
Research Article
J Bacteriol Mycol. 2015;2(1): 1011.
Intestinal Dysbiosis Following Cholestasis is Reduced by Active Immunization with a Detoxified Endotoxin Vaccine
Samuel M. Alaish¹*, Emmanuel F. Mongodin², Lei Zhang³, Ebony Murphy¹ and Alan Cross³
1Department of Surgery, University of Maryland School of Medicine, Baltimore
2Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore
3Center for Vaccine Development, University of Maryland School of Medicine, Baltimore
*Corresponding author: Samuel M. Alaish, M.D. Assistant Professor of Surgery, Department of Surgery, University of Maryland School of Medicine, 29 South Greene St, Suite 110 Baltimore, MD 21201
Received: January 22, 2015; Accepted: April 16, 2015; Published: April 17, 2015
Introduction
Background: Cholestasis inevitably leads to intestinal barrier loss resulting in endotoxemia and sepsis. Using a cholestatic mouse model, we have previously reported decreased intestinal barrier resistance, increased rates of bacterial translocation, endotoxemia and increased mortality following cholestasis. Furthermore, we found an intestinal dysbiosis, an altered gut microbiota, with increased numbers of virulent bacterial species 2 weeks after cholestatic injury as compared to sham-operated mice. We hypothesize that active immunization with the detoxified endotoxin vaccine, J5dLPS, will prevent the dysbiosis seen following cholestasis.
Methods: C57Bl/6J mice were vaccinated with a detoxified endotoxin vaccine, J5dLPS. Saline-injected mice served as controls. Following confirmation of serum antibody titers, mice then underwent either a Common Bile Duct Ligation (CBDL) as a model for cholestatic injury or a sham operation. Both before surgery and one week following surgery, stool specimens were collected for bacterial DNA isolation and sequencing.
Results: In this pilot study, J5dLPS vaccination was highly immunogenic and well-tolerated in C57Bl/6J mice. No dysbiosis was seen following active immunization. Moreover, J5dLPS-vaccinated mice which underwent CBDL demonstrated reduced intestinal dysbiosis and a strong trend [p=0.07] toward no dysbiosis compared to the dysbiosis seen in saline-injected mice after CBDL.
Conclusions: The detoxified endotoxin vaccine, J5dLPS, does not cause an intestinal dysbiosis in mice which have been actively immunized. This finding provides additional evidence of safety supporting future clinical use of this vaccine. Moreover, this same vaccine greatly reduces the gut dysbiosis seen in mice following cholestatic injury. Future studies are warranted to determine if maintenance of the gut microbiota strengthens the intestinal barrier following cholestasis and prevents gut colonization with opportunistic pathogens.
Keywords: Cholestasis; Endotoxin; Vaccine; Microbiota
Introduction
Infants can develop Short Bowel Syndrome (SBS) following an extensive bowel resection for either a congenital anomaly or a postnatal infection [1]. Consequently, they develop intestinal failure and require prolonged intravenous nutrition [1]. Moreover, these infants can experience diminished bile flow from the liver into the intestine, known as cholestasis. Cholestasis leads to liver damage and also to intestinal barrier breakdown with increased rates of systemic infection. The organisms often responsible for these infections arise from the gut and coincide with intraluminal bacterial overgrowth, which follows both cholestasis [2] and SBS [3]. Moreover, a vicious cycle is created as intestinal permeability increases with bacterial overgrowth [4].
Failure of the intestinal barrier has been shown to be a characteristic feature of cholestatic injury. Decreased intestinal resistance, increased bacterial translocation and increased episodes of sepsis have been well described [5-7]; however, the exact mechanisms remain poorly understood. Using Common Bile Duct Ligation (CBDL) in a mouse as a model for cholestatic intestinal injury and bacterial overgrowth, our lab has previously found that C57BL/6J mice developed an intestinal dysbiosis, an altered gut microbiota, as demonstrated on a Principal Coordinates Analysis 14 days following cholestasis; however, a different strain of inbred mice, the A/J strain, failed to develop a dysbiosis [8]. Importantly, the dysbiosis positively correlated with the decreased intestinal resistance, increased bacterial translocation and increased mortality observed in the cholestatic C57BL/6J mice as compared to the cholestatic A/J mice [8]. We found a relative increase in the number of both Clostridiae and Proteobacteriae organisms and a relative decrease in Lactobacillae in the cholestatic C57BL/6J mice compared to their non-cholestatic counterparts [8]. This may be indicative of a transition towards a more pathogenic microbiota following cholestasis which three weeks later contributes to the significant mortality difference between the strains. Furthermore, we found that soluble CD14 levels, a marker of the monocytic response to LPS, were dramatically increased in all mice fourteen days following CBDL compared to shams (p<0.000002), thus representative of LPS translocation from the gut into the circulation [8].
