
Special Article - Dengue Virus Infection
J Bacteriol Mycol. 2016; 3(5): 1039.
The Remodeling of the Cytoplasm by Dengue Virus
Chatel-Chaix L*
Institut National de la Recherche Scientifique, Institut Armand-Frappier, Université du Québec, 531 Boulevard des Prairies, Laval, H7V 1B7, Québec, Canada
*Corresponding author: Laurent Chatel-Chaix, Institut National de la Recherche Scientifique, Institut Armand-Frappier, Université du Québec, 531 Boulevard des Prairies, Laval, H7V 1B7, Québec, Canada
Received: November 22, 2016; Accepted: December 16, 2016; Published: December 19, 2016
Abstract
Infection with Dengue Virus (DENV) causes the most prevalent arthropodborne viral disease worldwide. Since there are no antivirals available, there is an urgent medical need to discover novel therapeutic targets, implying a better understanding of the cellular and molecular events governing DENV life cycle. Following infection, DENV induces a massive remodeling of the cytoplasm including the endoplasmic reticulum from which DENV replication factories are formed. In addition, DENV alters the morphology of other organelles such as mitochondria to hijack their functions and to create a cellular environment favorable to viral replication. This review summarizes our current knowledge about the cytoplasmic remodeling activities of DENV.
Keywords: Dengue virus; Replication factories; Autophagy; Mitochondria; Innate immunity
Abbreviations
CM: Convoluted Membranes; DENV: Dengue Virus; DRP1: Dynamin-Related Protein-1; dsRNA : Double-stranded RNA; eIF2α: Eukaryotic initiation factor 2 alpha; ER: Endoplasmic Reticulum; G3BP: Ras-GAP SH3-domain-Binding Protein; IFN: Interferon; IP3R: Inositol Trisphosphate Receptor; GRP75: Glucose- Regulated Protein 75 Kda; ISG: Interferon Stimulated Genes; LC3: Microtubule associated protein 1 Light Chain 3; LD: Lipid Droplets; MAM: Mitochondria-Associated Membranes; MDA5: Melanoma Differentiation-Associated Gene 5; MFN2: Mitofusin-2; mRNA: messenger RNA; PKR : Protein Kinase R; RF : Replication Factories; RIG-I: Retinoic Acid-Inducible Gene-I; sfRNA: Sub-flaviviral RNA; SG: Stress Granules; Spautin1: Specific And Potent Autophagy Inhibitor 1; STING: Stimulator of Interferon Genes; TCA: Tri Carboxylic Acid; TIA-1: T-Cell Intracellular Antigen Protein-1; TIAR: T-Cell Restricted Intracellular Antigen 1-Related Protein; VB: Virus Bags; VDAC-1: Voltage-Dependent Anion Receptor-1; VP: Vesicle Packets; vRNA: Viral RNA; WNV: West Nile Virus; YFV: Yellow Fever Virus; ZIKV : Zika Virus
Introduction
Infection with Dengue Virus (DENV) causes the most prevalent arthropod-borne viral disease worldwide. Humans are infected by the bite of female Aedes mosquitoes (Aedes aegypti and Aedes albopictus) during their blood meal [1]. Occurring in over 100 countries, the prevalence of this emerging disease has increased by 30 fold over the last 50 years. It has been estimated that 400 million individuals are infected annually [2], among which 25% are reported to develop symptoms typical of dengue fever. In some cases, the infection causes dengue hemorrhagic fever, dengue shock syndrome and eventually death. A prophylactic tetravalent vaccine has been recently developed by Sanofi (Dengvaxia®) [3]. However, it provides limited efficacy and has been approved only in 6 countries. Importantly, despite current efforts, antiviral drugs are still not available. Hence, DENV infection represents an important global burden and there is an urgent medical need to develop novel therapeutic approaches. This has been hampered by the fact that our knowledge about the molecular events governing DENV life cycle remains limited.
