
Research Article
Austin J Biotechnol Bioeng. 2016; 3(1): 1055.
Rapid Detection of Cactus virus X in Pitaya by Efficient Reverse Transcription Loop-Mediated Isothermal Amplification
Zhang Y¹, Liu Z¹, Huang Q¹, Luo Y¹, Pennerman KK², Lai D1,3, Bai H4, Lin Y¹ and Yin G1,2*
¹Key Laboratory of Biology and Genetic Resources of Tropical Crops, Ministry of Agriculture, Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, China
²Department of Plant Biology and Pathology, Rutgers, The State University of New Jersey, USA
³College of Environment and Plant Protection, Hainan University, China
4Department of Crop, Soil, and Environmental Sciences, University of Arkansas, USA
*Corresponding author: Yin G, Department of Plant Biology and Pathology, Rutgers, The State University of New Jersey, 59 Dudley Rd, New Brunswick, New Jersey, 08901, USA
Received: November 06, 2015; Accepted: January 18, 2015; Published: January 19, 2016
Abstract
Cactus virus X (CVX) is a recently reported virus in pitaya that can significantly reduce crop yield and quality. According to the CVX genome sequence in GenBank (accession no. NC_002815), primers for reverse transcription loop-mediated isothermal amplification (RT-LAMP) and RT-PCR were designed to detect this virus. Our results indicated that the best LAMP reaction conditions were as follows: 1.2-1.6 μM of internal primers, 0.2-0.25 μM of external primers, 0.4-0.8 μM of loop primers, and incubation at 62°C/63°C for 30 min. Our data also showed that the LAMP primers specifically targeted CVX and resulted in the typical waterfall-like bands after gel electrophoresis and sigmoidal amplification curves. Ten-fold serial dilutions of CVX cDNA indicated that LAMP is much faster and at least 10-100 times more sensitive than RT-PCR in detecting this virus. In field identification using either RT-LAMP or RT-PCR, the majority of samples (86.7%) was positive for CVX. This is the first report of CVX diagnosis in China and of the use of the efficient RT-LAMP technology to identify CVX in the field. RT-LAMP meets the requirements for rapid diagnosis analysis needed for control and management of this emerging pathogen.
Keywords: Cactus virus X; Pitaya; Reverse transcription loop-mediated isothermal amplification; RT-PCR
Abbreviations
CVX: Cactus virus X; ELISA: Enzyme-linked immunosorbent assays; PRSV: Papaya ring spot virus; RRSV: Rice ragged stunt virus; RT-LAMP: Reverse transcription loop-mediated isothermal amplification; RT-PCR: Reverse transcription polymerase chain reaction; SRBSDV: Southern rice black-streaked dwarf virus; SrMV: Sorghum mosaic virus
Introduction
Pitaya (also called dragon fruit; of the Cataceae family) is usually reproduced by asexual propagation from the branches, allowing viruses to easily spread and accumulate. Cactus virus X (CVX) is a cylindrical virion with a length of 480 to 520 nm and a member of the genus Potexvirus. CVX can infect many species belonging to Cactaceae family such as Cereus spp., Carnegiea spp., Opuntia spp., Schlumbergera spp. and Hylocereus spp., and appears to be gaining a worldwide distribution. It was first reported in pitaya in Taiwan in 2001 [1]. The complete nucleotide sequence of this virus was later cloned [2]. CVX is now an emerging threat to pitaya in the state of California in the United States [3]. Infected plants show symptoms on the epidermis of the fleshy stems, often including faded spots, pale yellow-green mosaics, deformed spines and necrosis. It is mainly spread by mechanical sap inoculation, grafting and vegetative propagation.
The methods used to detect plant viruses include indicator plants, electron microscopy, enzyme-linked immunosorbent assays (ELISA), RT-PCR and LAMP [4,5]. Loop-mediated isothermal amplification (LAMP) is a novel isothermal nucleic acid amplification technology that has been widely used to detect a variety of pathogenic microorganisms and genetic modifications in agricultural crops, livestock, aquatic organisms and humans. It has especially been used in plant virus detection and diagnosis [5-11]. There is currently no established protocol for the detection and diagnosis of CVX infection. In this study, we aimed to establish an efficient RT-LAMP method to detect CVX in pitaya in the field and compared its efficiency and sensitivity with an RT-PCR method. The RT-LAMP method described here may serve as a basis for future disease diagnosis and seedling or propagule quarantine.