Translocation of gut bacteria and endotoxin into the systemic circulation may contribute to the morbidity and mortality of cholestasis; therefore, we immunized mice with a detoxified endotoxin vaccine that was previously shown to elicit antibodies against the core region of LPS that is highly conserved among Enterobacteriaceae. Antibodies generated against the LPS core are highly protective in animal models of sepsis when induced actively or administered passively [9-12]. This vaccine has been well-tolerated when given to human subjects [13].
We hypothesize that a vaccine directed against endotoxin will offset some of the morbidity and mortality associated with cholestasis. In this study, we examine the effects of the detoxified endotoxin vaccine, J5dLPS, on the intestinal microbiota before and after cholestasis. The findings from this study shed light on potential safety concerns regarding the vaccine as well as an additional, potential therapeutic indication.
Materials and Methods
Reagents
The J5dLPS vaccine was developed as previously described [13].
Animals
Male C57BL/6J (B6) mice (8 weeks old) were obtained from the Jackson Laboratory (Bar Harbor, ME) and maintained in a pathogenfree animal facility with 12-hour light-dark cycles. All mice weighed 18 to 25 g at the time of operation. Animal studies were conducted according to protocols reviewed and approved by the University of Maryland School of Medicine Institutional Animal Care and Use Committee and adhered to guidelines promulgated by the National Institutes of Health.
Common Bile Duct Ligation (CBDL) model of cholestasis
Mice were anesthetized by inhaled isoflurane anesthesia. The abdomen was clipped and then prepared in sterile fashion with 70% ethyl-ethanol followed by betadine. A transverse upper abdominal incision was performed. The Common Bile Duct (CBD) was dissected away from the portal vein and was ligated near its junction with the duodenum using aneurysm clips engineered with a precisely standardized opening/closing mechanism. The abdominal wall was then closed in a two-layer fashion using absorbable sutures. Shamoperated mice were treated identically but without dissection or ligation of the CBD. Postoperatively, animals were resuscitated with warmed subcutaneous injections of saline (1 mL) to replace losses. Mice were returned to clean cages where food and water were provided ad libitum.
Vaccine schedule and measurement of antibody levels
The J5dLPS vaccine was administered with an intraperitoneal injection using a dose of 20μg (based on dLPS content) at 0, 2, and 4 weeks. Saline-injected mice were immunization controls. After an additional 4 weeks, the CBDL was performed. Sham-operated mice served as controls. Blood samples were collected 1 month after the final immunization (before the CBDL or sham operation). Levels of antibodies against the core glycolipid of LPS were measured using a standard ELISA method as previously described and expressed as Optical Density Units (ODUs) [13].
Gut Bacterial Community Profiling
DNA extraction: Profiling of the bacterial communities inhabiting the gut of male C57BL/6J (B6) mice was performed as earlier described [14]. Total genomic DNA was extracted from fecal pellets using the protocol previously described by Zupancic and colleagues [14]. Briefly, 1 ml of phosphate-buffered saline was added to the mouse stool aliquots, and cell lysis was performed using an enzymatic cocktail composed of lyzosyme, mutanolysin and lysostaphin. After 1-hour incubation at 37oC, samples were further lysed by addition of proteinase K and 10% SDS, followed by incubation at 55oC for 45 minutes. The samples were then disrupted by bead beating using a FP120 Fast Prep instrument and 0.1 mm silica spheres. The resulting crude lysate was processed using the ZYMO Fecal DNA Kit (Zymogen) according to the manufacturer–s recommendations. Negative extraction controls, where stool samples were omitted, were performed to ensure the samples were not contaminated by exogenous bacterial DNA during the extraction process.