DENV is an enveloped positive-strand RNA virus belonging to the Flavivirus genus within the Flaviviridae virus family [4]. In addition to DENV, flaviviruses comprise over 70 members including Yellow Fever Virus (YFV), Zika Virus (ZIKV) and West Nile Virus (WNV). Following receptor-mediated endocytosis and envelop fusion, the unique DENV RNA species, namely the viral genome (vRNA) is uncoated and released into the cytosol to be translated at the Endoplasmic Reticulum (ER). The vRNA contains one single open reading frame which encodes a viral transmembrane polyprotein. It is co- and post-translationally processed by cellular and viral proteases, generating 10 mature viral proteins. The nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) are indispensable for vRNA replication. Notably, NS5, through its RNA-dependent RNA polymerase and methyltransferase enzymatic activities, replicates and caps the vRNA. NS3 with its co-factor NS2B harbors a protease activity responsible for the processing of the polyprotein at the junctions located on the cytosolic side of the ER. In addition, NS3 also contains RNA helicase, NTPase and RNA triphosphatase activities, all indispensable for efficient vRNA replication. The structural proteins capsid (C), prM and envelop (E) are responsible for assembling and enveloping novel viral particles which are immature. During egress through the secretory pathway, viral particles undergo maturation and become infectious through conformational changes following the processing of prM by cellular furin [4-7].
Like most positive strand RNA viruses [6], DENV induces massive rearrangements of the ER in the infected cell to create organellelike membranous structures generically called Replication Factories (RF). These ultrastructures showing remarkable morphologies are believed to be required for the DENV life cycle. The viral and cellular determinants of DENV RF morphogenesis remain enigmatic. In the recent years, numerous studies have revealed that other cytoplasmic organelles than ER are also hijacked by DENV. The co-opting of their activities by DENV is often associated with noticeable changes in their morphologies. This cytoplasmic remodeling by DENV creates a cellular environment which is optimal not only for virus replication but also for evading antiviral defenses such as innate immunity.
DENV Replication Factories
Shortly after infection, DENV induces massive ER reorganization to generate three typical ultrastructures (Figure 1, left panel) which can be readily detected by Transmission Electron Microscopy (TEM).
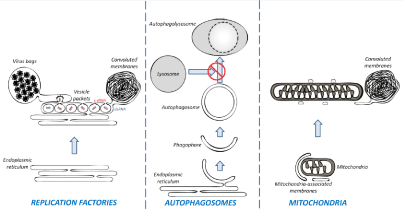
Figure 1: The remodeling of membranous organelles in DENV-infected cells. Left panel: DENV induces alterations of the ER to generate replications factories.
They include three typical ultrastructures: Vesicle packets (believed to be the site of vRNA replication), virus bags (in which immature viruses accumulate in regular
arrays) and convoluted membranes (which might be involved in the modulation of cellular processes and hence, in DENV pathogenesis). Middle panel: DENV
stimulates the biogenesis of autophagosomes but inhibits their fusion with lysosomes. Right panel: DENV induces the elongation of mitochondria which make
contacts with convolutes membrane via altered mitochondria-associated membranes.
Vesicle packets
DENV induces invaginations of the ER resulting in the formation of multiple vesicles with an average diameter of about 90 nm which align within a single plane in mammalian cells [8]. Immunogold labeling combined to TEM showed that these ultrastructures, called Vesicle Packets (VPs) contains NS1, NS2B3 protease, NS4A, NS4B, NS5 polymerase and double-stranded RNA (dsRNA), the replication intermediate [8,9]. Hence, VPs are believed to be the site of vRNA replication. This spatial segregation would allow concentrating the metabolites required for RNA replication such as NTPs but also importantly to protect vRNA from degradation by nucleases and detection by innate immunity sensors like RIG-I and MDA5. VPs are observed in both mammalian and mosquito infected cells implying a conserved virus-mediated regulation of VP morphogenesis [8,9]. Nevertheless, the viral and cellular determinants controlling VP morphogenesis are completely unknown. Transmembrane viral proteins NS4A and NS4B might be involved in this process since they are absolutely required for RNA replication through unknown mechanisms [10-13]. Very interestingly, several small molecules DENV inhibitors developed following phenotypic high-throughput screening campaigns selected for resistance in NS4B [14-18]. This highlights the critical role of NS4B during DENV life cycle. Considering that NS4B interacts with NS4A and NS2B-3 protease [10,11,13], VP morphogenesis and/or replication complexes might be the targets of these antivirals. Three-dimensional reconstruction of electron tomograms revealed that 50% of VPs have a homogenous 10 nm-wide pore connecting the interior of the VPs with the cytosol [8]. First, this pore would allow metabolite exchange between the replication complexes and the cytosol. Second, neosynthesized vRNA would exit VPs through this pore to be translated or encapsidated. Very interestingly, VP pore is sometimes juxtaposed to an electrondense structure budding into the ER whose size (~60nm) would be consistent with the one of a virus being enveloped [8]. This close relationship between replication and assembly compartments would allow coordinating in time and space steps of DENV life cycle while conferring specificity to the RNA packaging process.