Material and Methods
Samples and reagents
Pitaya branches suspected to be infected with CVX were collected and provided by the Changjiang Nafeng Jinsheng Green Fruits and Vegetables Cooperative (Hainan, China). Positive controls were provided by the Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences. An E.Z.N.A. RNA Isolation Kit and DNA markers were purchased from Omega Bio-Tek (USA). Bst DNA polymerase, betaine solution, dNTPs, fluorescent dyes and DL2000 DNA marker were purchased from the Guangzhou Gene Deaou Company (China). PrimeScript RT-PCR kit and RNase Inhibitor were purchased from Takara Bio (Japan). Papaya ring spot virus (PRSV), Rice ragged stunt virus (RRSV), Sorghum mosaic virus (SrMV) and Southern rice black-streaked dwarf virus (SRBSDV) were maintained on plants in the greenhouse at Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Haikou, Hainan, China
Primers
According to the sequence of CVX from GenBank (accession no. NC_002815), we performed homology analysis by DNAMAN 7.0 (Lynnon Biosoft) to identify unique sequences for primer design using Primer3 Input [12] for LAMP and PCR reactions (Table 1). All the primers were synthesized by Shanghai Lifei Biotechnology (China).
Primers
Sequences (5′-3′)
CVX-F3
TGCGACTCACTGGAGAGG
CVX-B3
ACCGGTTTGGCTTTGAGG
CVX-FIP
GCTGAGCAGTGCCGATTGGTATCACTTTCGACGCCAACACT
CVX-BIP
CGGGTGATGACTGCGCCTTTGAGACCTCGGTCTCGATCA
CVX-FLP
GCGAAGGCGATGTTGCATTC
CVX-BLP
GACTACGCCCCAGAGGATAA
CVX-D-F
GCAGACCATCGCCTCTTTCCAACAA
CVX-D-R
AAGGGGGGTAACTAACATCCCACAA
Table 1: Primers used for CVX detection by LAMP and PCR.
Total RNA extraction, RT-LAMP and RT-PCR
Total RNA was extracted from 200 mg of fresh healthy and symptomatic pitaya branches using the E.Z.N.A. RNA Isolation Kit according to the manufacturer’s instructions and was stored at -80°C for later use. LAMP was performed in a 25 μL system including 0.2 μM of external primers CVX-F3 and CVX-B3, 1.6 μM of internal primers CVX-FIP and CVX-BIP, 0.8 μM of loop primers CVX-FLP and CVX-BLP, 1.6 mM dNTPs, 1 M betaine, 6 mM MgSO4, 2.5 μL 10X ThermoPol II (Mg-free) Reaction Buffer (20 mM Tris–HCl, 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Triton X-100), 8 U Bst DNA polymerase (New England Biolabs, USA), 5 U AMV reverse transcriptase XL (Takara, Japan), 0.5 μL total RNA, and added nuclease-free water to a final volume of 25 μL. The mixture was covered with 20 μL mineral oil and was loaded on a 308C thermostat fluorescence detection system (Deaou Biological Technology, China). The reaction was performed at a single temperature 50-70°C for 60 min, and then heated at 80°C for 10 min to end the reaction. The real-time amplification curve was checked to determine if there was CVX in the plant sample. The products were inspected on a 1.5% agarose gel after electrophoresis at 120 V for 15 min.
