
Research Article
J Bacteriol Mycol. 2023; 10(2): 1207.
Mechanisms of Antimicrobial Resistance and Its Diagnostic Techniques in Nontyphoidal Salmonella Infections
Hambisa EM*; Umer AA
Animal Health Institute (AHI), PO Box: 04, Sebeta, Ethiopia
*Corresponding author: Hambisa EM Animal Health Institute (AHI), P.O.Box: 04, Sebeta, Ethiopia Tel: +251 932320809 Email: [email protected]
Received: June 01, 2023 Accepted: June 29, 2023 Published: July 06, 2023
Abstract
Antimicrobial Resistance (AMR) is the decreased sensitivity of microbes to drugs that are capable of causing cell death or inhibition of growth. It is one of the key issues linked with foodborne infections caused by various resistant pathogenic microorganisms. Nontyphoidal Salmonella (NTS) is one of the leading causes of food-borne infections in which its serotypes are usually zoonotic. The source of this resistance is classified as natural and acquired and the mechanism of AMR emergence in NTS can also be divided into two broad categories: biochemical and genetic mechanisms. The biochemical mechanisms are enzymatic inactivation, antimicrobial permeability reduction, active efflux pumps, and others; while the genetics mechanisms include mutation and horizontal and vertical resistant gene transfer. The techniques of AMR diagnosis are conducted by conventional and non-conventional methods. The utilization of antimicrobials for growth promotion, prophylaxis and its misuse in humans or animals, and contamination of the environment are some of the factors that attribute to AMR development. NTS infection is common throughout the world including Ethiopia and developed resistance to different antimicrobials, and the mechanism of AMR development and its diagnosis is not common in the developing countries including our country. The mechanisms by which NTS develop resistance to antimicrobials should be known very well and its dissemination to human, animals, and the environment could be managed, and methods for the diagnosis of AMR in NTS species should be available for the identification of resistant antimicrobials to give proper treatments and other measures. Additionally, proper policies and regulation systems on antimicrobial use and its distribution should be developed and implemented properly.
Keywords: Antimicrobial resistance; Diagnosis; Mechanism; Nontyphoidal salmonella
Abbreviations: ABC: ATP-Binding Cassette; AMR: Antimicrobial Resistance; ARGs: Antimicrobial Resistant Genes; ATP: Adenosine Triphosphate; BLAST: Basic Local Alignment Search Tool; DNA: Deoxyribonucleic Acid; FTIR: Fourier Transform Infrared; HGT: Horizontal Gene Transfer; HT-qPCR: High Throughput Quantitative PCR; iNTS: Invasive Nontyphoidal Salmonellae; LM PCR: Ligation Mediated Polymerase Chain Reaction; LPS: Lipopolysaccharide; MALDI-TOF: Matrix Assisted Laser Desorption/Ionization Time-of-Flight; MDR: Multi-Drug Resistant; MIC: Minimum Inhibitory Concentrations; MLST: Multi Locus Sequence Typing; NGS: Next Generation Sequencing; NTS: Nontyphoidal Salmonella; PCR: Polymerase Chain Reaction; PMQR: Plasmid Mediated Quinolone Resistance; QCM: Quartz Crystal Microbalance; QRDRs: Quinolone Resistance Determining Regions; RNA: Ribonucleic acid; RT-PCR: Reverse Transcriptase Polymerase Chain Reaction; SERS: Surface Enhanced Raman Spectroscopy; SGI1: Salmonella Genomic Island 1; T4SS: Type IV Secretion System; WGS: Whole Genome Sequencing; WHO: World Health Organization; WMS: Whole Metagenome Sequencing
Introduction
The global increment of foodborne infections linked with antimicrobial-resistant pathogenic microorganisms and the dissemination of Antimicrobial Resistance (AMR) is one of the key issues in developing and developed countries. The emergence of multi-antimicrobial resistant Salmonella strains and the continuous spread of its clones is one related concern for human health in different countries. Incidents of multidrug resistance in Salmonella species, and other bacterial pathogens causing enteric diseases, have been reported in different countries and became a main health concern as they can spread worldwide [1].
Salmonella is a genus of the Enterobacteriaceae family which is characterized as a gram-negative, rod-shaped, non-spore-forming, and facultative anaerobic bacterium and moves using a peritrichous flagellum. Based on the serotype, its genome ranges from 4460 to 4857 Kilobase (kb) [2]. Salmonella can be divided into typhoid and Non-Typhoid Salmonella (NTS) regarding their ability to develop specific pathologies in humans. Typhoid serovars are a subcategory of serovars known as specialists (adapted), capable of infecting and colonizing only a very narrow range of hosts, and include the Typhi, Sendai, and Paratyphi A, B, and C serovars, highly adapted to humans and presenting only higher primates and humans as reservoirs. Generalist serovars are capable of triggering infections in both humans and animals, including NTS Enteritidis and Typhimurium, known mainly for their epidemiological relevance. These serovars are mainly transmitted by animal-based foods, such as beef, pork, poultry, and contaminated raw eggs, fruits, and vegetables can also serve as vehicles. Poor access to improved water supplies and inadequate sanitation facilities, combined with growing urbanization, favor the transmission of NTS serovars [3].
Some NTS serovars demonstrate evident host predilection and usually are associated with a specific animal host, such as Choleraesuis and Dublin, which prefer pigs and cattle, respectively, but may also infrequently cause disease in other mammals, including humans. These NTS can cause invasive infections, and invasive NTS (iNTS) in humans [4].
Nontyphoidal Salmonella is a leading cause of foodborne infection worldwide in which its serotypes are usually zoonotic and have a wide range of animal reservoirs [5]. Some serotypes of Salmonella enterica (S. enterica) are considered emerging zoonotic pathogens, generating outbreaks worldwide in the human population. It is estimated that S. enterica gastroenteritis is responsible for about 93.8 million illnesses and 155,000 deaths worldwide each year, and of these, 80.3 million cases are estimated to be foodborne [6]. It is a facultative intracellular pathogen that is capable of causing different disease syndromes in a wide range of hosts. S. Typhimurium (S. Typhimurium) and Salmonella Enteritidis (S. Enteritidis) are the most frequently isolated serovars throughout the world, leading to severe economic losses [7].
Poultry and other food animals are considered the common reservoirs of S. enterica and undercooked poultry products are the major sources of human infection with NTS. Several studies on Salmonella isolates from poultry products and farms in the past studies were found to be resistant to several antimicrobials. Information on farm-level prevalence and antimicrobial susceptibility status of isolates can explain the level of public health risk associated with poultry products [8]. Antimicrobial Resistance is the decreased sensitivity of microbes to antibiotics that are capable of causing cell death or inhibiting growth [9]. This can be determined through antimicrobial sensitivity testing of Salmonella isolates to determine their susceptibility or resistance to the antibiotics. Resistance in Salmonella is encoded by genes that are present on either chromosome or extra-chromosomal DNA (plasmid) or transferable genetic materials (transposons, integrons), which is determined by genetic or molecular methods. Although resistance may occur due to mutation in key genetic loci in the bacterial genome, most resistance to antimicrobial agents mediated by genes is acquired via mobile genetic elements such as plasmids and transposons [10].
The mechanisms of antimicrobial development to NTS can result from enzymatic inactivation, decreased permeability, and development of efflux pump systems, alteration of target sites, and in most cases in many serovars the overproduction of target sites to overwhelm the used antibiotics. In several cases investigated, antibiotic resistance can be acquired through natural selection or mutation (induced or spontaneous); this however can be chromosomal mutation by the production of chromosomally mediated inducible enzymes or acquisition of plasmid-resistant genes which is the most common genetic basis of antibiotic resistance [11].
