxacin; ERY: Erythromycin; CMN: Clindamycin; SXT: Sulfamethoxazole-trimetroprime; VAN: Vancomycin
Table 2: Distribution of antibiotic-resistant Staphylococcus strains.
Methicillin Resistant Staphylococcus Aureus (MRSA) was observed in 31.25% (5/16). Among Staphylococcus Aureus; 12.50% (n=2) were resistant to gentamycin, 12.50% (n=2) to levofloxacin, and 18.75% (n=3) to ciprofloxacin. Also, SA was resistant to erythromycin (n=5, 31.25%) and vancomycin (n=2, 12.5%). All strains of SA were resistant to sulfamethoxazole-trimetroprime.
Moreover, all strains of Streptococcus agalactiae and Streptococcus pneumoniae were sensitives.
All of Gram-negative bacilli, the Extended Spectrum Beta Lactamase (ESBL) producing Enterobacteria represented 80.56% (29/36) of cases. The Acinetobacter baumannii strain also produced an ESBL. All Gram-negative bacilli resistances are listed in (Table 3).
Species
Resistance to Antibiotics tested n (%)
AMC
TCC
CRO
CTZ
IMP
ETP
CN
AMK
CIP
SXT
K. pneumoniae
(N=22)19 (86.36)
NT
22 (100)
NT
0
5
(22.72)20
(90.90)0
9
(40.90%)21
(95.45)E. cloacae
(N=7)6
(85.71)NT
2
(28.57)NT
1
(14.28)2
(28.57)3
(42.85)0
1
(14.28)6
(85.71)E. coli
(N=5)0
NT
3
(60)NT
0
1
(20)4
(80)0
0
5
(100)A.baumannii
(N=1)NT
1
(100)NT
1
(100)0
0
1
(100)0
1
(100)K. pneumoniae: Klebsiella pneumonia; E. cloacae: Enterobacter cloacae; E. coli: Escherichia coli; AMC: Amoxicillin-Clavulanic Acid; TCC: Ticarcillin-Clavulanic Acid; CRO: Ceftriaxone; CTZ: Ceftazidime; CN: Gentamycin; AMK: Amikacin; CIP: Ciprofloxacin; SXT: Sulfamethoxazole-Trimetroprime
Table 3: Distribution of antibiotic-resistant Gram-negative bacilli strains.
Multi-Drug Resistant Bacteria
Multi-Drug Resistant (MDR) bacteria (resistant to at least 3 families of antibiotics). are showed by (Figure 1). They were mainly represented by ESBL producing Enterobacteria (n=27; 32.93%). The others BMR were represented by MR-CoNS (n=9; 10.98%), MRSA (n=4, 4.87%), and cephalosporinase-hyper producing enterobacteria (n=1; 1.22%).
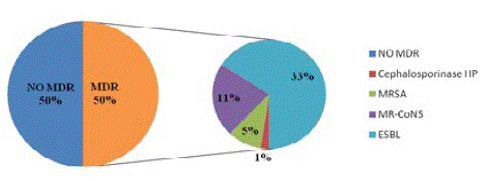
Figure 1: Proportion of multi-drug resistant bacteria.
Discussion
Diagnosis of neonatal sepsis is a challenge. Signs and symptoms of neonatal sepsis are non specific especially in the first days of life and difficult to differentiate from other neonatal pathologies [7,8]. Blood cultures are the « gold standard » for the detection of bacteraemia. The objective of this study was therefore to determine the bacteria responsible of neonatal sepsis and to describe their antibiotic susceptibility patterns. In our study, the prevalence of neonatal bacteraemia was 37.10% (82/221). This result is comparable to those of some studies from West African countries: 37.6% in Nigeria [9], 38.95% in Benin [10], and 34.7% in Cameroon [11]. However; lower prevalences were found in 2016 in Cocody, Ivory coast (22.7%) [5], Ghana (17.3%) [12], and Johannesburg, South Africa (8.5%) [13]. In developed countries, the prevalence of neonatal sepsis range at 0.5% to 1% [14]. Generally, these high rates of neonatal bacteraemia observed in our countries contrast with those in developed countries. This could be explained by real differences in the qualification of maternal and neonatal care, and neonatal units’ types [15]. and the lack of hygiene in ours hospitals. Furthermore, not all negative blood cultures exclude sepsis [16]. Indeed, a low rate of blood culture positive could be due to the administration of an antibiotic before blood collection or to the possibility of infection with other microorganisms. Adenovirus, enterovirus, coxsackievirus, rubella virus, and toxoplasma species have been implicated in neonatal sepsis [17]. Also, about 26% of all neonatal sepsis cases could be attributed to anaerobes [18]. Unfortunately, there is no uniform consensus definition for neonatal sepsis [19-21]. Many neonates are therefore diagnosed with "probable or possible" sepsis or "presumed symptomatic infection but without an identified bacterial cause" [22]; conditions often referred to as "culture-negative sepsis" [23]. Therefore, there is an urgent need to unify guidelines for the identification and management of neonatal sepsis.The main isolated bacteria were Gram-positive cocci (n=44, 53.65%). This is in accordance with findings from Saudi Arabia (27/40; 67.5%) [24], and China (57,49%) [25]. However, Gram-negative bacilli were the predominant bacteria isolatedin anothers countries like Congo [15], Nigeria [9], Cameroun [26], and Nepal [27]. CoNS (28.04%) was the predominant bacteria identified in our study followed by Klebsiella pneumoniae (26,83%).