RT-PCR was performed in a 20 μL system containing 1.25 μM of random primers, 1.25 μM of oligo (dT)18, 0.25 mM of dNTPs, 1 μL total RNA (50 ng) and added nuclease-free water to a final volume of 15 μL. First, the mixture was incubated at 65°C for 5 min and chilled on ice for 2 min. Next, 0.5 μL MLV reverse transcriptase (100 U), 0.5 μL RNase Inhibitor (20 U) and 4 μL of 5X M-MLV buffer were added. Then, the mixture was incubated at 42°C for 1h followed by inactivation of the reverse transcriptase at 75°C for 15 min and stored at -80°C for later use. PCR was performed in a 25 μL reaction system including 1 μL cDNA template, 0.5 μL Pfu Taq DNA polymerase (2.5 units/μL), 0.4 μM CVX-D-F primer, 0.4 μM CVX-D-R primer, 1 μL dNTPs, and added ddH2O to a final volume of 25 μL. The thermocycler program was performed as follows: pre-heated at 94°C for 3 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min, with a final extension at 72°C for 10 min. Eight microliters of the PCR product was mixed with 1 μL Gold View dye for electrophoresis on a 1.5% agarose gel at 120 V for 15 min to visualize PCR products.
Optimization of LAMP
To optimize the LAMP reaction, we selected eight different reaction temperatures (50°C, 60°C, 61°C, 62°C, 63°C, 64°C, 65°C and 75°C), four reaction times (30 min, 45 min, 60 min and 75 min) and four different concentrations of primers (0.4 μM, 0.8 μM, 1.2 μM and 1.6 μM for internal primers; 0.1 μM, 0.15 μM, 0.2 μM and 0.25 μM for external primers; 0.1 μM, 0.2 μM 0.4 μM and 0.8 μM for loop primers) to compare the effects of different reaction conditions on LAMP.
Comparison of the specificities and sensitivities of LAMP and RT-PCR
The cDNA from CVX and four other plant viruses including PRSV, RRSV, SrMV and SRBSDV were used to determine the specificity of LAMP. The cDNA from pitaya branches infected with CVX was 10-fold serially diluted in DEPC water (10-1, 10-2, 10-3, 10-4, 10-5, 10-6, 10-7 and 10-8) to compare the sensitivities of LAMP and RTPCR for virus detection.
Results
Infections in pitaya showed diverse symptoms ranging from none to distorted areoles, deformed spines, chlorosis, mottling and necrosis (Figure 1).

Figure 1: Typical symptoms on pitaya infected with CVX.
Optimization of the LAMP reaction to detect CVX
The reaction temperature, time and suitable concentrations of primers were optimized using total RNA extracted from healthy plants and those suspected to be CVX-infected. Successful LAMP reactions produced waterfall-like bands of different sizes by gel electrophoresis. The results showed that no bands could be produced at 50°C and 75°C, and successful LAMP reactions occurred from 60°C to 65°C. LAMP at 62°C and 63°C could both produce very clear and stepwise bands and a typical sigmoidal amplification curve (Figure 2).
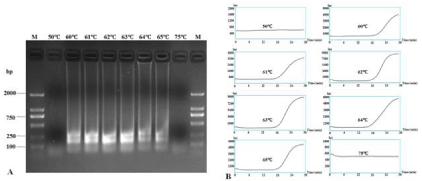
Figure 2: RT-LAMP detection of CVX under different reaction temperatures. RT-LAMP amplification products were subjected to 1.5% agarose gel electrophoresis
(A) and their amplification curves were monitored under different temperatures (B). Lane M: DL2000 DNA Marker; other lanes were LAMP products from different
reaction temperatures: 50°C, 60°C, 61°C, 62°C, 63°C, 64°C, 65°C and 75°C.
Different reaction times from 30 to 75 min were also assayed to check the speed of RT-LAMP. Results showed that 30 min was enough to obtain clear bands and the typical sigmoidal curve even though other times (45, 60 and 75 min) also produced very clear bands (Figure 3). Thus, 30 min is adequate for quick diagnosis of CVX, compared to the 70+ min required for the PCR reaction.
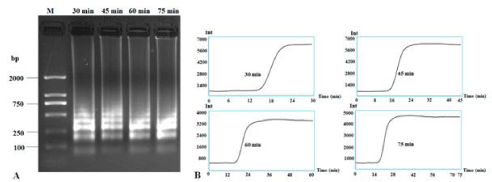
Figure 3: RT-LAMP detection of CXV under different reaction times. RT-LAMP amplification bands were subjected to 1.5% agarose gel electrophoresis (A) and
the application curves under different reaction times (B). Lane M: DNA Marker DL2000; other lanes were LAMP products from different reaction times: 30 min, 45
min, 60 min and 75 min.