The diagnosis of resistance genotype is accomplished through the detection of novel genetic materials and characterization of mutations in specific genes through Polymerase Chain Reaction (PCR), Deoxyribonucleic Acid (DNA) probes, and other amplification techniques [12]. Genotypic analysis of the antibiotic-resistant Salmonella species by use of real-time-polymerase chain reaction and molecular fingerprinting of DNA has been used to good effect. Plasmid gene profile analysis is a quick and relatively easy method to fingerprint strains and has been used in both human and veterinary medicine to study the spread of AMR Salmonella. Phage typing or alternative genetic techniques and full DNA sequencing are increasingly used to study genetic variations in AMR Salmonella species chiefly because of their low-cost automated methods [13].
However, the development of resistance in the responsible pathogens has worsened the situation often with very few resources to investigate and provide reliable susceptibility data on which rational treatments can be based as well as means to optimize the use of antimicrobial agents in most of the developing countries [14]. NTS is one of these resistance-developed microbes having major public health concerns and economic importance throughout the world, and the mechanism of AMR development and its diagnosis is not common in developing countries including Ethiopia.
In light of the above background information and existing facts, this review paper is prepared with the following objectives:
To review the mechanisms by which NTS develops resistance to antimicrobials
To describe the techniques that are used for the diagnosis of AMR in NTS species
Origins of Antimicrobial Resistance
Bacteria as a group or species are not necessarily uniformly susceptible or resistant to any particular antimicrobial agent. Levels of resistance may vary greatly within related bacterial groups. Susceptibility and resistance to antimicrobial agents are often measured as a function of Minimum Inhibitory Concentration (MIC), the minimal concentration of a drug that will inhibit the growth of the bacteria. The susceptibility is a range of the average MICs for any given drug across the same bacterial species. If that average MIC for a species is in the resistant part of the range, the species is considered to have intrinsic resistance to that drug. Bacteria may also acquire resistance genes from other related organisms, and the level of resistance will vary depending on the species and the genes acquired [15].
Natural Resistance
Intrinsic resistance: Intrinsic resistance may be defined as a trait that is shared universally within a bacterial species, is independent of previous antibiotic exposure, and is not related to horizontal gene transfer. The most common intrinsic resistance forms are reduced permeability of the outer membrane (most specifically the Lipopolysaccharide (LPS), in gram-negative bacteria) and the natural activity of efflux pumps [16].
Induced resistance: Induced resistance means that the genes are naturally occurring in the bacteria, but are only expressed to resistance levels after exposure to an antibiotic. Multidrug-efflux pumps are one of the common mechanisms of induced resistance [17].
Acquired Resistance
The genetic material that confers resistance can be acquired through all of the main routes by which bacteria acquire any genetic material: transformation, transposition, and conjugation; all termed Horizontal Gene Transfer (HGT); and, the bacteria may experience mutations to its chromosomal DNA. The acquisition may be temporary or permanent. Plasmid-mediated transmission of resistance genes is the most common route for the acquisition of outside genetic material; bacteriophage-borne transmission is fairly rare. Bacteria can be naturally competent and as a result capable of acquiring genetic material directly from the outside environment. Internally, insertion sequences and integrins may move genetic material around, and stressors (starvation, ultraviolet radiation, chemicals, etc.) are the common causes of genetic mutations (substitutions, deletions, and so on) [18].
Mechanisms of Antimicrobial Resistance In Nontyphoidal Salmonella Infections
Various mechanisms of AMR have been reported and these mechanisms lead to the emergence of multidrug resistance in Salmonella species. These mechanisms can be classified into two broad categories which involve biochemical mechanisms and genetic mechanisms [19].
Biochemical Mechanisms of Antimicrobial Resistance
Enzymatic inactivation: The enzymatic mechanisms of antibiotic resistance include hydrolysis, group transfer, and redox processes [20]. In terms of diversity, evolution, and spread, antibiotic resistance enzymes contribute remarkably to the bacterial ability to overcome antibiotic pressure. The Β-lactamases are the oldest known and the most diverse antibiotic-degrading enzymes that cleave the Β-lactam ring of the penicillin group of antibiotics and render them ineffective. Scientific evidence suggests the existence of Β-lactamases before penicillin was clinically employed, emphasizing that the production of antimicrobial compounds and the mechanisms to endure them occur in parallel in the environment [21].
Antimicrobial permeability reduction: The other mechanism of resistance to antimicrobial agents involves preventing drug permeability and access to the internal milieu of the pathogenic cells [22]. The molecular systems involved in the reduced permeability of antimicrobial agents include resistance mechanisms at the bacterial cell wall. The extensive structural nature of the LPS layer constitutes a formidable barrier to the passage of small molecules, especially those that are growth inhibitory in their properties [23]. Another important molecular mechanism for conferring resistance via permeability reduction involves porins, which are integral outer membrane proteins with water-filled pore-like channels that permit the passage of molecules with definitive sizes and charges. The relationship between bacterial AMR and the outer membrane porins can take one of several ways. A wide-type porin can be highly selective towards the entry of certain nutrients, like sugars, and not permit the passage of many antimicrobial agents. However, for those porins for which no such highly selective properties are a problem, then in such cases, the porin molecules may be depleted from the membrane or functionally disrupted by mutation. In other cases, permissive porins may be regulated by channel blockers or by Ribonucleic Acid (RNA) specific antisense modulators [24].
Active efflux pumps of antimicrobial agents: Efflux pumps are present in all bacteria and are integral parts of bacterial physiology, being involved in diverse functions such as the expulsion of toxic products of metabolism, and the maintenance of homeostasis. However, antibiotics as incidental substrates of efflux pumps have resulted in them being viewed largely as bacterial mechanisms of antimicrobial resistance, and have critical roles in ensuring bacterial survival and evolution into resistant strains. These bacterial multidrug efflux pump systems are energetically driven by Adenosine Triphosphate (ATP) hydrolysis, called primary active transport, and by electrochemical ion gradients or ion motive forces, called secondary active transport [25].
Active transport of antimicrobial agents represents an essential resistance mechanism in bacterial pathogens. As multiple structurally distinct antimicrobial agents with disparate modes of action are exported to the extracellular milieu, their growth inhibitory properties towards bacteria are diminished, if not wholly circumvented. During the primary active transport of antimicrobial agents, bacteria exploit the biological energy stored in the form of intact ATP to export drugs against the drug concentration gradient by performing ATP hydrolysis [26]. During the export of antibacterial agents from bacterial cells, ATP is hydrolyzed to energize the drug translocation through the transporter in an outward direction across the membrane. Thus, as the transporter substrate actively accumulates outside the cell, AMR is conferred upon the bacterial pathogen. One of the best-studied of these primary active drug efflux systems is the ATP-Binding Cassette (ABC) efflux pump family. The ABC transporter represents one of the most abundant protein families known across all taxa of living organisms [27].
Antimicrobial targets alteration: Antimicrobial targets play vital roles in microbial growth or survival and, thus, serve as potentially useful targets for mitigating infection. These targets must differ or be completely absent from humans or the animal species being treated with an antimicrobial to allow for a selective mode of action. A classic example of such a target is peptidoglycan. Peptidoglycan is essential to the growth and survival of many bacterial species and has a chemical structure that is not present in the mammalian hosts they infect. This allows for the targeting of enzymes responsible for the synthesis and assembly of peptidoglycan. The function of proteins associated with these target sites makes it non-viable for a bacterium to evolve resistance by removing these proteins. However, mutations that allow for continued functionality while reducing the ability of an antimicrobial agent to bind them at the target site have been a veritable regularity in the arms race between antimicrobial substances and antimicrobial-resistant bacteria. In addition to peptidoglycan, alteration in target sites has been attributed to ribosomes, nucleic acid enzymes, and LPS [28].