Andrianarivelo in Madagascar [28], Li and Guo in China [25,29], and Tessema in Germany [30] also reported a predominance of coagulase-negative Staphylococcus in their respective studies. However, in anothers studies in Africa, Klebsiella pneumoniae and Escherichia coli were the main species. These are the cases of Nigeria [9], Congo [15], Cameroon [26], and India [31]. This disparity in the isolated bacteria is in accordance with the fact that the epidemiology of neonatal sepsis is extremely variable throughout the world [32], making it difficult to establish differences between countries.
Antibiotic resistance is a global problem. In our study, the rates of MRSA and methicillin-resistant CoNS were 31.25% and 39.13% respectively. This finding is similar to a study performed in Morocco [33] which observed a MRSA rate of 31.08%. Lower MRSA rates have been observed. This is the case of a study carried out by the Pasteur Institute of Madagascar in 2007, which reported an MRSA rate of 9% out of 54 isolated strains of Staphylococcus aureus [34]. These differences may be justified by the fact that our study is much more recent and that reports of multidrug-resistant bacteria causing neonatal sepsis in developing countries are increasing [35]. Elsewhere, the prevalence of MRSA is much higher. This is the case in Nepal, where Chaudhary et al. found a prevalence of MRSA of 60% [27]. In a study in Ethiopia, MRSA was found in 66.7% of cases [36]. Also, we estimated the proportion of ESBL producing Enterobacteriaceae at 80.56%. This is contrary in Madagascar where the proportion of ESBL producing Enterobacteriaceae was estimated at 100% [28]. Lower rates were observed in South Africa (60.9%) [37] and China (62%) [29]. In our study, the overall rate of MDR bacteria was 50% and was mainly represented by ESBL producing Enterobacteriaceae, MR CoNS, and MRSA. This rate is comparable to a study performed in Dessie (Ethiopia) [36]. Higher rates were found by Chaudhary [27], Agarwal [38], and Pokhrel [39].
Our study has several limitations. It was a retrospective study that did not allow for an exhaustive collection of all data. Some clinical files were missing data. It was therefore impossible to analyze the risk factors associated with neonatal bacteraemia, the acquisition of multidrug-resistant bacteria, and sepsis-related mortality. Also, the high rate of MDR bacteria makes us suspect a nosocomial character, which was not investigated in our study. Finally, the small size of the population studied, due to the fact that the CHU of Angré is a new hospital, does not allow us to generalize the results.
Conclusion
In our study, the prevalence of neonatal bacteraemia is high. The main bacteria were Coagulase negative Staphylococcus and Klebsiella pneumoniae. The increasing rate of multi-drug resistance bacteria makes the treatment of neonatal bacteraemia difficult. It is very important to respect the rules of antibiotic prescription and the need to establish new guidelines for the management of neonatal bacteraemia.
Acknowledgements
The authors would like to thank the bacteriology team of the medical biology laboratory and the Paediatrics and Specialties Department of the Angré University Hospital.
Funding
This research work did not benefit from any financial support.
References
- Hug L, Alexander M, You D, Alkema L. National, regional, and global levels and trends in neonatal mortality between 1990 and 2017, with scenario-based projections to 2030: a systematic analysis. Lancet Global Health. 2019; 7: e710–e720.