Different concentrations of primers were also assayed at 62°C for 30 mins: 0.4 μM, 0.8 μM, 1.2 μM and 1.6 μM for internal primers; 0.1 μM, 0.15 μM, 0.2 μM and 0.25 μM for external primers; 0.1 μM, 0.2 μM 0.4 μM and 0.8 μM for loop primers. Results showed that the suitable concentrations for internal primers, external primers and loop primers were 1.2-1.6 μM, 0.2-0.25 μM and 0.4-0.8 μM, respectively, to produce both clear band ladders and typical sigmoidal curves (Figure 4). For field diagnosis, we chose 1.6 μM of internal primers, 0.25 μM of external primers and 0.8 μM of loop primers to detect CVX infection.
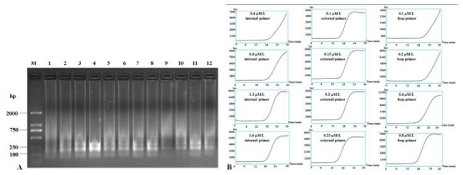
Figure 4: RT-LAMP detection of CXV by different concentrations of primers. RT-LAMP amplification products were subjected to 1.5% agarose gel electrophoresis
(A) and the application curves with different concentrations of primers were monitored (B). Lane M: DNA Marker DL2000; other lanes were LAMP products from
different concentrations of primers, lanes 1-4: 0.4 μM, 0.8 μM, 1.2 μM and 1.6 μM of internal primers; lanes 5-8: 0.1 μM, 0.15 μM, 0.2 μM and 0.25 μM of external
primers; lanes 9-12: 0.1 μM, 0.2 μM, 0.4 μM and 0.8 μM of loop primers.
Specificity of RT-LAMP to detect CVX
Total RNA extracted from fresh plant tissues respectively infected with CVX, PRSV, RRSV, SrMV and SRBSDV to evaluate the specificity of RT-LAMP CVX detection with our primers. Agarose gel electrophoresis and RT-LAMP results indicated that only CVXinfected tissues produced ladder-like bands and an amplification curve. These were not detected with the other four plant viruses (Figure 5).
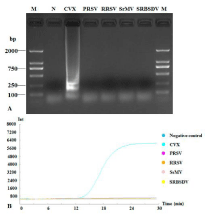
Figure 5: Specificity of RT-LAMP to detect CVX. RT-LAMP amplification
products were subjected to 1.5% agarose gel electrophoresis (A) and the
amplification curves were monitored (B). Lane M: DNA Marker DL2000; lane
N: negative control; other lanes were LAMP products with different plant
viruses: Cactus virus X (CVX), Papaya ringspot virus (PRSV), Rice ragged
stunt virus (RRSV), Sorghum mosaic virus (SrMV) and Southern rice blackstreaked
dwarf virus (SRBSDV).
Comparison of sensitivity of RT-PCR and RT-LAMP for CVX
To compare the sensitivities of RT-PCR and RT-LAMP, the cDNA was serially 10-fold diluted (50 ng to 5.0 x 10-8 ng). RT-PCR could detect 10-1, 10-2, 10-3 and 10-4 (very weak band, almost invisible) diluted cDNA (Figure 6A) while RT-LAMP could clearly detect 10-1 to 10-5 diluted cDNA (Figures 6B and 6C). Therefore, RT-LAMP was ten to one hundred times more sensitive than RT-PCR.
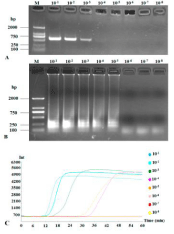
Figure 6: Sensitivities of RT-PCR and RT-LAMP to detect CVX. RT-PCR (A)
and RT-LAMP (B) amplification bands were subjected to 1.5% agarose gel
electrophoresis and RT-LAMP application curves in a 10-fold serial dilution
(B). Lane M: DNA Marker DL2000; other lanes are serial dilution: 10-1, 10-2,
10-3, 10-4, 10-5, 10-6, 10-7 and 10-8.