Biofilm formation: A biofilm is a structured consortium of bacteria embedded in a self-produced polymer matrix consisting of polysaccharides, proteins, and DNA. Bacterial biofilms cause chronic infections because they show increased tolerance to antibiotics and disinfectant chemicals as well as resisting phagocytosis and other components of the body's defense system. Biofilm production occurs in many loci, including teeth plaque, water environments, medical catheters, and trauma wounds, and those microorganisms that are found in biofilms are protected from the entry of multiple antimicrobial agents [29]. Thus, biofilms are increasingly becoming a challenge in the human clinical medicine arena when considering potential chemotherapies with antibacterial agents, and this is recognized as one mode of resistance [30].
Antimicrobial targets protection: One of the significant lines of defense against an antimicrobial is the bacterial cell wall. This structure also acts as a physical barrier to encase the cytoplasm and cell membrane from the external world [31]. Prokaryotic cell walls are made up of linear glycan strands cross-linked by small peptides and this peptidoglycan helps to limit which substances can continue towards the cell membrane and ultimately into the cytoplasm. Peptidoglycan also plays an essential role in bacterial growth and proliferation. While the cell wall helps protect cytoplasmic antimicrobial targets, it also ended up being the target for the first natural antibiotic, penicillin, which prevents the complete formation of this barrier by inhibiting peptide cross-linking to occur [32].
With this mechanism of protection compromised due to the advent of Β-lactam antibiotics, prokaryotes began to synthesize another tier of protection: Β-lactamases. These enzymes help to protect the peptidoglycan cell wall from Β-lactam antibiotics, precisely. Β-lactamase enzymes help confer resistant bacterial phenotypes, as their mechanism of action hydrolyzes the Β-lactam ring of such antibiotics, and the resulting chemical structure can no longer hinder bacterial cell wall synthesis [21] (Figure 1) and (Table 1).
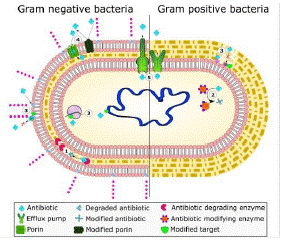
Figure 1: The common mechanisms of AMR in bacteria.
Includes: enzymatic hydrolysis (1), enzymatic modifications of antibiotics by group transfer and redox process (2), modifications of antibiotic targets (3), reduced permeability to antibiotics by modifications of porins (4), and active extrusion of antibiotics by membrane efflux pumps (5).
Source: [21]
Mechanism
Bacterial proteins/targets
responsibleAntibiotic targets
Enzyme
Hydrolysis
β-lactamases
Esterase
C-P lyase complexβ-lactams
Macrolide
FosfomycinGroup transfer
Acetyltransferase
Phosphotransferase
Nucleotidyltransferase
Glycosyltransferase
Ribosyltransferase
Thiol transferaseStreptogramins, aminoglycosides, chloramphenicol
Aminoglycosides, macrolides
Lincomycin,clindamycin, aminoglycosides
Macrolides
Rifampin
FosfomycinRedox process
TetX
Tetracyclines
Target modification
Structural alterations/modifications
Mutations in genes
Amino acid substitutions
Methylation
MutationPenicillin-binding proteins
Cell wall precursors
Ribosomal subunits
RNA polymerase
DNA gyrase/topoisomerase
16S rRNA
23S rRNA
23S rRNAβ-lactam antibiotics
Vancomycin
Streptomycin
Rifamycin
Quinolones
Aminoglycosides
Macrolide
OxazolidinonesReduced
permeabilityReduced expression/defective protein
Porins
β-lactams, fluoroquinolones, aminoglycosides, chloramphenicol
Target Protection
Ribosome protection
Ribosome protection proteins
Tetracycline
Efflux
Active extrusion
Membrane proteins
All major antibiotics
Source: [20]
Table 1: Different Mechanisms of antimicrobial resistance.
Genetics Mechanisms of Antimicrobial Resistance
The genes coding for AMR and virulence at times share common features of both being located in the bacterial chromosome, as well as on plasmid. They are associated in gene clusters to form resistance or Pathogenicity Island, which are transferred by mobile genetic elements such as integrons, transposons, or phage. The major genetic mechanisms of resistance include Mutation, and horizontal and Vertical resistant gene transfer [33].
Mutation: Mutation is one of the mechanisms by which resistance to antimicrobials can be developed. For example, mechanisms of resistance utilized by S. Typhimurium to resist quinolones include the occurrence of point mutations in the Quinolone-Resistance Determining Regions (QRDRs) of topoisomerase genes, efflux pump genes, and plasmids and ciprofloxacin modifying enzymes. The study conducted by [34] revealed that point mutations in the QRDRs of 31 ciprofloxacin-resistant S. Typhimurium were detected.
Mutations that aid in antimicrobial resistance usually only occur in a few types of genes; those encoding drug targets, transporters, regulators that control drug transporters, and those encoding antibiotic-modifying enzymes [16]. Bacteria have an average mutation rate of 1 for every 106 to 109 cell divisions, and most of these mutations will be deleterious to the cell [18]. One huge conundrum of AMR is that the use of these drugs leads to increased resistance. Even the use of low or very low concentrations of antimicrobials (sub-inhibitory) can lead to a selection of high-level resistance in successive bacterial generations, may select for bacteria that are hypermutable strains (increase the mutation rate), may increase the ability to acquire resistance to other antimicrobial agents, and may promote the movement of mobile genetic elements [17].
Horizontal and vertical resistant gene transfer: Horizontal gene transfer has occurred throughout evolutionary history, and one can consider two independent sets of events, largely differentiated by their time and the strength of selection pressure. The horizontal transfer of genetic determinants from a donor bacterium into recipient bacteria is mediated by the Type IV Secretion System (T4SS) encoded on conjugative elements. The presence of several T4SS-associated genes in the corresponding isolates suggests the ability of these isolates to acquire and disseminate genetic determinants such as AMR and virulence genes [35]. In HGT, the genes involved must either have come from commensal or environmental bacteria, since they were not present in human pathogens before the use of antibiotics. There is, in fact, compelling evidence that environmental bacteria are their origin [18].
Diagnostic Techniques of Antimicrobial Resistance
Conventional Methods
Conventional methods of AMR diagnosis mainly include culture-based and molecular-based approaches. Recently, microscopy-based and spectrometry-based approaches have also been incorporated into the tools for developing diagnostics [36].
Phenotypic methods: Culture-based methods can be divided into two categories, manual and automated, and it depends on phenotypic resistance detection by evaluating bacterial growth in the presence of antibiotics. Manual tests include disk diffusion, agar dilution, gradient test, and broth microdilution antimicrobial susceptibility testing methods. Broth dilution-based platforms typically use ready-made cartridges or plates including positive controls and gradient concentrations of antibiotics. Sensititre panels belong to the category of microdilution methods, and typically, such panels are plastic multi-well micro-titer plates precision-dosed with dried antimicrobial agents. Commercial platforms like VITEK COMPACT, Sensititre ARIS 2X, and Alfred 60AST system are some of the automated methods [37].
Such platforms usually offer real-time growth monitoring and MIC analysis through their comprehensive databases which include a broad spectrum of organisms. These methods provide qualitative and quantitative data for the strain under investigation. Disk diffusion provides a zone of inhibition. Epsilometer tests (E-tests) belong to the gradient test methods and are especially useful for fastidious microorganisms. Dilution methods and E-tests provide quantitative values [38] for the MIC, as the lowest concentration of a given antimicrobial which prevents the visible overnight growth of a culture [39].
Molecular methods
PCR-based methods: A PCR is the most commonly used nucleic acid amplification technique for the detection of Antimicrobial Resistant Genes (ARGs) [40], and other techniques like real-time [41], quantitative [42], digital [43], and multiplex [44] PCR assays have further boosted clinical acceptance of genetic testing. The changes in Next Generation Sequencing (NGS) and Whole Genome Sequencing (WGS) have impacted the availability of ARG targets, paving the way for high throughput quantitative PCR (HT-qPCR), which is comparatively fast, convenient, and allowing for simultaneous investigation of a large number of ARGs [45], and it has been successfully used with various sample types, such as animal feces, soil, and surface water [46]. In the past decades, Ligation-Mediated PCR (LM PCR) coupled with a low denaturation temperature method has been proposed leading to specific melting-profile DNA patterns, both fungal and bacterial isolates. This method is suitable for strain characterization and differentiation [47].
