
Mini Review
Austin Biochem. 2017; 2(1): 1006.
Targetomics - More than a Drug Race on Protein Libraries
Qing Y¹, Stahl F² and Zeilinger C¹*
¹Institute of Technical Chemistry, Leibniz University Hanover, Germany
²Institute of Biophysics, Leibniz University Hanover, Germany
*Corresponding author: Zeilinger C, Center of Biomolecular Drug Research (BMWZ), Leibniz University Hanover, Schneiderberg 38, 30167 Hannover, Germany
Received: January 02, 2017; Accepted: February 20, 2017; Published: February 21, 2017
Abstract
Nature and chemists produced a huge arsenal of small molecules that are able to inhibitor manipulate target proteins. To identify preferred selectivity and activity on targets, different screenings have been developed with the aim to examine influences of small molecules on targets. This requires an understanding of the function of the and often a special adaptation of the experimental control and is often accompanied with material consumption. The development of a highly miniaturized microarray based screening method allows us to verify new inhibitors based on a target-oriented screening. Targeting Hsp90 with geldanamycin as lead structure to compete ATP from the binding site enabled screenings of different Hsp proteins against in-house drug library using inhibitors from a predicted structure. It was possible to identify Gda derivatives with this in vitro assay with affinity in the nM range for human Hsp90 or against pathogens. We can show that such a microarray-assay is a powerful multiplex screening tool for inhibitor libraries versus protein libraries. This assay strategy disclosed later also that full-length target proteins can be used for protein interaction studies and additionally is also suitable for diagnostic improvements.
Keywords: Heat shock proteins; Target-oriented screening; Protein libraries
Abbreviations
Hsp: Heat shock proteins
Introduction
High-throughput technologies have been available to biological research with the turn of the century, which enable the various biomolecule species of the cell (e.g., genes, proteins, metabolic products, etc.) to be collected in their entirety. These high-throughput technologies are known as “omics” technologies and are primarily based on so-called microarrays, which can be used to carry out several thousand individual observations in parallel, requiring only a very small amount of sample. Although many technical aspects concerning microarray have been addressed in great detail it remains still crucial to establish this technology in new research fields such as target identification. Microarrays consist of modified glass surfaces on which high spot densities (up to 120,000) can be applied with very small spot diameters (smaller than 250 μm). The use of this technology leads to a quantum leap in the information content of the individual experiments. In order to store this information, special, standardized databases have been created, e.g. NCBIs Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/).
A large number of DNA sequences stored in databases without corresponding knowledge of the function or functional integrity of the relevant genes / biomolecules [1], they now become filled with knowledge about the interactions of biomolecules. Because such knowledge is with respect to the interaction of clinically relevant targets and potential active compounds also essential for pharmaceutical research, to develop new medicines or to improve existing pharmaceutical research indirectly used the stored “omics” data to perform predictions by virtual screenings. Here we report a novel strategy analyzing protein versus inhibitor libraries by spotting target proteins on microarrays multiplexing differences in the susceptibility of inhibitors or interaction candidates.
Targets are marker of key processes
Heat shock proteins (Hsp) are essential for the survival of an organism [2,3]. Contrary to the name, heat shock proteins are not only synthesized by heat, but also in other cellular stress situations, such as exposure to UV radiation, heavy metals, alcohol or oxygen deficiency. Due to these uncontrolled influences the accompanied genomic transformation triggers carcinogenic cells to survive and try to escape immunological sentinel. At this point different strategies required to diagnose the correct cancer type and to hinder the rapid and often aggressive growth of cancer cells [4,5]. In recent years, it has been shown that heat shock proteins play a key role in numerous pathologies and are therefore ideal targets for the development of drugs [6]. Single heat shock proteins like Hsp90, Hsp70 and other are embedded with co-chaperones in a machinery to maintain the functional proteome, whereas clients of the machinery become folded or degraded [7-9]. Hsp90 is of particular relevance since it leads nascent polypeptides through the folding process and thus supports maturation to a native protein having an intact three-dimensional structure, which is essential for the function of the protein. Thus, de novo synthesized proteins are folded, but also proteins denatured by cellular stress. The cell response to stress is highly conserved in prokaryotes and all species of eukaryotes and thus represents a general mechanism for the maintenance of cellular processes and protection against the formation of protein aggregates in the cytosol.