Field diagnosis of CVX by RT-PCR and RT-LAMP
In order to test the possibility of LAMP technology in practical applications, we collected 30 pitaya samples suspected to be infected with CVX from the main pitaya-growing areas in Hainan, China. Altogether, 26 out of 30 samples (86.7%) were diagnosed as being positively infected with CVX (Figure 7). The data from RT-LAMP exactly matched RT-PCR test results.
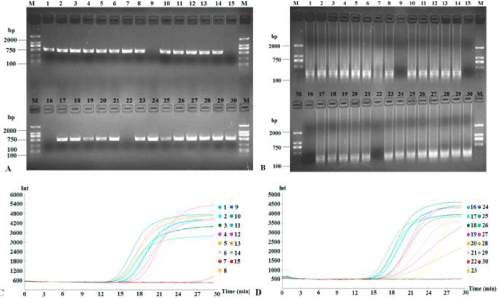
Figure 7: Detection of suspected CVX from thirty field samples. Products of field diagnosis of CVX by RT-PCR and RT-LAMP were subjected to 1.5% agarose gel
electrophoresis (A and B, respectively) and the application curves were monitored (C and D, respectively).
Discussion and Conclusion
There are many methods to detect plant virus, such as indicator plants, electron microscopy, ELISA and RT-PCR. However, indicator plants are time-consuming, electron microscopy requires a high concentration of virus, and ELISA is prone to producing false positive results. RT-PCR is generally sensitive and reliable, but it requires more time and is very expensive. Compared to these methods, RT-LAMP technology is very fast, simple and sensitive, and it can be conveniently and widely used in field diagnosis of plant viruses. Unlike RT-PCR that requires a longer time (~5 hours) for cycling amplification reactions, RT-LAMP does not need as many special reagents and can quickly detect plant viruses in a much shorter time (~2 hours) at a constant temperature (usually 60-65°C) and lower cost [13,14]. Thus, RT-LAMP saves a considerable amount of time and money over RT-PCR and qRT-PCR [11,15,16].
Previous studies showed that the sensitivity of LAMP was at least 10 to 1,000 times higher than RT-PCR: 1,000 timer higher for Bean pod mottle virus [17], 100 times higher for Grapevine rupestris stem pitting-associated [18], 10 times higher for Arabis mosaic virus [14]. In our study, the sensitivity of RT-LAMP technology was at least 10- 100 times higher than RT-PCR. Moreover, RT-LAMP also indicated high specificity in detection of plant viruses. Conventionally, gel electrophoresis and ultraviolet imaging are used to check the amplification products of LAMP. However, addition of ~0.1 μL SYBR Green I fluorescent dye allows formation of the products to be easily observed with the naked eye [19]. Furthermore, as a thermocycler is not needed, the LAMP method can be applied in the field with a simple heating apparatus [20].
In this study, we established an effective RT-LAMP detection approach to quickly detect CVX in the field. The method is very simple, fast, and specific, and is more sensitive than RT-PCR. It can be widely used in breeding, and field diagnosis and inspection. RTLAMP is quickly becoming an economical and convenient tool for pathogen detection by technical personnel and research institutes.
Acknowledgment
This work was funded by Central Public Research Institutes Fundamental Research Projects China (No. ITBB2015ZD09) and the Major Science and Technology Program of Hainan Province ZDZX2013023-1). The funders had no role in study design, data collection and analysis; decision to publish; or preparation of the manuscript.
References
- Liou MR, Hung CL, Liou RF. First Report of Cactus virus X on Hylocereus undatus (Cactaceae) in Taiwan. Plant Dis. 2001; 85: 229.
- Liou MR, Chen YR, Liou RF. Complete nucleotide sequence and genome organization of a Cactus virus X strain from Hylocereus undatus (Cactaceae). Arch Virol. 2004; 149: 1037-1043.