Isothermal amplification methods: It is another development, and several methods of isothermal nucleic acid amplification have been developed, which include strand displacement amplification, transcription-mediated amplification, nucleic acid sequence-based amplification, rolling circle amplification, recombinase polymerase amplification, loop-mediated isothermal amplification, and helicase-dependent amplification. These methods have paved the way for the implementation of rapid, next-generation molecular diagnostics [48].
DNA microarrays: A DNA microarray is a tool, which allows for the assessment of bacterial genomic diversity. This approach relies on the detection of the presence or absence of genes in a target organism when compared to a reference strain or genome [49]. There have been many advancements in DNA microarray technology during the past two decades [50]. A fast and simple DNA labeling system based on biotinylated primers specific to the linkers has been developed for disposable microarrays [51].
Non-Conventional Methods
This AMR diagnosing method consists of sequencing, Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS), and Fourier Transforms Infrared (FTIR) spectroscopy.
Genome Sequencing and Metagenomics in AMR Diagnosis: The major technical advancement of NGS was multiplexing, allowing for the simultaneous analysis of thousands of samples. Typically, an NGS workflow comprises DNA extraction and fragmentation, adaptor ligation, DNA amplification, and sequencing. The Illumina platforms, which use synthesis technology where reversible terminator nucleotides labeled with fluorescence are incorporated into DNA strands and visualized via their fluorophore excitation, were subsequently incorporated with the same aim [52].
Oxford Nanopore Technologies developed another approach relying on DNA molecule's movement through a nanopore and measuring an electrical signal changing analogously to the base presently passing the pore [53]. The introduced second and third-generation sequencing approaches have paved the way for single-genome sequencing, as well as for the characterization of complex microbial communities and the identification of antibiotic resistance determinants [54]. Whole Metagenome Sequencing (WMS) and analysis of genetic material in patient samples allow for the identification of ARG directly from clinical specimens without the need for prior isolation or identification of specific bacteria.
Numerous methods, tools, and databases have been reported in past years for the detection of genetic determinants related to AMR from WGS [55] and WMS data [56]. These evolving methods and technologies act as complementary tools to traditional culture-based methods, providing opportunities for rapid and sensitive resistance determination in uncultivable and cultivable bacteria. More information on the use of databases for AMR detection can be found in two recent reviews. The organization of sequencing data is considered a crucial pre-processing step before ARG analysis. Short reads, produced by technologies like Illumina, could be processed employing assembly-based methods (sequencing reads are initially assembled into contiguous fragments followed by annotation where comparison takes place with public or custom reference databases), or directly analyzed utilizing read-based methods where resistance determinants are forecasted by mapping reads to a reference database [57].
The MinION nanopore system (Oxford Nanopore) is a portable, palm-sized, real-time device for DNA and RNA sequencing, able to detect changes in ionic current upon DNA or RNA passing through the nanopores.
Pyrosequencing
The detection and identification based on virulence genes, which led to an assay based on pyrosequencing for characterizing ARG profiles were developed [58]. The efficiency of the pyrosequencing was evaluated based on the rapid detection of resistance to Fluoroquinolones (FQs), Rifampicin (RIF), Kanamycin (KAN), and Capreomycin (CAP) in tuberculosis clinical isolates. The sensitivity of the assay for detecting the resistance to RIF, FQs, CAP, and KAN was 100%, 100%, 40%, and 50%, respectively, with 100% specificity. This assay was considered as a fast and effective method for the detection of mutations [59]; but, it has been superseded by other sequencing technologies.
Whole genome sequencing
Whole genome sequencing is a powerful, inexpensive open accessible epidemiological tool that can predict the genotypic and phenotypic resistance of a suspected bacterium in just a few days. Multi-locus sequence typing, multiple-locus variable number of tandem repeat analysis, single-nucleotide polymorphism analysis, CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-multi-locus virulence sequence typing, and NGS are some of the techniques used in the sequencing of NTS antibiotic-resistant gene clusters [60].
Upon acquisition, sequences are then compared to reference sets sourced from databases such as ResFinder, Basic Local Alignment Search Tool (BLAST), and ARG- ANNOT, followed by a phenotypic test to validate the predicted accuracy. Typically, an isolate will be genotypically resistant if a suspected AMR gene is 75% identical to a reference sequence [61].
Whole genome sequencing for predicting AMR in NTS was evaluated in human and food isolates employing v2 or v3 chemistry with paired-end 2- by 25- or 2- by 300-bp reads on the MiSeq platform (Illumina). The data suggested that acquired resistance is highly correlated with the presence of known resistance determinants, useful for risk assessment linked to drug use in food animal production (Figure 2) [62].
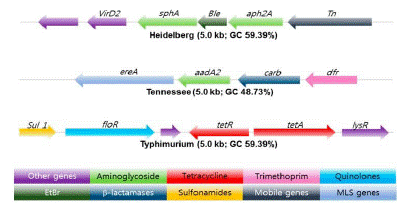
Figure 2: Antibiotic resistance gene clusters in three Salmonella serotypes in Swine.
Bleomycin and EtBr resistance gene (Green) flanked by antibiotic resistance genes marked in green. The arrow shows the orientation of the genes and genes are color coded to define different classes of antibiotic resistance, mobile, and other genes categories
Source: [81].
Combination of Short and Long Read WGS Sequencing
Plasmids are capable of transferring ARGs among bacterial isolates. Nonetheless, they are difficult to assemble from short-read WGS data. Short and long-read WGS sequencing was used to characterize ARGs on plasmids as well as establish their localization. Because of the rising concern about the spread of ARGs, it is of crucial importance to establish their location, especially when they are in mobile elements. Risk assessment of AMR spread was feasible by overcoming the challenges of plasmid reconstruction when employing the combination of long and short-read sequencing [63].
Nanopore Sequencing
Nanopore sequencing has been widely used for performing de novo bacterial assembly [64] undertaking metagenomics studies [65], and detecting ARGs [66]. The MinION nanopore sequencer was implemented to resolve the structure as well as the chromosomal insertion site of a composite antibiotic resistance island in Salmonella Typhi [67]. Long-read analysis of WGS data facilitated the identification of mobile genetic elements where AMR determinants were positioned and revealed the combination of various AMR determinants co-located on the same mobile element. The MinION could successfully identify bacterial pathogens as well as acquired resistance genes without culturing directly from urine samples within 4 hours. This study highlights the importance of WMS-based diagnosis in adjusting antimicrobial therapy [68].
The Oxford Nanopore MinION long-read DNA sequencing device was exploited for the detection of ARGs, the assessment of ARGs' taxonomic origin as well as decoding of their genetic organization and possible correlation with mobilization markers. Based on the findings, targeted intervention measures could be implemented to mitigate the risks of ARGs transferring among sites and, thus, improve biosecurity practices in hospitals and other environments [69]. MinION nanopore sequencing was also employed for the rapid pathogen, plasmids, and ARG identification in bacterial DNA extracted from positive blood cultures [70].
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI- TOF MS) in AMR diagnosis: Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI- TOF MS) can be used for the detection of AMR, alternatively to traditional genotypic or phenotypic bacterial characterization [71]. It relies on the cellular proteome and is capable of profiling proteins (mainly ribosomal, 2–20 kD) from whole bacterial cell extracts creating a bacterial spectral fingerprint or profiles that discriminate microorganisms at a genus, species, and subspecies level [72]. MALDI-TOF MS has also allowed the detection of AMR mechanisms, (for example, carbapenemases) [73]. It is considered a reliable, rapid (within minutes), accurate, easy-to-use, cost-effective, and environmentally friendly methodology [74].
