
Review Article
Austin J Biotechnol Bioeng. 2016; 3(3): 1067.
Saltol QTL and Their Role in Salinity Tolerance in Rice
Waziri A, Kumar P and Purty RS*
University School of Biotechnology, Guru Gobind Singh Indraprastha University, India
*Corresponding author: Purty RS, University School of Biotechnology, Guru Gobind Singh Indraprastha University, Sector 16C, Dwarka, New Delhi 110078, India
Received: March 18, 2016; Accepted: July 18, 2016; Published: July 21, 2016
Abstract
Salinity is one of the major abiotic stresses that reduce the yield of several crop species including rice. However, there are several traditional cultivars such as Pokkali, Nona Bokra, Cheriveruppu and Getu that are saline tolerant but possess poor agronomic characteristics. Pokkali is widely used as a donor in many breeding program and salt tolerance related studies. A major Quantitative Trait Loci (QTL), Saltol has been mapped on chromosome 1 in one of the Recombinant Inbred Lines(RILs) i.e., FL478, obtained from a cross between Pokkali and IR29, was responsible for maintaining low Na+, high K+, and Na+/ K+ homeostasis in shoots of rice. Since salinity stress is a multigenic trait and involves activity of many genes working in co-ordination. In order to provide salinity tolerance to some elite rice varieties plant breeders have transferred whole Saltol QTL using Marker Assisted Backcrossing and Marker Assisted Selection, whereas molecular biologists have attempted to find candidate genes within Saltol QTL that play crucial role in providing salinity tolerance. This paper provides a comprehensive review of Saltol QTL and their role in providing tolerance to salinity stress.
Keywords: Saltol; Rice; FL478; Pokkali; Salinity; Marker Assisted Selection; QTL
Introduction
Rice (Oryza sativa L.) is a one of the most important cereal crops and serves as the staple food for over one-third of the world’s population [1]. However the productivity of rice is greatly affected due to soil salinity which is the second most widespread soil problem next to drought in rice growing areas of the world [2,3]. There are two types of salinity, inland salinity which is due to irrigation practices with sloppy water and coastal salinity which is mainly due to high tides of ocean in the coastal region [4]. Approximately 21.5 million hectares of arable land in Asia are facing salinity problem and estimated to loss crop up to 50% of fertile land by the 21st midcentury [5,6]. It is important to increase rice production by at least 25% by 2030 to keep pace with predicted population growth [7].
Salinity affects rice growth in all growth stages with varying degrees starting from germination through maturation [8]. Rice is very sensitive during early seedling and later at reproductive stages, however, it is comparatively tolerant during germination, active tillering and at maturity [9]. Salinity affects yield components such as panicle length, spikelet number per panicle, grain yield and also delays panicle emergence and flowering [10-12]. There are some traditional cultivars and landraces which are naturally tolerant to salt stress due to their adaptation to thrive on salt affected land for generations. However, they generally have poor agronomic characteristics such as tall plant stature, poor grain quality, low yield, and photosensitivity [13,14]. One of the traditional cultivar, Pokkali has been recognized for providing more salt tolerance when compared to other tolerant cultivar and thus used as high potential salt tolerant donor. Although salinity tolerance is a polygenic trait, consequently it involves many physiological activities.
Recently, in order to understand the molecular mechanism of salinity tolerance in Pokkali remarkable effort has been made for the identification of candidate genes localized within the Saltol QTL by genome wide transcriptome analysis between contrasting rice genotypes [18,19]. On the other hand, for plant breeder’s identification of this QTL, has become a major breakthrough for salinity tolerance breeding program. Several salt tolerant rice lines has been developed by incorporating Saltol QTL into modern high yielding, but salt-sensitive rice varieties through a targeted marker assisted backcrossing and marker assisted selection approach [6,20].