- Arnaud I, Jarlier V, Carbonne-Berger A, Maugat A, Bajolet O, et al. Multi-drug resistant bacteria at hospital : Extended Spectrum Beta Lactamase (ESBL) producing Enterobacteria and Methicillin Resistant Staphylococcus aureus (MRSA), BMR-Raisin Network. 2002-2010. BEH. 2012; 42-43: 472-476.
- Laxminarayan R, Matsoso P, Suraj P, Broyer C, Rottingen JA, et al. Access to effective antimicrobials: a worldwide challenge. Lancet. 2016; 387: 168–75.
- Lawn JE, Wilczynska-Ketende K, Cousens SN. Estimating the causes of 4 millionneonataldeaths in the year 2000. Int J Epidemiol. 2006; 35: 706-718.
- Folquet-Amorissani M, Dainguy ME, Diomande D, Kouakou C, Kamenan M, et al. Actualization of the profile of new-borns’bacterial infections at the CHU of Cocody in Abidjan. Journal of paediatrics and child care. 2016; 29: 8-14.
- Subspecialty Group of Neonatology Paediatrics Society Chinese Medical Association; Editorial Board Chinese Journal of Paediatrics. Protocol for diagnosis and treatment of neonatal septicaemia. Zhonghua. Er Ke Za Zhi. 2003; 41: 897-899.
- Hedegaard SS, Wisborg K, Hvas AM. Diagnostic utility of biomarkers for neonatal sepsis - a systematicreview. Infect Dis (Lond). 2015; 47: 117–124.
- Shah BA, Padbury JF. Neonatal sepsis: an oldproblemwith new insights. Virulence. 2014; 5: 170–178.
- Olorukooba AA, Ifesumu WR, Ibrahim SM, Jibril MB, Awadu L, et al. Prevalence and factors associated withneonatal sepsis in a tertiary hospital, North West Nigeria. Nigerian Medical Journal. 2020; 61: 60-66.
- Omoregie R, Egbe CA, Dirisu J, Ogefere HO. Microbiology of neonatalsepticemia in a tertiaryhospital in Benin City, Nigeria. Biomarkers Genomic Med. 2013; 5: 142 146.
- Chiabi A, Djoupomb M, Mah E, Nguefack S, Mbuagbaw L, et al. The clinical and bacteriological spectum of neonatal sepsis in a tertiaryhospital in Yaounde Cameroon. Iran J Pediatr. 2011; 21: 441-448.
- Aku FY, Akweongo P, Nyarko K, Sackey S, Wurapa F, et al. Bacteriological profile and antibiotic susceptibility pattern of commonisolates of neonatal sepsis, Ho Municipality, Ghana-2016. Matern Health Neonatol Perinatol. 2018; 4: 2.
- Motara F, Ballot DE, Perovic O. Epidemiology of neonatal sepsis at Johannesburg Hospital. S Afr J Epidemiol Infect. 2005; 20: 90–93.
- Aujard Y. Neonatal infections. Medical and surgical encyclopaedia. Elsevier Masson. 2015.
- Mulongo Mbarambara Ph, Wakeka Kabyuma C, Mudimba Lamata M, Bahati Maheshe JP, Kyambikwa Bisangamo C. Frequency and risk factors of neonatal infections to the General Reference Hospital of Uvira (East DR Congo). Technologies laboratory. 2015; 9.
- Tewabe T, Mohammed S, Tilahun T, Melaku B, Fenta M, et al. Clinical outcome and risk factors of neonatal sepsis among neonates in FelegeHiwot referral Hospital, Bahir Dar, Amhara Regional State, North West Ethiopia 2016: a retrospective chart review. BMC Res Notes. 2017; 10: 265.
- Glaser MA, Hughes LM, Jnah A, Newberry D. Neonatal sepsis: a review of pathophysiology and current management strategies. Advances in Neonatal Care. 2021; 21: 49–60.
- Klingenberg C, Kornelisse RF, Buonocore G, Maier RF, Stocker M. Culture-negative early-onset neonatal sepsis at the crossroad between efficient sepsis care and antimicrobial stewardship. Front Pediatr. 2018; 6: 285.
- Wynn JL, Polin RA. Progress in the management of neonatal sepsis: the importance of a consensus definition. Pediatr Res. 2018; 83: 13–15.
- Haque KN. Definitions of bloodstream infection in the new-born. Pediatr Crit Care Med. 2005; 6: 45–49.
- Wynn JL, Wong HR, Shanley TP, Bizzarro MJ, Saiman L, Polin RA. Time for a neonatal-specific consensus definition for sepsis. Pediatr Crit Care Med. 2014; 15: 523–528.