Fourier Transform Infrared (FTIR) spectroscopy in AMR diagnosis: FTIR spectroscopy is a phenotypic method that has emerged as an attractive and dynamic weapon enriching the tools employed for biochemical analysis, owing to the detailed information it can provide on the chemical composition at the molecular level. FTIR spectroscopy allows for the quantification of the Infrared light absorption by molecules such as lipids, LPS, carbohydrates, proteins, and nucleic acids, resulting in a characteristic FTIR spectrum that represents the complete composition of the sample [75]. These characteristic spectra of the cell biomolecules offer ample functional and structural information. Infrared spectroscopy has been applied to differentiate the molecular changes associated with the development of AMR in prokaryotes [76]. In 2017, the detection of structural molecular changes linked to AMR by employing FTIR microscopy coupled with a novel statistical classification approach for spectral analysis was developed [77].
Microfluidics and Lab-on-a-Chip Technologies towards Rapid Diagnosis
Lab-on-a-Chip (LoC) technology has been used in the detection of antibiotic-resistant bacteria [78]. Some of the advantages offered by the LoC technology compared to macro-scale methods are fast and high throughput analysis, accurate fluid manipulation, low cost, low reagent, and power consumption, smaller sample volume, automation, integration, compactness, and portability [79].
Genotypic and phenotypic assays are the two main categories of microfluidic-based detection methods. Genotypic microfluidic assays (for example PCR, LAMP) target genetic markers (for example ARG), thus circumventing bacterial growth and allowing for shorter TAT (several hours) [80]. The implementation of microfluidics combined with isothermal DNA amplification protocols offers enhanced features due to the elimination of thermal cycling [81]. This approach is highly promising for the development of cheap, convenient, and efficient diagnostic tools for food safety, clinical, and environmental applications [82]. On the other hand, phenotypic microfluidic assays monitor the growth of bacteria in the presence of antibiotics, thus offering accurate antimicrobial susceptibility test results.
Optical biosensors: Different biosensors based on fluorescence, absorption, refractive index, Raman, and Surface Enhanced Raman Spectroscopy (SERS) techniques have been used in the identification of AMR Salmonella (Table 2) [83].
Salmonella Serotypes
Sensing Method
Sample Matrix
Analysis Time (min)
Detection Limit (CFU/mL)
S. Typhimurium DT104
SERS
Assay media
30
105
S. Typhimurium DT104
SERS
Assay media
15
105
S. Typhimurium
SERS
H2O and milk
120
20
S. Enterica
Raman Spectroscopy
Urine
150
N/A
S. Enteritidis
Fluorescence
Water, milk, and beef
30
2.0 × 102
S. Typhimurium DT104and S. Typhi
SERS
Assay media
120
100
Source: [60].
Table 2: Biosensors used in the detection of antibiotic-resistant Salmonella serotypes.
Surface Enhanced Raman Spectroscopy is considered a main biochemical fingerprinting approach since it can precisely reflect the macromolecular profiles as well as the changes occurring within the bacterial cells as a result of antibiotic action [84]. SERS has been applied for the investigation of the resistance or susceptibility to antibiotics of bacteria, as well as for studying the working mechanism of antibiotics relying on the whole cells' spectral fingerprint. SERS is capable of providing rapid, accurate, and ultra-sensitive detection of resistant bacteria with minimum requirements for sample preparation and handling [85].
Colorimetric-based approaches: The development of colorimetric-based microfluidic platforms addresses the pathogen identification and antimicrobial susceptibility test. On-chip determination of MIC is also provided via a colorimetric assay using a pH-dependent colorimetric broth. The total TAT of the on-chip microfluidic assay is 16–24h, approximately. Automated fluidic control, for example, transportation, and mixing is achieved using a pneumatically controlled custom-made module connected to the microfluidic chip. The initial loading of all samples [250μL of bacterial suspension (106 CFU/mL) /chamber] and reagents was performed manually [86]. The micro-device consisted of an array of incubation microchambers loaded with chromogenic medium and antibiotics.
pH-based approaches: A microfluidic device integrating polymer-based microfluidic channels with a pH-sensitive chitosan hydrogel was proposed for the detection of small pH changes for rapid antimicrobial susceptibility tests. Fourier-Transformed Reflectometric Interference Spectroscopy (FT-RIFS) was used for the real-time observation of the changes in the pH. The Turnaround Time (TAT) for the detection of whole bacterial growth was less than 2 hours [87]. The multiplexed colorimetric assay was facilitated via the use of paper substrates within the cell culture microchambers, allowing for a versatile combination of the antimicrobial agents and the chromogenic media. The assay was completed within 15 hours and the outcome of the chromogenic reaction was monitored via a camera Snapshots were taken every 30 min and analyzed with image analysis software [88].
Quartz-Crystal Microbalance (QCM)-based approaches: A quartz-crystal microbalance is a physical nanogram-sensitive device with a piezoelectric sensor and it facilitates the real-time, rapid, on-site detection of AMR bacteria [89]. It has been described that ARG detection employs a Magnesium Zinc Oxide (MZO) nanostructure-modified quartz crystal microbalance (MZOnano-QCM) biosensor [90]. The main advantages of this method are low cost, low demand in clinical samples volume, and it has rapidity (within 10 minutes) [91].
Point-of-Care (POC) approaches: A POC system is used for AMR diagnosis and phenotypic antimicrobial susceptibility tests of bacteriuria and urinary tract infection [91]. The TAT is 2 hours with the ability of detection and quantification of bacterial concentrations ranging from 50 to 105 CFU/ml. The detection is based on a portable particle-counting instrument comprising a miniature confocal microscope coupled with software for real-time data analysis. The detection system allows for growth curve measurements of fluorescently stained bacterial cells in control and antibiotic-treated samples. The main advantages of the proposed POC lie in the elimination of pre-processing steps example: pre-culture, enrichment, and centrifugation of urine samples as well as in the sensitivity of the instrument [92]. One limitation of this method is the negative effect of mixed cultures both on the specificity and the sensitivity of the results.
Multiplex approaches: A multiplex (eight samples) microfluidic chip for high throughput rapid phenotypic Antimicrobial susceptibility test was proposed. A mix of bacterial isolates and agarose was loaded in an array of microchambers within the chip. The growth rate of bacterial colonies under antibiotic gradients is monitored with the aid of a custom-built dark-field microscope coupled to a motorized camera (taking snapshots every 10 min) followed by automated image analysis. The TAT is 5 hours and the method achieves stable MIC values showing 100% agreement with reference (broth microdilution) MIC values. The key advantage of this system is the ability to simultaneously and rapidly analyze eight samples on a single chip, which can also allow for parallel testing of several antibiotics [93].
Single-cell or single-molecule approaches: A rapid antimicrobial susceptibility test system based on a microfluidic agarose channel with immobilized bacteria allows for single-cell growth and monitoring by microscopy [94]. The TAT for determining susceptibility, by monitoring the growth of the bacteria (single-cell level) in the presence of antibiotics, was 30 minutes. Moreover, the proposed system can be extensively applied for bacteria detection and complex (blood cultures, urine, whole blood) polymicrobial samples analysis [95].
Factors for Antimicrobial Resistance Development, Its Consequences, and Strategies to Combat Resistance Problems Posed By Non-Typhoid Salmonella
Predisposing Factors for the Development of Antimicrobial Resistance
The factors identified as driving global AMR include misuse of antimicrobials in humans or animals, contamination of the environment (including sewage and heavy metals), healthcare transmission, travel, mass drug administration, and incorrect dosing of antibiotics [96] and it is mainly driven by inappropriate use. In many countries, antibiotics can be bought without a prescription or do not have underlying standard treatment guidelines. These factors increase antibiotic resistance because of a lack of knowledge of proper antibiotic use [97].