In this review, an attempt has been made to revisit the current knowledge about Saltol QTL, their origin, structure, candidate genes of Saltol QTL involved in salinity tolerance and their expression. Since the capacity to perceive and respond to salt stress has long been described as a quantitative genetic trait, present review also cover the current knowledge on molecular breeding approaches in which whole Saltol QTL has been incorporated for the development of salt tolerant rice lines using Marker Assisted Selection. Ultimately, this paper will explain the most recent data available for improvement of salinity tolerance in rice to the researchers, physiologists and breeders.
Saltol QTL: Origin and Structure
Mapping of QTL has enabled us to understand the genetic control of the salt tolerance mechanism with possibilities to develop salt tolerant varieties by precisely transferring QTL into popular elite varieties. In earlier studies several QTL for salinity tolerance has been mapped on different chromosomes in rice [21-27]. One of them, i.e., Saltol has been mapped on chromosome 1 in an F8 Recombinant Inbred Line (RIL) population obtained by a cross between Pokkali (salt tolerant) and IR29 (salt sensitive) at the International Rice Research Institute (IRRI) in their salt stress tolerance breeding program. A total of 78 putative Recombinant Lines (RILs) were developed and used to map Na+/K+ selectively with AFLP markers. One of the lines identified from the RIL population, FL478 also known as IR 66946- 3R-178-1-1, showed salt tolerance higher than or comparable to the tolerant parent, Pokkali. The Saltol QTL was found to be associated with Na+/K+ ratio and seedling stage salinity tolerance [16,25] and accounted for low Na+ absorption, high K+ absorption, and low Na+-to-K+ ratio in rice shoots under salinity stress [28]. In Pokkali, it has been found that Saltol QTL explains about 64.3-80.2 % of the variability in shoot Na+/K+ ratio [15-17]. Its location was confirmed on chromosome1after the analysis that has been done for 100 Simple Sequence Repeat (SSR) markers on 140 IR29/Pokkali Recombinant Inbred Lines (RILs), and also additional QTLs were identified associated with salt tolerance [15].
In a study Saltol QTL has been mapped between RM23 and RM140 (10.7–12.2 Mb), and the effect of the shoot Na+/K+ ratio with an LOD of 6.6 and R2 of 43% using 54 RILs was also confirmed [25]. Recently, the expression profiling of genes localized within the Saltol QTL i.e., between SSR marker RM 1287 and RM 6711(10.8 MB to 16.4Mb) (Figure 1A and 1B) has been carried out in the two contrasting genotypes [19]. The source of the Saltol region in FL478 is still in question. It is not yet clear whether the positive allele in FL478 is derived from Pokkali, IR29 or both. One study says that Pokkali is the source of positive alleles in FL478 [25], while another study indicates that IR29 was the contributor of the Saltol region in FL478 [29]. Moreover, another study says that FL478 contained a <1 Mb DNA fragment from Pokkali at 10.6–11.5 Mb on chromosome 1, flanked by IR29 alleles [30].
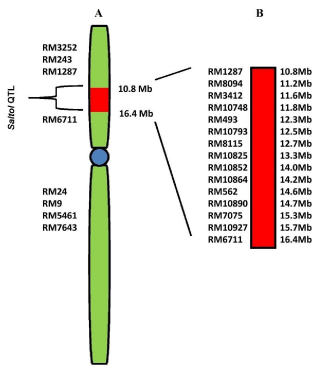
Figure 1: A) Location of Saltol QTL (Red square) on short arm of chromosome
1 flanked by SSR markers RM1287 and RM6711. Centromere is shown in
blue. B) Enlarged view of Saltol region, 15 SSR markers are shown with the
physical map position in megabases [19].