Virus bags
In DENV-infected cells, 45 nm-wide electron dense particles which resemble viruses accumulate in highly regular arrays within enlarged ER cisternae, called Virus Bags (VBs) [8]. Immunogold labeling experiments confirmed they are indeed viral particles since they contain DENV envelop protein E [8]. Consistently, treatment with the capsid inhibitor ST-148 reduced the number of VBs per cell [19]. Because of their spiky morphology, these accumulated viruses are believed to be immature and hence, non-infectious. This is consistent with the fact that furin-mediated virus maturation is believed to occur downstream the ER, namely in the trans-Golgi network in which the pH is more acidic. It remains unclear whether the transit through VBs is absolutely required for the release of infectious particles, implying in that case that virus bags contain viruses awaiting to egress. An alternative model could be that VBs serve as a quality control to get rid of particles wrongly assembled, implying a dead-end pathway for virus assembly/release. Further studies are needed to clarify these questions.
Convoluted membranes
DENV infection also induces the accumulation of large ER-derived membranous areas called Convoluted Membranes (CM) whose exact functions are still debated [8]. While NS4A overexpression induces CMs in some extent [20], their biogenesis is not understood. Since NS3 protease accumulates within CMs [8,21,22], they were initially proposed to be the site of polyprotein translation and processing. More recently, CM were shown to contain large amounts of NS4B but to be devoid of the replication intermediate dsRNA [21], implying that they are involved in other processes than vRNA replication. In support to this, CMs, in contrast to VPs, were not detected in infected mosquito cells by electron microscopy [9] implying that they are not absolutely required for vRNA replication. Moreover, this suggests that CM morphogenesis requires species- and/or cell type-specific cellular factors and that they influence cellular processes that do not exist in insects. NS3 was shown to inhibit host factors STING and 14-3-3epsilon, resulting in the inactivation of MAVS-dependent innate immunity (absent in insects) through proteolytic and nonproteolytic activities, respectively [23-25]. Hence, one attractive model could be that some host factors are recruited by DENV into CM in which NS3 is enriched and are subsequently inactivated within this compartment. Transmission electron microscopy analyses suggested that CMs originate at least in part from the ER tubules in contact with mitochondria [21]. As explained below, this compartment, called Mitochondria-Associated Membranes (MAMs) is critical for RIG-I-dependent interferon induction [26]. Hence, the alteration of MAMs by CM biogenesis would contribute to DENVmediated interference with early innate immune responses. To sum up, through the hijacking of antiviral responses, CMs would contribute to the establishment of a cellular environment favorable to the infection rather than directly to RNA replication.