16S rRNA gene sequencing: Following DNA extraction, the universal primers 27F and 338R were used for PCR amplification of the V1–V3 hypervariable regions of 16S rRNA genes. The 338R primer included a unique sequence tag to barcode each sample. Using 96 barcoded 338R primers, the V1–V3 regions of 16S rRNA genes were amplified in 96-well microtiter plates using AmpliTaq Gold DNA polymerase (Applied Biosystems) and 50 ng of template DNA in a total reaction volume of 50 mL, using the cycling conditions described by Zupancic and colleagues [14]. Negative controls without a template were included for each barcoded primer pair. PCR products were quantified using the Quant-iT PicoGreen dsDNA assay, and equimolar amounts (100 ng) of the PCR amplicons were mixed in a single tube. The purified amplicon mixture was then sequenced by 454 FLX Titanium pyrosequencing using 454 Life Sciences primer A by the Genomics Resource Center at the Institute for Genome Sciences, University of Maryland School of Medicine, using protocols recommended by the manufacturer as amended by the Center.
16S rRNA gene sequences statistical analysis: Processing (sequence binning and trimming) and analysis of the 16S rRNA reads was performed using the CLoVR system (CloVR 1.0-RC5; https:// clovr.org/) and the CloVR-16S protocol (https://clovr.org/methods/ clovr-16s; version 1.1), and the output of the CLoVR pipeline processed further using the Qiime software package, as well as the Phyloseq package implemented in R. Phylogenetic trees in Figures 2 & 3 were created using the Phyloseq plot tree function from the tree generated in Qiime using the make_phylogeny.py command. The tree was constructed with a set of sequences representative of the OTUs, using the defaults Qiime parameters (Fast Tree for tree building). Sobs analysis of the number of Observed Operational Taxonomic Units (OTUs) was performed with p<0.05 considered significant. The Shannon Diversity index and the Chao1 richness estimate were calculated using Phyloseq to determine species diversity and richness, respectively.
Phylogenetic trees: The phylogenetic trees were created using the Phyloseq plot tree function, from the tree generated in Qiime using the make_phylogeny.py command. The Qiime tree was constructed with a set of sequences representative of the OTUs aligned against a database of reference 16S sequences (in this case, Green genes), using the defaults Qiime make_phylogeny.py parameters, i.e. using Fast Tree for tree building. Fast Tree infers approximately-maximumlikelihood phylogenetic trees from alignments of nucleotide sequences, and can handle alignments with up to a million of sequences in a reasonable amount of time and memory. Using the default parameters, Fast Tree estimates the reliability of each split in the tree by computing local support values with the Shimodaira- Hasegawa test, and does not use traditional bootstrapping: this is why there are no bootstrap values for these trees.
Statistical analysis
The experiment was replicated once. A paired Student–s t-test was used to measure antibody levels with p<0.05 considered significant. The Wilcoxon rank sum test was used to determine significance when comparing OTU–s, Chao1 richness estimates and Shannon diversity estimates. P<0.05 was considered significant.
Results
Immunogenicity of J5dLPS vaccine
A strong antibody response was demonstrated in 7 of the 8 mice one month following 3 doses of 20μg of J5dLPS vaccine given 2 weeks apart as shown in Figure 1. Saline-injected mice served as controls and had no detectable antibody response. The IgG response in the mice treated with J5dLPS had a mean of 6395 +/- 2286 (SE) ODUs/ mL; whereas, the saline-treated mice had a mean IgG response of 0 +/- 0 ODUs/mL; the p-value between the two groups was p< 0.0062.
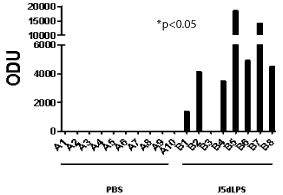
Figure 1: IgG response in mice to J5dLPS vaccine vs. saline control. A
denotes samples from saline-injected mice; whereas, B denotes samples
from J5dLPS-vaccinated mice. There is a significant antibody response in
the J5dLPS-vaccinated mice as compared to the saline-injected mice with
*p<0.05.
No dysbiosis seen following active immunization with J5dLPS
Stool samples were collected from C57Bl/6J mice with strong antibody responses following 3 doses of J5dLPS vaccine. Stool samples were also collected from control C57Bl/6J mice following 3 injections of saline. Following DNA isolation and characterization of the gut microbiota using 16S rRNA gene sequencing, no difference in species diversity and richness was noted between the groups (Figure 2). Thus, no dysbiosis was seen following the vaccination.