The utilization of antimicrobials in agriculture for the growth promotion of animals, for prophylaxis, and the treatment of diseases caused by bacterial pathogens can lead to the development of AMR in pathogens. In different studies, both pig and chicken meats have been documented as reservoirs for drug-resistant Salmonella species. This transmission of drug resistance through the food chain is considered a major public health concern [1].
Antibiotic resistance in NTS strains can be also due to genetic mutations or through the acquisition of resistance encoding genes on mobile elements [98]. Plasmids are typical carriers of determinants that confer resistance against conventional antibiotics such as ampicillin, chloramphenicol, and tetracycline. However, the chromosome can also harbor these determinants, for example, on the multidrug resistance region of Salmonella Genomic Island 1 (SGI1) [99].
Consequences of Antimicrobial Resistance in Nontyphoidal Salmonella Species
There are various clinical and public health consequences associated with AMR in NTS species. These involve increased mortality and morbidity, thereby posing threats to public health, and failure in therapy, thereby resulting in limitations in the choice of treatment after the establishment of microbial diagnosis and increased burden of illness [100]. The dissemination of Multi-Drug Resistant (MDR) Salmonella from animals to humans is a major adverse consequence for public health which leads to treatment failure in human infectious diseases [99].
The other consequences may involve outbreaks in settings where patients are treated with antimicrobial drugs, increased transmission of resistant Salmonella strains, longer stay in the hospital, which increases the risk of acquisition of nosocomial infections [100]; increased virulence of Salmonella species as a result of ‘drug-bug combination’ that poses selective pressure on the microorganism [101] and increased cost of treatments [102].
Strategies to Combat Resistance Problems Posed By Nontyphoidal Salmonella
Several efforts have been adopted by several organizations, governments, and researchers to combat AMR imposed by some pathogenic organisms that are of public health significance including Salmonella species [103]. The efforts to address threats posed by AMR include monitoring programs for AMR microbes that integrate human, animal, and food sampling schemes, modification of drugs that led to the production of third and fourth-generation cephalosporins, and the use of medicinal plants also known as herbal medicine [19].
Additionally, the following are also needed to be done to track down the current rise in the spread of resistant S. Enterica: intensive surveillance of vended foods in developing countries to reduce the microbial risk associated with their consumption, public enlightenment to discourage the patronage of vended foods should be intensified as vended foods which is a potential vector responsible for the spread of resistant Salmonella species, or high level of hygiene practice should be maintained by food vendors under strict supervision and monitoring by food regulatory authorities [104].
Conclusion and Recommendations
Antimicrobial resistance is one of the key issues that are linked with food-borne infections caused by resistant pathogenic microorganisms in developing and developed countries. NTS is one of these food-borne illnesses, causing salmonellosis which can be transmitted through the ingestion of contaminated food of animal origin and vegetables. The emergence of AMR is developed by biochemical and genetic mechanisms, and it is diagnosed by conventional and non-conventional methods. In another way, the factors such as misuse of antimicrobials in humans or animals, contamination of the environment, utilization of antimicrobials for growth promotion of animals, for prophylaxis, and incorrect dose of treatment predispose the occurrence of AMR. As a result, the development of AMR poses serious public health and economic impacts and various efforts are adopted by different organizations and researchers to address the threats posed by AMR.
Therefore, based on the above facts, the following recommendations are forwarded:
The mechanisms by which NTS develops resistance to antimicrobials should be known and its dissemination to humans, animals, and the environment should be prevented and controlled.
The methods and techniques that are essential for the diagnosis of AMR in NTS species and the important related technologies should be available as much as possible by concerning bodies for the identification of resistant genes and resistant antimicrobials and the use of proper drugs.
Additionally, proper policies and regulation systems for antimicrobial use and distribution should be developed and implemented.
References
- Xu X, Biswas S, Gu G, Elbediwi M, Li Y. Characterization of Multidrug Resistance Patterns of Emerging Salmonella enterica Serovar Rissen along the Food Chain in China. MDPI. 2020; 9: 1-15.
- Gut AM, Vasiljevic T, Yeager T, Donkor ON. Salmonella infection, prevention and treatment by antibiotics and probiotic yeasts: a review. Microbiology (Reading). 2018; 164: 1327-44.
- Haselbeck AH, Panzner U, Im J, Baker S, Meyer CG, et al. Current perspectives on invasive nontyphoidal Salmonella disease. Curr Opin Infect Dis. 2017; 30: 498-503.
- Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012; 379: 2489-99.
- El-ghany WAA. Salmonellosis: A food borne zoonotic and public health disease in Egypt. Jour Infect Dev Count. 2006; 14: 674-8.
- Lapierre L, Cornejo J. Virulence Factors and Susceptibility to Antibiotics in Salmonella Infantis Strains Isolated from Chicken Meat: First Findings in Chile. MDPI. 2020; 10: 1-15.
- Ammar AM, Mohamed AA, Abd El-Hamid MI, El-Azzouny MM. Virulence genotypes of clinical SalmonellaSerovars from broilers in Egypt. J Infect Dev Ctries. 2016; 10: 337-46.
- Eguale T. Non-typhoidal Salmonella serovars in poultry farms in central Ethiopia: prevalence and antimicrobial resistance. BMC Vet Res. 2018; 14: 217.
- TM Al-sanouri, Paglietti B, Haddadin A, Murgia M, Bacciu D, et al., Emergence of plasmid-mediated multidrug resistance in epidemic and non-epidemic strains of Salmonella enterica serotype Typhi from Jordan Jour Infec Dev Count. 2008; 2: 295-301.
- De Toro M, Sáenz Y, Cercenado E, Rojo-Bezares B, García-Campello M, Undabeitia E et al. Genetic characterization of the mechanisms of resistance to amoxicillin/clavulanate and third-generation cephalosporins in Salmonella enterica from three Spanish hospitals. Int Microbiol. 2011; 14: 173-81.
- Singh S, Yadav AS, Singh SM, Bharti P. Prevalence of Salmonella in chicken eggs collected from poultry farms and marketing channels and their antimicrobial resistance. Food Res Int. 2010; 43: 2027-30.
- Browne AJ, Kashef Hamadani BH, Kumaran EAP, Rao P, Longbottom J, et al. Drug-resistant enteric fever worldwide, 1990 to 2018: a systematic review and meta-analysis. BMC Med. 2020; 18: 1.
- Afolami OI, Onifade AK. Antibiotic resistant Salmonella spp: mechanism of drug resistance, gene variations and clinical implications. AJRIMPS. 2018; 4: 1-6.
- Byarugaba DK. A view on Antimicrobial resistance in developing countries and responsible risk factors. Int J Antimicrob Agents. 2004; 24: 105-10.
- Coculescu BI. Antimicrobial resistance induced by genetic changes. J Med Life. 2009; 2: 114-23.
- Martinez JL. Drug resistance General principles of antibiotic resistance in bacteria. Dru Discov Tod Technol. 2014; 11: 33-9.
- Cox G, Wright GD. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol. 2013; 303: 287-92.
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010; 74: 417-33.
- Akinyemi KO, Ajoseh SO. Factors contributing to the emergence and spread of antibiotics resistance in Salmonella species. Cur Topi Salm Salmon. 2017; 6: 97-114.
- Varela MF, Kumar S. Molecular mechanisms of bacterial resistance to antimicrobial agents,’ MICR path Strat Combat them sci. J Technol Educ. 2014; 522-34.
- Varela MF et al. Bacterial Resistance to Antimicrobial Agents. MDPI. 2021; 10: 1-22.
- Nikaido H, Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003; 67: 593-656.
- Ghai IGS, Ghai S. Understanding antibiotic resistance via outer membrane permeability. Infect Drug Resist. 2018; 11: 523-30.
- Pagès JM, James CE, Winterhalter M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008; 6: 893-903.
- Shi Y. Common folds and transport mechanisms of secondary active transporters. Annu Rev Biophys. 2013; 42: 51-72.