Autophagosomes
Several studies reported that DENV infection induces autophagy in numerous cell lines [27-31] as well as in vivo in infected mice [32] (Figure 1, middle panel). Autophagy is a cellular process allowing to get rid of damaged organelles or specific proteins. The isolation membrane, called the phagophore originates from ER protrusions, elongates and selects cargos through interactions with the receptor LC3. The phagophore closes on itself to form a double-membrane vesicle called autophagosome. The autophagosomes eventually fuse with lysosomes to form autolysosomes in which content is degraded [33]. Autophagy induction is important for DENV life cycle since viral replication is inhibited by the treatment of infected cells with autophagy inhibitors 3-methyladenine or by knocking down genes important for autophagosome biogenesis such as Beclin-1, ATG4B and ATG12 [27]. Very recently, we have demonstrated that while DENV stimulates autophagosome biogenesis, their fusion with lysosomes is actually inhibited by the virus [29]. This suggests that DENV reprograms the autophagy process for the benefit of replication. Considering that the morphogenesis of RFs relies on the expansion and remodeling of the ER as described above, one can speculate that this requires a DENV-dependent modulation of ER turnover by autophagy (called ER-phagy or reticulophagy) [34,35]. In addition to replication, virus particle egress and infectivity relies on autophagy. Indeed, the use of Spautin-1, a specific inhibitor of beclin1- Vps34-Atg14 complex activation (required for autophagy induction) significantly reduced DENV specific infectivity [28], suggesting that autophagy is required for virus maturation. More recently, Wu and colleagues demonstrated that DENV particles exploit the autophagy pathway as a non-classical secretory pathway to egress out of the infected cell and to favor cell-to-cell transmission [36]. DENVinduced autophagy was demonstrated to target lipid droplets, the intracellular storage of neutral lipids such as triglycerides and free cholesterol [27]. This results in a decrease of the number and size of lipid droplets in infected cells. Very interestingly, a sub-population of the structural protein C was shown to localize at the surface of lipid droplets [37-39]. Hence, this sub-population of C, which is important for particle assembly [37,39,40], might be mobilized through autophagy. How DENV induces and regulates autophagy remains mostly enigmatic. DENV NS4A overexpression is sufficient to induce autophagy [41]. Such strategy seems to be conserved among flaviviruses since WNV NS4A also induces autophagy in addition to NS4B [42]. More recently, it was shown for ZIKV that these two proteins cooperate to stimulate autophagy [43]. Considering that DENV CM components NS4A and NS4B physically interact [13] and that NS4A induces CM biogenesis in some extent [20], one can speculate that both autophagosomes and CM morphogenesis might be interdependent and rely on several viral proteins. While it seems clear that autophagy impacts on several steps of DENV life cycle, further studies are required to clearly establish the underlying molecular mechanisms governing this virus/host interplay.
Mitochondria
Very recently, we and others reported that DENV infection induces a drastic elongation of mitochondria (Figure 1, right panel) [21,44]. This modulation of mitochondria morphodynamics resulted at least in part from the downregulation of the mitochondria fission factor DRP1 notably at the level of its activation by phosphorylation on serine 616 [21, 44]. The inhibition of DRP1 translocation to mitochondria perturbs the fusion/fission equilibrium in favor of fusion, resulting in an increased elongation of the mitochondria. This phenotype was attributed to NS4B whose transient overexpression was sufficient to induce mitochondria elongation. Very interestingly, elongated mitochondria make contacts with CMs via MAMs which are altered in DENV-infected cells. Enforced mitochondria elongation favored DENV replication supporting the idea that DENV modulates mitochondria morphology for the benefit of its replication. In contrast, induction of mitochondria fragmentation reduced DENV replication and inhibited CM formation and maintenance [21]. This shows that mitochondria elongation is important for CM biogenesis. Very importantly, mitochondria elongation alleviates RIG-I-dependent innate immune response resulting in a decrease of type-I and -III interferon expression. This correlated with a loss of RIG-I translocation to MAMs, which have been shown to be critical for MAVS-dependent signaling [21]. This is consistent with an overall alteration of ER-mitochondria contacts in DENV-infected