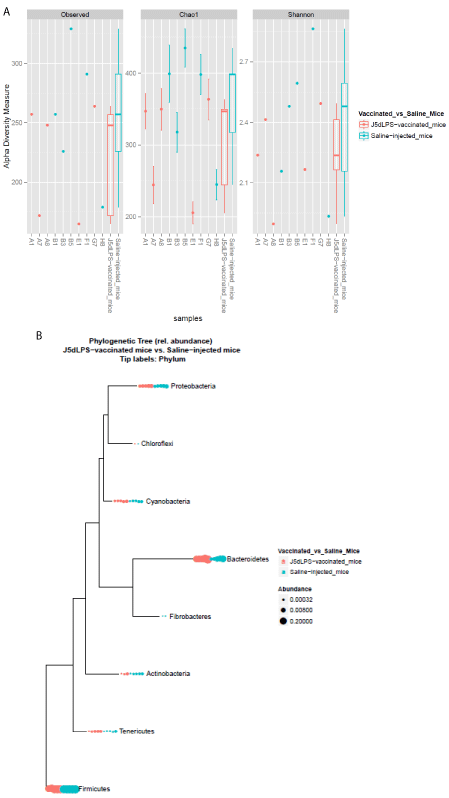
Figure 2: A) Characterization of the structure of the fecal microbiota collected
from J5dLPS-vaccinated and saline-injected C57Bl/6J mice. A) Alphadiversity
measures of the fecal microbiota: Observed (left panel) refers to the
observed number of species-level OTU, Chao1 (middle panel) refers to the
Chao1 richness estimate, and Shannon (right panel) refers to the community
diversity. The number of species is plotted against the number of samples
analyzed. Note that there is no significant difference between the two groups
[p=0.35 for OTU, p=0.22 for Chao1 and p=0.55 for Shannon by Wilcoxon rank
sum test]. B) Phylum-level phylogenetic tree demonstrating similar relative
abundance of bacteria in the two groups.
Dysbiosis associated with cholestasis greatly reduced in J5dLPS-vaccinated mice
Gut microbiota sequenced from DNA isolated from stool samples obtained 1 week following CBDL in non-immunized C57Bl/6J mice demonstrated a dysbiosis with a relative increase in proteobacteria species compared to that from sham-operated C57Bl/6J mice, which is consistent with our previous published findings [8]. Interestingly, the gut microbiota from stool samples from C57Bl/6J mice which were vaccinated with J5dLPS prior to CBDL exhibited a greatly reduced dysbiosis and a clear trend [p=0.07] towards not developing any dysbiosis. Therefore, the vaccine may maintain the species diversity and richness similar to the mice which did not undergo CBDL (Figure 3).
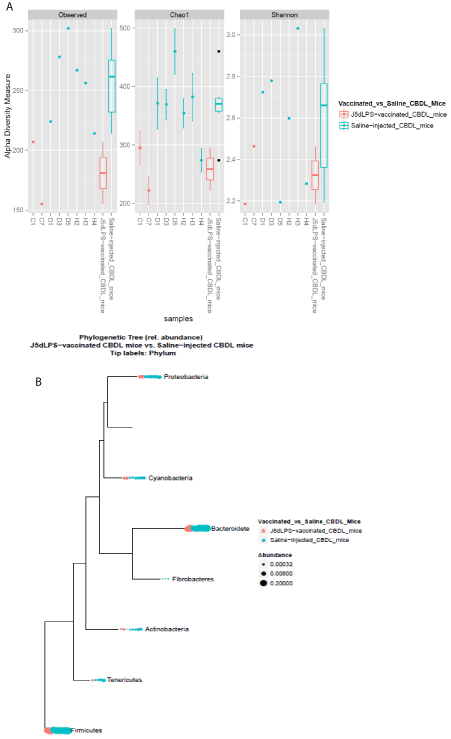
Figure 3: A) Characterization of the structure of the fecal microbiota
collected from J5dLPS-vaccinated and saline-injected C57Bl/6J mice which
then underwent CBDL. A) Alpha-diversity measures of the fecal microbiota:
Observed (left panel) refers to the observed number of species-level OTU,
Chao1 (middle panel) refers to the Chao1 richness estimate, and Shannon
(right panel) refers to the community diversity. The number of species is
plotted against the number of samples analyzed. Note that the salineinjected
group develops a dysbiosis following CBDL with increased diversity
[secondary to increased numbers of pathogenic species which have not yet
overwhelmed the benign species to decrease the diversity]. The J5dLPSvaccinated
group has reduced dysbiosis and demonstrates a clear trend
toward not developing this dysbiosis [p=0.07 for OTU, p=0.14 for Chao1
and p=0.29 for Shannon by Wilcoxon rank sum test]. B) Phylum-level
phylogenetic tree demonstrating increased relative abundance of bacteria
including proteobacteria in the saline-injected CBDL mice as compared to the
J5dLPS-vaccinated CBDL mice.