Annotation of Genes of Saltol QTL Loci
For the identification of loci within the Saltol QTL annotation has been done using Rice Genome Annotation Project - MSU Rice Genome Annotation (Osa1) Release 7. It was found that approximately 783 loci are present within this QTL. Out of which, 25% genes codes for proteins of unknown function (PUFs), while 31 % genes encodes for retro-transposons and transposons. A total of 14 different categories were made for the retrieved genes using the MIPS Functional Catalogue Database (FunCatDB). These categories included genes involved in diverse cellular activities such as metabolism, development and DNA processing etc (Table 1). It indicates that the genes present within Saltol QTL control versatile aspects of cell survival both under salt stress and non-stress conditions [19].
S.No.
Functional Categories
Percentage
1
Retro-transposons and transposons
31%
2
PUF
25%
3
Development
12%
4
Metabolism
7%
5
Cellular transport related genes
5%
6
Cell rescue, defense and virulence related genes
4%
7
Protein with binding function
3%
8
Cell cycle and DNA processing
2%
9
Protein fate
2%
10
Signaling
2%
11
Interaction with environment
2%
12
Transcription factor
2%
13
Protein synthesis
1%
14
Energy
1%
Table 1: Functional classification of annotated gene within Saltol QTL [19].
Gene Expression Studies within Saltol QTL
Saltol QTL is associated with providing seedling stage salinity tolerance. In order to identify the contribution of salinity tolerance related candidate genes localized within the Saltol QTL, comparative transcriptome study has been done. For that differential regulation of salinity-responsive genes among tolerant and sensitive rice cultivar has been studied in time course experiments [18,19,29].
In a genome scale gene expression analysis between IR29 and FL478 it was found that the two genotypes are strikingly different at their transcriptional perspective, under salinity stress. IR29 showed expression of relatively large number of genes as compared to the tolerant FL478, including the genes of Saltol QTL region. The transcript level of gene present at locus Os1g20160 (11.46Mb), codes for cation transporter was induced by salt stress in both the genotype but its expression was at found to be higher in FL478. Salt stress responsive gene salT at locus Os01g25280 was induced in IR29 but not in FL478 [29].
In a similar study, IR64 (salt sensitive) and Pokkali (salt tolerant) were used for gene expression analysis using subtractive cDNA library [19]. A few genes within Saltol QTL which were differentially regulated in these contrasting genotypes were mapped. Expression profile of few genes was compared in a time course analysis study. Salt stress-induced protein (EF576533) present at a 13.9-Mbp region and tetracopeptide repeat domain containing protein (EF575991) at 14.48-Mbp region showed almost negligible transcript level in IR64, however in Pokkali it consistently maintained a moderate amount under unstressed condition. Magnesium protoporphyrin IX monomethyl esterase (EF576502) mapped at a 9.87-Mbp and deleted in split hand/split foot (EF576234) protein at 11.95-Mbp, showed the presence of high constitutive levels under unstressed conditions in sensitive cultivar IR64 with expression level falling gradually, while Pokkali tends to show an increase in transcript level until 24 h of salt stress, thereafter reduced sharply. However, another geneSINE element (EF575921) mapped at 10.49-Mbp region maintained higher transcript in IR64 as compared to Pokkali with little variation in transcript amount [18].
In another study, to find the candidate genes localized within Saltol QTL expression profiling has been done, for a set of genes coding for signaling related proteins(SRPs) in contrasting rice genotypes (IR64 and Pokkali) [19]. By the qRT-PCR analysis it was found that genes encoding Signaling Related Protein (SRPs) are differentially regulated. Genes encoding Proteins of Unknown Function (PUFs) also termed SIFs (Salinity Induced Factors) within this QTL were also studied for their role in providing salinity tolerance. It was proposed that these SIFs may have a putative function in Vegetative Growth (SIFVG), Fertility (SIFF), Viability (SIFV) or Early Flowering (SIFEF) based on the phenotypes of insertional mutants [19].