- National Institute for Health and Care Excellence (NICE). Antibiotics for early-onset neonatal infection: antibiotics for the prevention and treatment of early-onset neonatal infection. London Clinical guideline [CG149]. 2012.
- Cantey JB, Baird SD. Ending the culture of culture-negative sepsis in the neonatal ICU. Pediatrics. 2017; 140: e20170044.
- Alharbi AS. Common bacterial isolates associated with neonatal sepsis and their antimicrobial profile: A retrospective study at king Abdulaziz University Hospital, Jeddah, Saudi Arabia. Cureus. 2022; 14: e21107.
- Li X, Ding X, Shi P, Zhu Y, Huang Y, Li et al. Clinicalfeatures and antimicrobialsusceptibility profiles of culture-provenneonatal sepsis in a tertiary children’s hospital, 2013 to 2017. Medicine. 2019; 98: e14686.
- Kemeze S, Moudze B, Chiabi A, Eposse C, Kaya A, et al. Profilclinique et bactériologique des infections néonatales bactériennes à l’HôpitalLaquintinie de Douala, Cameroun. Pan African Medical Journal. 2016; 23: 97.
- Chaudhary BR, Malla KK, Poudel S, Jha BK. Study of antibiotic susceptibility among bacterial isolates in neonatal intensive care unit of a tertiary care hospital: a descriptive cross-sectional study. J Nepal Med Assoc. 2020; 58: 893-899.
- Andrianarivelo AM, Rafaravavy NE, Rafalimanana C, Andriantahiana TN, Robinson AL. Bacteriological profile of neonatal infections in the neonatal intensive care unit of the Maternity Hospital of Befelatanana. Journal of Anaesthesia and Emergency Medicine. 2010; 2: 1-4.
- Junfei Guo, Yasha Luo, Yongbing Wu, Weiming Lai, Xiaoping Mu.Clinical characteristic and pathogen spectrum of neonatal sepsis in guangzhou city from June 2011 to June 2017. Med Sci Monit. 2019; 25: 2296-2304.
- Tessema B, Lippmann N, Knüpfer M, Sack U, König B. Antibiotic resistance patterns of bacterial isolates from neonatal sepsis patients at University Hospital of Leipzig, Germany. Antibiotics. 2021; 10: 323.
- Report of the National Neonatal Perinatal Database.Neonatal morbidity and mortality. Indian Pediatr. 1997; 34: 1039-1042.
- Vergnano S, Sharland M, Kazembe P, Mwansambo C, Heath PT. Neonatal sepsis: An international perspective. Arch Dis Child Fetal Neonatal. 2005; 90: 220-224.
- Elmouali A. Blood culture: bacteriological profile and antibiotic resistance at the IBN SINA hospital in Rabat. Pharmacy thesis. Mohamed V University. 2012. Thesis N°64.
- Institut Pasteur of Madagascar (IPM), Ministry of Health and Family Planning (MIN/SAN/P-F), French cooperation. Bacterial resistance to antibiotics. PM 2007.
- Jyothi P, Basavaraj MC, Basavaraj PV. Bacteriological profile of neonatal septicemia and antibiotic susceptibility pattern of the isolates. Journal of Natural Science, Biology and Medicine. 2013; 4: 306-309.
- Fenta GM, Woldemariam HK, Metaferia Y, Seid A, Gebretsadik D. Admission outcome and antimicrobial resistance pattern of bacterial isolates among neonates with suspected sepsis in Neonatal Intensive Care Unit at Dessie Comprehensive Specialized Hospital, Dessie, Northeastern Ethiopia. Interdisciplinary Perspectives on Infectious Diseases. 2022; 2022: 1318295.
- Pillay D, Naidoo L, Swe-Han KS, Mahabeer Y. Neonatal sepsis in a tertiary unit in South Africa. BMC Infectious Diseases. 2021; 21: 225.
- Agarwal R, Sankar J. Characterisation and antimicrobial resistance of sepsis pathogens in neonates born in tertiary care centres in Delhi, India: a cohort study. Lancet Global Health. 2016; 4: 752-760.
- Pokhrel B, Koirala T, Shah G, Joshi S, Baral P. Bacteriological profile and antibiotic susceptibility of neonatal sepsis in neonatal intensive care unit of a tertiary hospital in Nepal. BMC paediatrics. 2018; 18: 208.