Discussion
Despite recent advances in intestinal rehabilitation, young children diagnosed with SBS continue to succumb not infrequently, with a mortality rate of 11% [17]. Their deaths are attributable to stagnant bile flow, termed cholestasis, and infectious complications whose source is usually the babies– own gut. Those infants fortunate to survive will still suffer considerable morbidity from these same complications. The mechanisms underlying the intestinal barrier breakdown in SBS patients remain incompletely understood, resulting in the current practice of prophylactic antibiotic therapy to combat bacterial overgrowth and prevent future infections. Unfortunately, this preventive measure has had two unintended but devastating complications: 1) the emergence of antibiotic-resistant organisms and 2) an increase in fungal infections. Delineation of the specific mechanisms responsible for this barrier loss is the first step towards designing focused, effective prophylactic therapies void of antibiotics. We hypothesize that cholestasis leads to bacterial overgrowth of gramnegative organisms, impaired gut barrier function and increased levels of endotoxin which continuously activate the TLR4 pathway, overwhelm the host–s regulation, and cause further intestinal barrier breakdown and the systemic spread of non-microbial tissue-injurious factors(“alarmins” or DAMPs), as well as endotoxin and bacteria. We posit that a vaccine directed against endotoxin, such as J5dLPS, could break this deleterious cycle and be therapeutic against cholestatic injury.
The detoxified endotoxin vaccine, J5dLPS, has previously been shown to be protective in neutropenic rat cecal ligation and puncture (polymicrobial) models of lethal sepsis [11,13] and lethal respiratory challenges with Francisella tularensisin mice [9], as well as Klebsiella pneumonia [12]. The present study provides a foundation on which to assess whether this vaccine can limit the morbidity and mortality resulting from intestinal barrier breakdown secondary to cholestasis. We reported earlier that cholestasis resulted in a gut dysbiosis with an increase in the number of virulent strains such as Proteobacteria and Clostridiae as compared to Lactobacillae [8]. We speculate that our time point represents the window when new pathogenic species appear and increase the diversity of the microbiota, but before the time point at which the pathogenic species overwhelm the host flora and decrease the diversity. The asymptomatic nature of the mice at our time point supports this notion. In this manuscript, our data also shows a greatly reduced dysbiosis in the ligated mice immunized with J5dLPS, as compared to the ligated mice injected with saline. In fact, there is a clear trend toward the J5dLPS vaccine preventing the development of this pathologic microbiota. Given the dynamic relationship between the gut microbiota and the intestinal epithelium, this “maintenance” of the microbiota may translate into “maintenance” of the strength of the intestinal barrier.
Recent literature has highlighted the interplay between the intestinal microbiota and vaccine responses. Antibody responses to seasonal influenza vaccination in both germ-free and antibiotictreated mice were impaired, but restored with a flagellated strain of Escherichia coli [18]. Moreover, a study of infants found that oral polio, bacilli Calmette-Guerin tetanus toxoid and hepatitis B virus vaccine responsiveness were all improved in those with increased Bifido bacteria in their stool microbiota [19]. These previous two reports focus on the effect of the gut microbiota on the vaccine response, but what about the vaccine–s effect on the gut microbiota? We speculate that immunization may induce a Th17 response in the gut epithelia that contributes to the maintenance of the gut microbiota [20,21]. A perception has arisen that an endotoxin vaccine may alter the gut microbiota, and this effect, itself, may have pathogenic consequences. Quelling that concern, the present study found no significant change in the gut microbiota following administration of the vaccine. This strengthens the safety profile for the vaccine, as it does not appear to select for a pathogenic microbiota. Indeed, by maintaining the normal gut flora, the vaccine may reduce the likelihood of intestinal colonization with opportunistic pathogens, such as Pseudomonas aeruginosa.
An effective anti-LPS response may be lacking in premature and very young infants. For this reason, we believe passive immunization with anti-LPS antibodies will be required early on for these young infants. We envision the infants would receive both active and passive immunization initially, and then later, the treatment would simply be additional doses of the J5dLPS vaccine.
In summary, this brief report has two notable findings: 1) the detoxified endotoxin vaccine, J5dLPS, does not, by itself, cause an intestinal dysbiosis, but rather, 2) the vaccine reduces the dysbiosis seen following cholestasis. Continued research is warranted to determine if this antibiotic-free modality carries therapeutic benefit against cholestatic intestinal injury.