This process, which is vital in “healthy” cells, is contra productive in tumor cells: if essential proteins are not introduced into the folding cycle, the formation of protein aggregates in the activation of programmed cell death (apoptosis) occurs in eukaryotes. The anti-apoptotic nature of Hsp90 is that cells can survive stress. If the protein aggregates are not degraded, however, apoptosis can be initiated. An approach of the cancer therapy suppresses the Hsp function in the tumor cells by inhibitors. Because heat shock proteins are over expressed in tumor cells, they occur in higher concentrations and activity than normally and thus prevent the cancer cell from being damaged by the above-mentioned protective mechanisms of apoptosis. Therefore the hope is that specific inhibitors can be accumulated cancer specific [10]. In cancer cells or other pathogenic cells, however, apoptosis and the associated cell death is quite desirable. With the identification of Hsp90 or Hsp70 as targets for anti-tumor therapy, the search for suitable inhibitors began. In the development of inhibitors for Hsp90, new biologically active substances are repeatedly discovered and synthesized since these natural products are designed by evolutionary selection steps to provide survival in environments [10]. The first inhibitor for Hsp90 was Geldanamycin, which is a secondary metabolite of the actinomycetes Streptomyces hygroscopicus and belongs to the benzochinoidansamycins. However, since ansamycin has low solubility, high hepatotoxicity and limited bioavailability, or is rapidly unstable, alternatives are sought [6,11].
Development of target-oriented screenings
Since only small quantities of both the heat shock proteins and derivatives were available, a miniaturized screening of microarrays was developed. For this purpose, the protein of interest was immobilized on chip surfaces made of nitrocellulose to maintain its native conformation as a so-called capture molecule. If this capturing molecule now binds a colour marker, which is usually a molecule from the cellular environment, this can be measured as an optical binding signal. This can be a ligand or an interacting molecule (Figure 1). Subsequently, the competing binding of more specific compounds to the ligand or interacting other substrates (e.g. ansamycin derivatives) to the binding site of the heat shock protein is examined in a so-called displacement assay [12-16]. A possible active substance candidate can now not only displace the marker, but also prevents new cellphysiologically relevant molecules from binding to the position again. This method involves different application potentials and can be used in the optimization of the active substance in order to clarify predictions from the predictive modelling and, on the other hand, to carry out screening of chemical libraries (small molecules) in a high throughput process against a defined target such as HSP90.
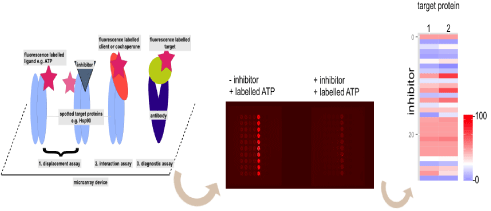
Figure 1: Principle of highly miniaturized microarray based screenings (left). Different applications are possible displacement assay of labelled ligand to test
competitive inhibitors (1) interaction assay (2) and diagnostic assay (3). Typical read out form of spotted proteins on a subpad from a microarray device. Proteins
spotted in a row of ten spots and incubated with or without corresponding heat map analysis (right, blue no label binding; 100 bound label).
Thus, specific interactions between target proteins (such as Hsp90 or Hsp70) and potential inhibitors can be investigated quickly and efficiently on the biochips [12-14]. Compound libraries or natural products have been screened with this method. Both tiny amounts of heat shock protein (300 pmol) and inhibitor are sufficient (Figure 1). The printed chips can be stored and used for several weeks at 4°C and with around 150 μg protein it is possible to generate more than 1,000 arrays. Due to the high heterogeneous multiplex, large substance banks can be screened quickly and effectively. Simultaneous is it possible to identify new inhibitors against human Hsp90 as well as against the prokaryotic homologue HtpG from H. pylori [13].