In addition, a set of shortlisted genes has been reported to be present within Saltol QTL (CaMBP, GST, LEA, V-ATPase, OSAP1 zinc finger protein and transcription factor HBP1b) are salinity inducible and differentially regulated within the contrasting rice genotypes [18,31]. Among the above set of reported genes, transcription factor HBP1b (histone gene binding protein-1b) belongs to bZIP family was found to be salinity inducible in rice seedlings [18]. Time course expression analysis of OsHBP1b in response to salt stress has been studied between contrasting rice genotype and found that it shows differential expression [31]. It is well known that bZIP transcription factor plays an important role in ABA signaling pathway of abiotic stress in plants [32,33].
Breeding Program and Saltol QTL
Several breeding approaches have been used for the improvement of salinity tolerance in different elite cultivars of rice such as conventional breeding, Marker Assisted Selection and Marker assisted Backcrossing.
Marker Assisted Backcrossing (MABC) for Salinity Tolerance in Rice
Backcrossing is a widely used technique in rice breeding for introgression or substitution of a target gene/QTL from donor parent to recipient. Generally, the backcrossing approach has been established by applying molecular markers such as Simple Sequence Repeats (SSRs) and Single Nucleotide Polymorphisms (SNP). Marker- Assisted Backcrossing (MABC) is an attractive tool for breeding and identifying genomic regions of interest. Molecular markers that are tightly linked with economically important traits have been identified and/or used for MABC in rice as foreground selection, recombinant selection and background selection, for tolerance to salinity and other abiotic and biotic stresses [34].
There are several successful examples where Saltol QTL has been precisely introgressed into elite rice cultivars through marker assisted breeding strategy to provide salt tolerance. FL478 has been promoted as an improved donor of Saltol QTL, as it has a high level of seedling stage salinity tolerance and is photoperiod insensitive, shorter and flowers earlier than the original landrace Pokkali (Table 2).
S.No.
Donor of Saltol QTL
QTL transferred
Method of transfer
Marker used
Recipient variety
Reference
1
FL478
Saltol QTL
MABC
SSR
PB1121and PB6
[35]
2
FL478
Saltol QTL
MABC
SSR
AS996
[36]
3
FL478
Saltol QTL
MABC
SSR
BT7
[37]
4
FL478
Saltol QTL
MABC
SSR
Bacthom 7
[38]
5
FL478
Saltol QTL
MABC
SSR
Q5DB
[6]
6
FL478
Saltol QTL
MABC
SSR
BRRI-dhan 49
[39]
7
Line IR61920-3B-22-2-1
Saltol QTL
MAS
SSR
Novator
[40]
Table 2: Successful examples of introgression of Saltol QTL into elite rice varieties for salt tolerance.
To develop salt tolerance in indica aromatic export quality rice varieties PB1121 and PB6, Saltol QTL has been transferred using MABC in two independent programs. Three SSR markers RM8094, RM3412 and RM493 linked with Saltol locus were used for foreground selection [35]. At least four popular rice cultivars in Vietnam were used as recipient for the introgression of Saltol QTL from FL478 parent line through MABC approach i.e. cultivar AS996 [36], cultivar BT7 [37], cultivar Bacthom 7 [38] and cultivar Q5DB [19]. Rice cultivar BRRI Dhan-49 from Bangladesh was used as recipient for the introgression of Saltol QTL from FL478 parent line through MABC approach [39].
Marker Assisted Selection (MAS) for Salinity Tolerance in Rice
The highly productive elite variety from Russia named Novator was used as a recipient for introgression of Saltol QTL from donor line IR61920-3B-22-2-1 (NSIC Rc106) using Marker Assisted Selection (MAS) [40]. Identification of Saltol QTL was done by closely linked microsatellite markers RM8094 and RM493 and RM493, and were used to control transfer the Saltol QTL genes into Novator [40].
QTL Mapping for Salinity Tolerance
QTL mapping for salt tolerance in rice has significantly increased in the last few years .There are several reports in which QTL mapping has been done for salinity tolerance using different rice genotypes. Attempts have been made to identify QTL associated with seedling stage salinity tolerance in rice [41-44].