cells. Hence, the functional and physical relationship between DENV replication factories, mitochondria and MAMs is important for DENV replication and for dampening antiviral responses. This suggests that some mitochondrial metabolic activities might be influenced by this DENV-dependent morphological change. Notably, it was shown that DENV increases the O2 consumption [44] by mitochondria implying an enhanced mobilization of the Tri Carboxylic Acid cycle (TCA) cycle and ATP production by the mitochondria. In addition, the takeover of LDs by the autophagy machinery described above results in increased levels of β-oxidation [27]. This process takes place in mitochondria and catalyzes the degradation of triglycerides and results in ATP synthesis independently of the TCA cycle. While it was shown that mitochondrial elongation can stimulate respiration [45-49], whether the DENV-induced increases in β-oxidation and respiration rely on the modulation of mitochondria morphodynamics remains to be determined. It was shown very recently that the mitochondrial VDAC1 is important for DENV replication [50]. VDAC1 is a bona fide component of MAM, contributing to tether mitochondria to ER through interactions with GRP75 and ER-resident protein IP3R. Moreover, this protein is important for the transfer of calcium from the ER to mitochondria at the MAM [51,52]. This suggests that DENV might influence calcium intracellular homeostasis through the alteration of MAMs. Finally, another MAM-resident protein, MFN2 is cleaved by DENV protease NS2B-NS3 [53]. MFN2 is involved in the fusion of mitochondria (leading to elongation) but also in the ER-mitochondria tethering. While MFN2 processing seems to not negatively impact on DENV-induced mitochondria elongation in normal infection conditions [53], it might rather constitute the molecular mechanism by which DENV alters MAMs although this hypothesis was never tested. To conclude, further studies are needed to determine whether DENV influences additional MAM functions and/or mitochondrial metabolic activities and how this is linked to CM biogenesis and mitochondria morphodynamics.
Peroxisomes
Recently, you and colleagues have demonstrated that the number of peroxisomes dramatically decreases in cells infected by DENV or WNV [54]. This alteration was recapitulated upon overexpression of the viral protein capsid which interacts with cellular peroxin Pex19 required for peroxisome biogenesis. Notably, Pex19 protein levels were significantly decreased following DENV infection accounting for peroxisome partial depletion. Very interestingly, Pex19 expression knockdown impaired type-III IFN transcription [54] suggesting that DENV modulates peroxisome biogenesis to dampen innate immunity. Nevertheless, this did not result in an increase in virus replication implying that peroxisome components are important for DENV life cycle.
Stress Granules
Under stress conditions like oxidative stress, hypoxia, heat shock or virus infection, specific ribonucleoparticles assemble into large structures called Stress Granules (SG) which contain mRNAs whose translation is blocked at the level of initiation [55]. This generally correlates with an activation of PKR resulting in the phosphorylationmediated inhibition of the translation initiation factor eIF2α. Intriguingly, Emara and colleagues have demonstrated that both DENV and WNV infections inhibit the formation of SGs in cells treated with sodium arsenite (which induces oxidative stress) [56]. Moreover, TIA-1 and TIAR, two components of SGs involved in their assembly, are relocalized to the replication complexes [56]. Later, Ruggieri and colleagues showed that DENV, as compared to other viruses such as Newcastle disease and Sendai viruses for instance, induces relatively few SGs. In that case, SGs showed a slow assembly/ disassembly frequency [57]. Another study showed that DENV co-opts the function of the SG assembly factors G3BP1, G3BP2 and CAPRIN1 [58]. Notably, these proteins were demonstrated to positively regulate the translation of several Interferon-Stimulated Genes (ISG) such as PKR and IFITM2. Very interestingly, the DENV sub-flaviviral RNA (sfRNA), a non-coding RNA resulting from the incomplete 5’-3’ degradation of vRNA by Xrn1 exonuclease, was shown to associate with G3BP1, G3BP2 and CAPRIN1 and to inhibit their function in ISG translation. Consistently, G3BP1, G3BP2 and CAPRIN1 combined knockdown increased DENV replication and resistance to IFN treatment while impairing ISG production [58]. Altogether, these studies support the idea that DENV developed strategies to hijack SG components and dampen the stress associated to infection, hence hampering the establishment of an antiviral cellular environment.