- Lobo R, Fernandez de Soto J, Aguiar JL, Mathews DM, Tanizaki G. Cactus virus X (CVX) a New Threat to Pitahaya/Dragon fruit (Hylocereus spp.) Production in California. 2014 ASHS Annual Conference; Orlando, FL. 2014.
- López MM, Bertolini E, Olmos A, Caruso P, Gorris MT, Llop P, et al. Innovative tools for detection of plant pathogenic viruses and bacteria. Int Microbiol. 2003; 6: 233-243.
- Kogovšek P, Hodgetts J, Hall J, Prezelj N, Nikolić P, Mehle N, et al. LAMP assay and rapid sample preparation method for on-site detection of flavescence dorée phytoplasma in grapevine. Plant Pathol. 2015; 64: 286-296.
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000; 28: E63.
- Kil EJ, Kim S, Lee YJ, Kang EH, Lee M, Cho SH, et al. Advanced Loop-Mediated Isothermal Amplification Method for Sensitive and Specific Detection of Tomato chlorosis virus Using a Uracil DNA Glycosylase to Control Carry-Over Contamination. J Virol Methods. 2015; 213: 68-74.
- Aydin-Schmidt B, Xu W, González IJ, Polley SD, Bell D, Shakely D, et al. Loop Mediated Isothermal Amplification (LAMP) Accurately Detects Malaria DNA from Filter Paper Blood Samples of Low Density Parasitaemias. PLOS ON. 2014; 9: e103905.
- Almasi MA, Erfan Manesh M, Jafary H, Dehabadi SM. Visual detection of Potato Leafroll virus by loop-mediated isothermal amplification of DNA with the GeneFinderâ„¢ dye. J Virol Methods. 2013; 192: 51-54.
- Kimura H, Ihira M, Enomoto Y, Kawada J, Ito Y, Morishima T, et al. Rapid detection of herpes simplex virus DNA in cerebrospinal fluid: comparison between loop-mediated isothermal amplification and real-time PCR. Med Microbiol Immunol. 2005; 194: 181-185.
- Shen W, Tuo D, Yan P, Li X, Zhou P. Detection of Papaya leaf distortion mosaic virus by Reverse-Transcription Loop-Mediated Isothermal Amplification. J Virol Methods. 2014; 195: 174-179.
- Rajkhowa S. Development of a novel multiplex PCR assay for rapid detection of virulence associated genes of Pasteurella multocida from pigs. Lett Appl Microbiol. 2015; 61: 293-298.
- Liu J, Huang CL, Wu ZY, Zhang XH, Wang YQ. Detection of Tomato aspermy virus Infecting Chrysanthemums by LAMP. Scientia Agricultura Sinica. 2010; 43: 1288-1294.
- Chen XF, Zhang JH, Cui JX, Zhang HL, Guo LX, Yu SQ. Detection of Arabis mosaic virus by RT-PCR. Acta Phytophylacica Sinica. 2013; 40: 189-190.
- Shen W, Tuo D, Yan P, Yang Y, Li X, Zhou P. Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Rapid Detection of Papaya ringspot virus. J Virol Methods. 2014; 204: 93-100.
- Peng J, Zhang J, Xia Z, Li Y, Huang J, Fan Z. Rapid and sensitive detection of Banana bunchy top virus by loop-mediated isothermal amplification. J Virol Methods. 2012; 185: 254-258.
- Wen W, Yang C, Cui J, Zhang Y. Detection of Bean pod mottle virus by RT-PCR. Plant Protection. 2010; 36: 139-141.
- Fan XD, Dong YF, Zhang ZP, Ren F, Hu GJ, Zhu HJ. RT-LAMP Assay for Detection of Grapevine rupestris stem pitting-associated virus. Acta Phytopathologica Sinica. 2013; 43: 286-293.
- Wang Y, Zhou Y, Li Z, Su H, Huang A, Tang K, et al. A RT-LAMP Assay for Detection of Citrus tristeza virus. Scientia Agricultura Sinica. 2013; 46: 517-524.
- Lu C, Dai T, Zhang H, Wang Y, Zheng X. Development of a Loop-Mediated Isothermal Amplification Assay to detect Fusarium oxysporum. J Phytopathol. 2015; 163: 63-66.