This target-oriented screening system, established recently has the advantage that it can identify and use active substances whose potential for drug development is not obvious and thus hardly recognizable in virtual screenings e.g. using natural raw extracts from fungi or isolated compounds from plants [15]. The development of an anti Hsp compound is also important for academic questions when it is used to understand and characterize the biological system. Thus, valuable functional and structural data on active compounds, which in turn can provide important insights for the design of new active compounds or libraries can be used to strobe highly homologues binding sites by weak differences in the sequence by selecting more specific derivatives [13]. This could lead to the identification of active compounds more specific for targets before the associated disease pattern is confirmed. By identifying new compounds, these active substances can be used not only in therapy, but also in diagnostics, so that the smallest amounts of tissue samples are sufficient to detect possible diseases. In this way, pharmaceutical research and the search for active substances will be massively altered by the interplay of molecular biology and medical chemistry as well as the modern possibilities of miniaturization, automation, combinatory, diagnostics and bioinformatics, leading to a more rational drug search and contributing to the personalization of medicine in the long term.
Conclusion
With the development of a highly miniaturized microarraybased strategy novel compounds were identified by a target-oriented screening. Further results indicate that this forward strategy has a broad application potential to filter active compounds on a broad target spectrum as well as interacting candidates depending on the sensing and visualization system.
References
- Ermolaeva O, Rastogi M, Pruitt KD, Schuler GD, Bittner ML, Chen Y, et al. Data management and analysis for gene expression arrays. Nat Genet. 1998; 20: 19-23.
- Lindquist S. The Heat-Shock Response. Ann Rev Biochem. 1986; 55: 1151- 1191.
- Velazquez JM, DiDomenico BJ, Lindquist S. Intracellular localization of heat shock proteins in Drosophila. Cell. 1980; 20: 679-689.
- Li Z, Srivastava P. Heat-Shock Proteins. Current Protocols in Immunology. 2004; 58: A.1T.1-A.1T.6.
- Srivastava P. Interaction of heat shock proteins with peptides and antigen presenting cells: chaperoning of the innate and adaptive immune responses. Annu. Rev. Immunol. 2002; 20: 395-425.
- Franke J, Eichner S, Zeilinger C, Kirschning A. Targeting Heat-shock-protein 90 (Hsp90) by natural products: geldanamycin, a show case in cancer therapy. Nat Prod Rep. 2013; 30: 1299-1323.
- Pearl LH. Review: The HSP90 molecular chaperone-an enigmatic ATPase. Biopolymers. 2016; 105: 594-607.
- Smith JR, Clarke PA, de Billy E, Workman P. Silencing the cochaperone CDC37 destabilizes kinase clients and sensitizes cancer cells to HSP90 inhibitors. Oncogene. 2009; 28: 157-169.
- Ou JR, Tan MS, Xie AM, Yu JT, Tan L. Heat shock protein 90 in Alzheimer’s disease. Biomed Res Int. 2014; 2014: 796869.
- Shrestha L, Bolaender A, Patel HJ, Taldone T. Heat Shock Protein (HSP) Drug Discovery and Development: Targeting Heat Shock Proteins in Disease. Curr Top Med Chem. 2016; 16: 2753-2764.
- Eichner S, Eichner T, Floss HG, Fohrer J, Hofer E, Sasse F, et al. Broad substrate specificity of the amide synthase in S. hygroscopicus--new 20-membered macrolactones derived from geldanamycin. J Am ChemSoc. 2012; 134: 1673-1679.
- Kirschning A, Walter J-G, Stahl F, Schax E, Scheper T, Aliuos P, et al. Molecular Survival Strategies of Organisms: HSP and Small Molecules for Diagnostics and Drug Development. In Springer International Publishing. 2015; 9: 323-344.
- Schax E, Walter J-G, Märzhäuser H, Stahl F, Scheper T, Agard DA, et al. Microarray-based screening of heat shock protein inhibitors. J Biotechnol. 2014; 180: 1-9.
- Hermane J, Bulyszko I, Eichner S, Sasse F, Collisi W, Poso A, et al. New, non-quinone fluorogeldanamycin derivatives strongly inhibit Hsp90. ChemBioChem. 2015; 16: 302-311.
- Sharma R, Mohammadi-Ostad-Kalayeh S, Stahl F, Zeilinger C, Dräger G, Kirschning A, et al. Two new labdanediterpenoids and one new β-lactam from the aerial parts of Royleacinerea. PhytochemLett. 2017; 19: 101-107.
- Schax E, Neunaber J, Stahl F, Walter J-G, Scheper T, Eichner S, et al. Multiplexed heat shock protein microarray as a screening platform for the selection of novel drug compounds. Biodiscovery. 2014; 14: 22.