Conclusion
As an obligate intracellular parasite, dengue virus has evolved to establish within the cells a cytoplasmic environment optimal for vRNA replication while antiviral responses are shut-off. The precise molecular mechanisms governing these remarkable remodeling activities remain mostly enigmatic. Theoretically, specific interactions between host factors and viral proteins critical for DENV life cycle could be targeted by small molecule inhibitors. Since the cellular proteins are not subjected to NS5-driven adaptation and resistance acquisition, the antiviral activity of such compounds is expected to be associated with a high barrier-to-resistance. If these DENV coopting activities are conserved among flaviviruses, anti-DENV drugs might actually constitute pan-flaviviral inhibitors. Given the recent outbreak of ZIKV in the Americas and the continuously growing burden of dengue worldwide, a considerable effort is being made to push Flavivirus research forward. Hence, one can be optimistic that antivirals and more efficient vaccines will be widely available in a close future to prevent these emerging diseases.
References
- World Health Organization. Dengue: Guidelines for Diagnosis,Treatment, Prevention and Control. 2009.
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013; 496: 504-507.
- Alagarasu K. Introducing dengue vaccine: Implications for diagnosis in dengue vaccinated subjects. Vaccine. 2016; 34: 2759-2761.
- Acosta EG, Kumar A, Bartenschlager R. Revisiting dengue virus-host cell interaction: new insights into molecular and cellular virology. Advances in virus research. 2014; 88: 1-109.
- Chatel-Chaix L, Bartenschlager R. Dengue virus- and hepatitis C virus-induced replication and assembly compartments: the enemy inside--caught in the web. Journal of virology. 2014; 88: 5907-5911.
- Paul D, Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World journal of virology. 2013; 2: 32-48.
- Paul D, Bartenschlager R. Flaviviridae Replication Organelles: Oh, What a Tangled Web We Weave. Annual review of virology. 2015; 2: 289-310.
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell host & microbe. 2009; 5: 365-375.
- Junjhon J, Pennington JG, Edwards TJ, Perera R, Lanman J, Kuhn RJ. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. Journal of virology. 2014; 88: 4687-4697.
- Chatel-Chaix L, Fischl W, Scaturro P, Cortese M, Kallis S, Bartenschlager M, et al. A Combined Genetic-Proteomic Approach Identifies Residues within Dengue Virus NS4B Critical for Interaction with NS3 and Viral Replication. Journal of virology. 2015; 89: 7170-7186.
- Zou J, Lee le T, Wang QY, Xie X, Lu S, Yau YH, et al. Mapping the Interactions between the NS4B and NS3 proteins of dengue virus. Journal of virology. 2015; 89: 3471-3483.
- Zou J, Xie X, Lee le T, Chandrasekaran R, Reynaud A, Yap L, et al. Dimerization of flavivirus NS4B protein. Journal of virology. 2014; 88: 3379-3391.
- Zou J, Xie X, Wang QY, Dong H, Lee MY, Kang C, et al. Characterization of dengue virus NS4A and NS4B protein interaction. Journal of virology. 2015; 89: 3455-3470.
- Wang QY, Dong H, Zou B, Karuna R, Wan KF, Zou J, et al. Discovery of Dengue Virus NS4B Inhibitors. Journal of virology. 2015; 89: 8233-8244.
- Xie X, Wang QY, Xu HY, Qing M, Kramer L, Yuan Z, et al. Inhibition of dengue virus by targeting viral NS4B protein. Journal of virology. 2011; 85: 11183-11195.
- Xie X, Zou J, Wang QY, Shi PY. Targeting dengue virus NS4B protein for drug discovery. Antiviral research. 2015; 118: 39-45.
- Cleef KW, Overheul GJ, Thomassen MC, Kaptein SJ, Davidson AD, Jacobs M, et al. Identification of a new dengue virus inhibitor that targets the viral NS4B protein and restricts genomic RNA replication. Antiviral research. 2013; 99: 165-171.
- Wispelaere M, LaCroix AJ, Yang PL. The small molecules AZD0530 and dasatinib inhibit dengue virus RNA replication via Fyn kinase. Journal of virology. 2013; 87: 7367-7381.
- Scaturro P, Trist IM, Paul D, Kumar A, Acosta EG, Byrd CM, et al. Characterization of the mode of action of a potent dengue virus capsid inhibitor. Journal of virology. 2014; 88: 11540-11555.
- Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. The Journal of biological chemistry. 2007; 282: 8873-8882.
- Chatel-Chaix L, Cortese M, Romero-Brey I, Bender S, Neufeldt CJ, Fischl W, et al. Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell host & microbe. 2016; 20: 342-356.
- Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. Journal of virology. 1997; 71: 6650-6661.
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS pathogens. 2012; 8: e1002934. PubMed PMID: 23055924. Pubmed Central PMCID: 3464218.
- Chan YK, Gack MU. A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity. Nature immunology. 2016 May; 17: 523-530. PubMed PMID: 26998762. Pubmed Central PMCID: 4837045.
- Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, et al. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS pathogens. 2012; 8: e1002780. PubMed PMID: 22761576. Pubmed Central PMCID: 3386177.
- Horner SM, Liu HM, Park HS, Briley J, Gale M, Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proceedings of the National Academy of Sciences of the United States of America. 2011; 108: 14590-14595. PubMed PMID: 21844353. Pubmed Central PMCID: 3167523.
- Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell host & microbe. 2010; 8: 422-432. PubMed PMID: 21075353. Pubmed Central PMCID: 3026642.
- Mateo R, Nagamine CM, Spagnolo J, Mendez E, Rahe M, Gale M, et al. Inhibition of cellular autophagy deranges dengue virion maturation. Journal of virology. 2013; 87: 1312-1321.
- Metz P, Chiramel A, Chatel-Chaix L, Alvisi G, Bankhead P, Mora-Rodriguez R, et al. Dengue Virus Inhibition of Autophagic Flux and Dependency of Viral Replication on Proteasomal Degradation of the Autophagy Receptor p62. Journal of virology. 2015; 89: 8026-8041. PubMed PMID: 26018155. Pubmed Central PMCID: 4505648.
- Lee YR, Lei HY, Liu MT, Wang JR, Chen SH, Jiang-Shieh YF, et al. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008; 374: 240-248. PubMed PMID: 18353420.
- Panyasrivanit M, Khakpoor A, Wikan N, Smith DR. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. The Journal of general virology. 2009; 90: 448-456. PubMed PMID: 19141455.
- Lee YR, Hu HY, Kuo SH, Lei HY, Lin YS, Yeh TM, et al. Dengue virus infection induces autophagy: an in vivo study. Journal of biomedical science. 2013; 20: 65. PubMed PMID: 24011333. Pubmed Central PMCID: 3848819.
- Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, et al. Mammalian Autophagy: How Does It Work? Annual review of biochemistry. 2016; 85: 685-713. PubMed PMID: 26865532.
- Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nature cell biology. 2016; 18: 1173-84. PubMed PMID: 27749824.
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015; 522: 354-358. PubMed PMID: 26040720.
- Wu YW, Mettling C, Wu SR, Yu CY, Perng GC, Lin YS, et al. Autophagy-associated dengue vesicles promote viral transmission avoiding antibody neutralization. Scientific reports. 2016; 6: 32243. PubMed PMID: 27558165. Pubmed Central PMCID: 4997566.
- Carvalho FA, Carneiro FA, Martins IC, Assuncao-Miranda I, Faustino AF, Pereira RM, et al. Dengue virus capsid protein binding to hepatic Lipid Droplets (LD) is potassium ion dependent and is mediated by LD surface proteins. Journal of virology. 2012; 86: 2096-2108. PubMed PMID: 22130547. Pubmed Central PMCID: 3302401.
- Martins IC, Gomes-Neto F, Faustino AF, Carvalho FA, Carneiro FA, Bozza PT, et al. The disordered N-terminal region of dengue virus capsid protein contains a lipid-droplet-binding motif. The Biochemical journal. 2012; 444: 405-415. PubMed PMID: 22428600.
- Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, et al. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS pathogens. 2009; 5: e1000632. PubMed PMID: 19851456. Pubmed Central PMCID: 2760139.
- Iglesias NG, Mondotte JA, Byk LA, De Maio FA, Samsa MM, Alvarez C, et al. Dengue Virus Uses a Non-Canonical Function of the Host GBF1-Arf-COPI System for Capsid Protein Accumulation on Lipid Droplets. Traffic. 2015; 16: 962-977. PubMed PMID: 26031340. Pubmed Central PMCID: 4543523.