- Davidson AL, Dassa E, Orelle C, Chen J, MIM Ol, BIOLR. Ev. Structure, function, and evolution of bacterial ATP-binding cassette systems Amer Soc Microbiol. 2008; 72: 317-64.
- Holland IB. Rise and rise of the ABC transporter families. Res Microbiol. 2019; 170: 304-20.
- Lambert PA. Bacterial resistance to antibiotics: modified target sites. Adv Drug Deliv Rev. 2005; 57: 1471-85.
- Roberts AP, Mullany P. Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev Anti-Infect Ther. 2010; 8: 1441-50.
- Ammons MCB. Anti-Biofilm Strategies and the Need for Innovations in Wound Care. Rec Pat Anti infec. Drug Discov. 2020; 5: 10-7.
- Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta. 2008; 1778: 1714-34.
- Pandey M, Cascella. Beta lactam antibiotics. Researchgate. 2019.
- Villa L, Carattoli A. Integrons and transposons on the Salmonella enterica serovar Typhimurium virulence plasmid. Antimicrob Agents Chemother. 2005; 49: 1194-7.
- Yuan J, Guo W. Mechanisms of resistance to quinolones in Salmonella Typhimurium from patients with infectious diarrhea,’ no. Microbiol Immunol. 2017; 61: 138-43.
- Dhanani AS, Block G, Dewar K, Forgetta V, Topp E. Genomic Comparison of Non-Typhoidal Salmonella enterica serovars Typhimurium, Enteritidis, Heidelberg, Hadar and Kentucky Isolates from Broiler Chickens. PLOS ONE. 2015: 1-24.
- Kaprou GD, Bergšpica I, Alexa EA, A álvarez-ord, M Prieto. Rapid methods for antimicrobial resistance diagnostics MDP. 2021; 10: 1-30.
- Richter SS, Karichu J, Otiso J, Heule HV, Keller G, et al. Evaluation of Sensititre Broth microdilution Plate for determining the susceptibility of carbapenem-resistant Klebsiella pneumoniae to polymyxins. Diagn Microbiol Infect Dis. 2019; 91: 89-92.
- Alvarez-Molina A, de Toro M, Alexa EA, Alvarez-Ordonez A. Applying genomics to track antimicrobial resistance in the food chain. Compreh Foodom. 2021: 188-211.
- Andrews J. Effects of using lignite mine drainage for irrigation on soils – A case study of perumal tank command area in Tamil Nadu State. J Ind Pollut Control. 2006; 22: 149-60.
- El Seedy FR, Samy AA, Salam HSH, Khairy EA, Koraney AA. Polymerase chain reaction detection of genes responsible for multiple antibiotic resistance Staphylococcus aureus isolated from food of animal origin in Egypt. Vet World. 2017; 10: 1205-11.
- Mackay IM. Real-time PCR in the microbiology laboratory. Clin Microbiol Infect. 2004; 10: 190-212.
- Böckelmann U, Dörries HH, Ayuso-Gabella MN, Salgot de Marçay M, Tandoi V, et al. Quantitative PCR monitoring of antibiotic resistance genes and bacterial pathogens in three European artificial groundwater recharge systems. Appl Environ Microbiol. 2009; 75: 154-63.
- Bogožalec Košir A, Cvelbar T, Kammel M, Grunert HP, Zeichhardt H, et al. Digital PCR method for detection and quantification of specific antimicrobial drug-resistance mutations in human cytomegalovirus. J Virol Methods. 2020; 281: 113864.
- Jousset AB, Bernabeu S, Bonnin RA, Creton E, Cotellon G, Sauvadet A et al. Development and validation of a multiplex polymerase chain reaction assay for detection of the five families of plasmid-encoded colistin resistance. Int J Antimicrob Agents. 2019; 53: 302-9.
- Waseem H, Jameel S, Ali J, Rehman HS, Tauseef I, et al. Contributions and challenges of high throughput qPCR for determining antimicrobial resistance in the environment: A critical review. MDPI. 2019; 24: 1-25.
- Xu L, Chen H, Canales M, Ciric L. Use of synthesized double-stranded gene fragments as qPCR standards for the quantification of antibiotic resistance genes. J Microbiol Methods. 2019; 164: 105670.
- Masny A, Plucienniczak A. Ligation mediated PCR performed at low denaturation temperatures--PCR melting profiles. Nucl Aci Res. 2003; 31: 1-6.
- Gill P, Ghaemi A. Nucleic acid isothermal amplification technologies: a review. Nucleosides Nucleotides Nucleic Acids. 2008; 27: 224-43.
- Call DR, Bakko MK, Krug MJ, Roberts MC. Identifying antimicrobial resistance genes with DNA microarrays. Antimicrob Agents Chemother. 2003; 47: 3290-5.
- Anjum MF, Zankari E, Hasman H. Molecular methods for detection of antimicrobial resistance. Microbiol Spectr. 2017; 5.
- Strauss C, Endimiani A, Perreten V. A novel universal DNA labeling and amplification system for rapid microarray-based detection of 117 antibiotic resistance genes in Gram-positive bacteria. J Microbiol Meth. 2015; 108: 25-30.
- Teng JLL, Yeung ML, Chan E, Jia L, Lin CH, et al. PacBio but not Illumina technology can achieve fast, accurate and complete closure of the high GC, complex Burkholderia pseudomallei two-chromosome genome. Front Microbiol. 2017; 8: 1448.
- Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009; 4: 265-70.
- Ruppe E, Iame I. How next-generation sequencing can address the antimicrobial resistance. AMR Cont. 2019; 60-5.
- Köser CU, Ellington MJ, Peacock SJ. Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 2014; 30: 401-7.
- Oniciuc EA, Likotrafiti E, Alvarez-molina A, Prieto M. The Present and Future of Whole Genome Sequencing (WGS) and Whole Metagenome Sequencing (WMS) for Surveillance of Antimicrobial Resistant Microorganisms and Antimicrobial Resistance Genes across the Food Chain. MDPI. 2018; 9: 1-28.
- Hendriksen RS, Bortolaia V, Tate H, Tyson G, Frank M, Mcdermott P. ’Using Genomics to Track Global Antimicrobial Resistance,’ front pub heal. 2019; 7: 2296-565.
- Amoako KK, Thomas MC, Kong F, Janzen TW, Hahn KR, et al. Rapid detection and antimicrobial resistance gene pro fi ling of Yersinia pestis using Pyrosequencing technology. J Microbiol Meth. 2012; 90: 228-34.
- Govindappa M, Farheen H, Chandrappa CP, Rai RV, Raghavendra VB. Mycosynthesis of silver nanoparticles using extract of endophytic fungi, Penicillium species of Glycosmis mauritiana, and its antioxidant, antimicrobial, anti-inflammatory. Advan Nat Sci Nanosci Nanotechnol. 2016; 7.
- Wu S, Hulme JP. Recent advances in the detection of antibiotic and multi-drug resistant salmonella: an update. Int J Mol Sci. 2021; 22.
- Almeida F et al. Phylogenetic and antimicrobial resistance gene analysis of Salmonella Typhimurium strains isolated in Brazil by whole genome sequencing. PLOSONE. 2018; 83: 1-16.
- McDermott PF, Tyson GH, Kabera C, Chen Y, Li C. The use of whole genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Am Soc Microbiol. 2016: 1-25.
- Berbers B, Saltykova A, Garcia-graells C, Philipp P, Arella F, et al. Combining short and long read sequencing to characterize antimicrobial resistance genes on plasmids applied to an unauthorized genetically modified Bacillus. Sci Rep. 2020; 10: 1-13.
- Loman NJ, Quick J, JT Simpson. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Meth. 2015: 11-4.
- Brown BL, Watson M, Minot SS, Rivera MC, Franklin RB. MinION TM nanopore sequencing of environmental metagenomes: a synthetic approach. Gig Sci. 2017; 6: 1-10.