Acknowledgment
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number K08GM081701. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Kurkchubasche AG, Rowe MI, Smith SD. Adaptation in short-bowel syndrome: reassessing old limits. J Pediatr Surg. 1993; 28: 1069-1071.
- Deitch EA, Sittig K, Li M, Berg R, Specian RD. Obstructive jaundice promotes bacterial translocation from the gut. Am J Surg. 1990; 159: 79-84.
- Dibiase JK, Young RJ, Vanderhoof JA. Enteric microbial flora, bacterial overgrowth, and short-bowel syndrome. Clin Gastroenterol Hepatol. 2006; 4: 11-20.
- Wells CL, Hess DJ, Erlandsen SL. Impact of the indigenous flora in animal models of shock and sepsis. Shock. 2004; 22: 562-568.
- Pascual S, Such J, Esteban A, Zapater P, Casellas JA, Aparicio JR, et al. Intestinal permeability is increased in patients with advanced cirrhosis. Hepatogastroenterology. 2003; 50: 1482-1486.
- Campillo B, Pernet P, Bories PN, Richardet JP, Devanlay M, Aussel C, et al. Intestinal permeability in liver cirrhosis: relationship with severe septic complications. Eur J Gastroenterol Hepatol. 1999; 11: 755-759.
- Frances R, Benlloch S, Zapater P, Gonzalez JM, Lozano B, Munoz C, et al. A sequential study of serum bacterial DNA in patients with advanced cirrhosis and ascites. Hepatology. 2004; 39: 484-491.
- Alaish SM, Smith AD, Timmons J, Greenspon J, Eyvazzadeh D, Murphy E, et al. Gut microbiota, tight junction protein expression, intestinal resistance, bacterial translocation and mortality following cholestasis depend on the genetic background of the host. Gut Microbes. 2013; 4: 292-305.
- Gregory SH, Chen WH, Mott S, Palardy JE, Parejo NA, Heninger S, et al. Detoxified endotoxin vaccine (J5dLPS/OMP) protects mice against lethal respiratory challenge with Francisella tularensis SchuS4. Vaccine. 2010; 28: 2908-2915.
- Opal SM, Palardy JE, Chen WH, Parejo NA, Bhattacharjee AK, Cross AS. Active immunization with a detoxified endotoxin vaccine protects against lethal polymicrobial sespsis: its use with CpG adjuvant and potential mechanisms. J Infect Dis. 2005; 192: 2074-2080.
- Bhattacharjee AK, Opal SM, Palardy JE, Drabick JJ, Collins H, Taylor R, et al. Affinity-Purified Escherichia coli J5 Lipopolysaccharide-Specific IgG Protects Neutropenic Rats Against Gram-Negative Bacterial Sepsis. J Infect Dis. 1994; 170: 622-629.
- Chen WH, Kang TJ, Bhattacharjee AK, Cross AS. Intranasal administration of a detoxified endotoxin vaccine protects against heterologous Gram-negative bacillary pneumonia. Innate Immunity. 2008; 14: 269-278.
- Cross AS, Opal SM, Palardy JE, Drabick JJ, Warren HS, Huber C, et al. Phase I study of detoxified Escherichia coli J5 lipopolysaccharide (J5dLPS)/group B meningococcal outer membrane protein (OMP) complex vaccine in human subjects. Vaccine. 2003; 21: 4576-4588.
- Zupancic ML, Cantarel BL, Liu Z, Drabek EF, Ryan KA, Cirimotich S, et al. Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome. PLoS One. 2012; 7: e43052.
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010; 7: 335-336.
- Chen H, Wu B, Nelson DR, Wu K, Liu C. Computational identification and systematic classification of novel cytochrome P450 genes in Salvia miltiorrhiza. PLoS One. 2014; 9: e115149.
- Modi BP, Langer M, Ching YA, Valim C, Waterford SD, Iglesias J, et al. Improved survival in a multidisciplinary short bowel syndrome program. J PediatrSurg. 2008; 43: 20-24.
- Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014; 41: 478-492.
- Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, et al. Stool microbiota and vaccine responses of infants. Pediatrics. 2014; 134: e362-372.
- Kumar P, Chen K, Kolls JK. Th17 cell based vaccines in mucosal immunity. Curr Opin Immunol. 2013; 25: 373-380.
- Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O–Leary CE, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014; 20: 524-530.