- McLean JE, Wudzinska A, Datan E, Quaglino D, Zakeri Z. Flavivirus NS4A-induced autophagy protects cells against death and enhances virus replication. The Journal of biological chemistry. 2011; 286: 22147-22159. PubMed PMID: 21511946. Pubmed Central PMCID: 3121359.
- Blazquez AB, Martin-Acebes MA, Saiz JC. Amino acid substitutions in the non-structural proteins 4A or 4B modulate the induction of autophagy in West Nile virus infected cells independently of the activation of the unfolded protein response. Frontiers in microbiology. 2014; 5: 797. PubMed PMID: 25642225. Pubmed Central PMCID: 4295549.
- Liang Q, Luo Z, Zeng J, Chen W, Foo SS, Lee SA, et al. Zika Virus NS4A and NS4B Proteins Deregulate Akt-mTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell stem cell. 2016; 19: 663-671. PubMed PMID: 27524440.
- Barbier V, Lang D, Valois S, Rothman AL, Medin CL. Dengue virus induces mitochondrial elongation through impairment of Drp1-triggered mitochondrial fission. Virology. 2016; 500: 149-160. PubMed PMID: 27816895.
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. The Journal of biological chemistry. 2005; 280: 26185-26192. PubMed PMID: 15899901.
- Mourier A, Motori E, Brandt T, Lagouge M, Atanassov I, Galinier A, et al. Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. The Journal of cell biology. 2015; 208: 429-442. PubMed PMID: 25688136. Pubmed Central PMCID: 4332246.
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007; 130: 548-562. PubMed PMID: 17693261.
- Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010; 141: 280-289. PubMed PMID: 20403324. Pubmed Central PMCID: 2876819.
- Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013; 155: 160-171. PubMed PMID: 24055366. Pubmed Central PMCID: 3790458.
- Jitobaom K, Tongluan N, Smith DR. Involvement of Voltage-Dependent Anion Channel (VDAC) in dengue infection. Scientific reports. 2016; 6: 35753. PubMed PMID: 27779201. Pubmed Central PMCID: 5078847.
- van Vliet AR, Verfaillie T, Agostinis P. New functions of mitochondria associated membranes in cellular signaling. Biochimica et biophysica acta. 2014; 1843: 2253-2262. PubMed PMID: 24642268.
- Vance JE. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochimica et biophysica acta. 2014;1841: 595-609. PubMed PMID: 24316057.
- Yu CY, Liang JJ, Li JK, Lee YL, Chang BL, Su CI, et al. Dengue Virus Impairs Mitochondrial Fusion by Cleaving Mitofusins. PLoS pathogens. 2015; 11: e1005350. PubMed PMID: 26717518. Pubmed Central PMCID: 4696832.
- You J, Hou S, Malik-Soni N, Xu Z, Kumar A, Rachubinski RA, et al. Flavivirus Infection Impairs Peroxisome Biogenesis and Early Antiviral Signaling. Journal of virology. 2015; 89: 12349-12361. PubMed PMID: 26423946. Pubmed Central PMCID: 4665241.
- Panas MD, Ivanov P, Anderson P. Mechanistic insights into mammalian stress granule dynamics. The Journal of cell biology. 2016; 215: 313-323. PubMed PMID: 27821493. Pubmed Central PMCID: 5100297.
- Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proceedings of the National Academy of Sciences of the United States of America. 2007; 104: 9041-9046. PubMed PMID: 17502609. Pubmed Central PMCID: 1885624.
- Ruggieri A, Dazert E, Metz P, Hofmann S, Bergeest JP, Mazur J, et al. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell host & microbe. 2012; 12: 71-85. PubMed PMID: 22817989. Pubmed Central PMCID: 3873964.
- Bidet K, Dadlani D, Garcia-Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS pathogens. 2014; 10: e1004242. PubMed PMID: 24992036. Pubmed Central PMCID: 4081823.