- Judge K, Harris SR, Reuter S, Parkhill J, Peacock SJ. Early insights into the potential of the Oxford nanopore MinION for the detection of antimicrobial resistance genes. Jour Antimicr Chemother. 2015:1-4.
- Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, et al. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol. 2015; 33: 296-300.
- Schmidt K et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. Jourof Antimicr Chem. 2016: 1-11.
- Kamathewatta KI, Bushell RN, Young ND, Stevenson MA, Billman-Jacobe H, et al. Exploration of antibiotic resistance risks in a veterinary teaching hospital with Oxford nanopore long read sequencing. PLOS ONE. 2019; 14: e0217600.
- Taxt AM, Ave E, Stephan AF, Na U, Rafi A. Rapid identification of pathogens, antibiotic resistance genes and plasmids in blood cultures by nanopore sequencing. Sci Rep. 2020; 10: 1-11.
- Liu N, Wang L, Cai G, Zhang D, Lin J. Establishment of a simultaneous detection method for ten duck viruses using MALDI-TOF mass spectrometry. J Virol Methods. 2019; 273: 113723.
- Welker M, Van Belkum A. One system for all: is mass spectrometry a future alternative for conventional antibiotic susceptibility testing? Front Microbiol. 2019; 10: 2711.
- Hoyos-Mallecot Y, Riazzo C, Miranda-Casas C, Rojo-Martín MD, Gutiérrez-Fernández J, et al. Rapid detection and identi fi cation of strains carrying carbapenemases directly from positive blood cultures using MALDI-TOF MS. J Microbiol Meth. 2014; 105: 98-101.
- Vrioni G, Tsiamis C, Oikonomidis G, Theodoridou K, Kapsimali V, et al. MALDI-TOF mass spectrometry technology for detecting biomarkers of antimicrobial resistance: current achievements and future perspectives. Ann Transl Med. 2018; 6: 240.
- Vogt S, Löffler K, Dinkelacker AG, Bader B, Autenrieth IB, et al. Fourier-transform infrared (FTIR) spectroscopy for typing of clinical Enterobacter cloacae complex isolates. Front Microbiol. 2019; 10: 2582.
- Novais Â, Freitas AR, Rodrigues C, Peixe L. Fourier transform infrared spectroscopy: unlocking fundamentals and prospects for bacterial strain typing. Eur Jourof Clin Microbiol Infect Dis. 2018.
- Salman A, Sharaha U, Rodriguez-diaz E, Shufan E. Detection of antibiotic resistant Escherichia coli bacteria using infrared microscopy and advanced multivariate analysis. Roy Soc Chem. 2017.
- Leonard H, Colodner R, Halachmi S, Segal E. Recent advances in the race to design a rapid diagnostic test for antimicrobial resistance. ACS Sens. 2018; 3: 2202-17.
- Kaprou GD, Papadopoulos V, Papageorgiou DP, Kefala I, Papadakis G, et al. Ultrafast, low-power, PCB manufacturable, continuous-flow microdevice for DNA amplification. Anal Bioanal Chem. 2019; 411: 5297-307.
- Zhang G, Zheng G, Zhang Y, Ma R, Kang X. Evaluation of a Micro/nanofluidic chip platform for the high-throughput detection of bacteria and their antibiotic resistance genes in post-neurosurgical meningitis. Int J Infect Dis. 2018; 70: 115-20.
- Gupta SK, Sharma P, McMillan EA, Jackson CR, Hiott LM, et al. Genomic comparison of diverse Salmonella serovars isolated from swine. PLOS ONE. 2019; 14: e0224518.
- Giuffrida MC, Spoto G. Integration of isothermal amplification methods in microfluidic devices,’ Biosen and Bioelect. 2016.
- Wang D, He P, Wang Z, Li G, Majed N, Gu AZ. Advances in single cell Raman spectroscopy technologies for biological and environmental applications. Curr Opin Biotechnol. 2020; 64: 218-29.
- Wang K, Li S, Petersen M, Wang S, Lu X. Detection and Characterization of Antibiotic-Resistant Bacteria Using Surface-Enhanced Raman Spectroscopy. MDPI. 2018; 8: 1-21.
- Chen X, et al. Surface-enhanced Raman scattering method for the identification of methicillin-resistant Staphylococcus aureus using positively charged silver nanoparticles. Microc Act. 2019; 2: 1-8.
- Lee WB, Chien CC, You HL, Kuo FC, Lee MS, et al. An integrated microfluidic system for antimicrobial susceptibility testing with antibiotic combination. Lab Chip. 2019; 19: 2699-708.
- Tang Y, Zhen L, Liu J, Wu J. Rapid antibiotic Susceptibility Testing in a Micro fl uidic pH Sensor. Anal Chem. 2013; 85: 2787-94.
- Xu B, Du Y, Lin J, Qi M, Shu B, et al. Simultaneous identification and antimicrobial susceptibility simultaneous identification and antimicrobial susceptibility testing of multiple Uropahtogens on a microfluidic chip with paper-supported cell culture arrays. Anal Chem. 2016: 1-24.
- Bragazzi NL, Amicizia D, Panatto D, Tramalloni D, Valle I, Gasparini R. Quartz-crystal microbalance (QCM) for public health: an overview of its applications. Adv Protein Chem Struct Biol. 2015; 101: 149-211.
- Ivanoff P, Yang K, Zheng A, Li R, Li G. Magnesium zinc oxide nanostructure-modified quartz crystal microbalance for dynamic monitoring of antibiotic effects and antimicrobial resistance. Process Technol. 2017; 27: 46-7.
- Ivanoff P, et al. Dynamic monitoring of antimicrobial resistance using magnesium zinc oxide nanostructure-modi fi ed quartz crystal microbalance. Biosen Bioelectr. 2016; 1-9.
- Toosky MN, Grunwald JT, Pala D, Shen B, Zhao W, et al. A rapid, point- -care antibiotic susceptibility test for urinary tract infections. J Med Microbiol. 2020; 69: 52-62.
- Yuan PW, Malmberg C, Kavalopoulos NF, Lubke M. A multiplex fluidic chip for rapid phenotypic antibiotic susceptibility testing. Am Soc Microbiol. 2020; 11: 1-14.
- Choi J, Jung Y-G, Kim J, Kim S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Roy Soc Chem. 2012.
- Li H, Torab P, Mach KE, Surrette C, England MR, et al. Adaptable microfluidic system for single-cell pathogen classification and antimicrobial susceptibility testing. Proc Natl Acad Sci USA. 2019; 116: 10270-9.
- Castro-Sánchez E, Moore LSP, Husson F, Holmes AH. What are the factors driving antimicrobial resistance? Perspectives from a public event in London, England. BMC Infect Dis. 2016; 16: 465.
- WHO, WIPO, WTO. Antimicrobial resistance – a global epidemic. WHO; 2016.
- Fluit AC. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol Med Microbiol. 2005; 43: 1-11.
- Djeghout B, Ayachi A, Paglietti B, Langridge GC, Rubino S. An Algerian perspective on non-typhoidal Salmonella infection. J Infect Dev Ctries. 2017; 11: 583-90.
- Mølbak K. Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin Infect Dis. 2005; 41: 1613-20.
- Soto SM, Rodríguez I, Rodicio MR, Vila J, Mendoza MC. Detection of virulence determinants in clinical strains of Salmonella enterica serovar Enteritidis and mapping on macrorestriction profiles. J Med Microbiol. 2006; 55: 365-73.
- Bisi-Johnson MA, Obi CL, Vasaikar SD, Baba KA, Hattori T. Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape, South Africa. Gut Pathog. 2011; 3: 9.
- Borck Hog B, et al. DANMAP – Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Res Gat. 2016; 17-31.
- Akinyem KO, Fashola MO, Habib N, Akinwande E. Vended foods in Lagos, Nigeria: A potential reservoir for the spread of emerging strains of drug resistant bacteria. Health. 2013; 05: 675-80.