Apart from Saltol QTL, other QTLs have been identified which is responsible for providing seedling stage salinity tolerance. By crossing Milyang 23 (japonica/indica) and Gihobyeo (japonica) several recombinant inbred lines (MG RILs) were produced and used to evaluate their salt tolerance capability at the young seedling stage of rice at 0.7% NaCl concentration. Out of 164, 22 MG RILs showed visual scores ranging 3.5-5.0 and were classified as tolerant. Interval mapping of QTLs for salt tolerance at the young seedling stage were done by their visual scores. Two QTL, qST1 and qST3 conferring salt tolerance was detected on chromosome 1 and 3, respectively. The major QTLqST1 was flanked by RG179-RZ596 explained 27.8% of the total phenotypic variation [41]. In another study, 164 recombinant inbred lines (MG RILs) were subjected to two different concentration of NaCl i.e., 0.5% and 0.7% NaCl. Two QTLs qST1 and qST3, were mapped on chromosome 1 and 3, respectively conferring salt tolerance at the young seedling stage with phenotypic variation 35.5–36.9% in 0.5% and 0.7% NaCl. The positive allele of qST1 was contributed by Gihobyeo, and that of qST3 by Milyang 23. The results obtained in 0.5% and 0.7% NaCl for 2 years were similar in flanked markers and phenotypic variation [42].
Detection of QTLs for salinity tolerance at the seedling stage identified in a F2 breeding population derived from the cross between BRRI dhan40, a moderately tolerant female parent with IR61920- 3B-22-2-1 (NSIC Rc106), a highly tolerant male parent. The position of QTL on chromosome 1 was flanked by RM8094 and RM3412 marker which is in the same region as a previously identified major QTL designated as Saltol. However, two other QTLs with relatively large effects were flanked by RM25 and RM210 on chromosome 8, and RM25092 and RM25519 on chromosome 10, and appear to be novel QTLs. The markers flanking these QTLs should be useful for molecular marker assisted breeding for salinity tolerance [3]. In the related study, investigation for QTL analysis of physiological traits related to salt tolerance was carried out using F2population of rice derived from a cross between a salt-tolerant variety, Gharib (indica), and a salt-sensitive variety, Sepidroud (indica) [43].
A study employing a F5population of 300 recombinant inbred lines RILs, derived from a cross between IR29 indica (sensitive) and Hasawi Saudi cultivar (tolerant), evaluated at young seedling stage under hydroponic conditions at an EC of 12 dsm-1. From this study seven significant QTL related to 4 different traits associated with salt tolerance at the young seedling stage were mapped on chromosomes 1, 2 and 6. The QTL qDW1.1, qDW2.1, qDW2.2 and qDW6.1 cosegregated with shoot dry weight and accounted for 10.6 and 42.3% of its phenotypic variation; for plant height qPH1.1 and qPH1.2 were found to be associated, explaining its phenotypic variation between 12.7 and 13.8% and qF2.1 co-segregated with shoot fresh weight 10.6% of trait variation. These QTLs confirms that Hasawi alleles contributed to enhancing traits related to salt tolerance at young seedling stage [44].
Conclusion
Rice feeds more than half of the world population, but its sensitivity towards salinity results in significant yield loss. Rice is more sensitive during the early seedling and later at reproductive stage. However, there are several traditional varieties which are salt tolerant, but they have poor agronomic traits. Several works has been done so far for the improvement of salt tolerance in elite cultivars around the globe using Pokkali, a salinity tolerant traditional cultivar with considerable results. Also, few other genotypes have been recently screened that might have Saltol QTL or other salt tolerance related QTL sand can be used as an alternative donor in salt tolerant rice breeding program. This may be somewhat helpful to the breeders that offer a wider optionto combine superior QTLs into one genetic background using gene pyramiding technique. Working in this direction may certainly help to combat with salinity stress problem at a higher level. With various increased genomic resources, significant achievements have been made for the development of salt tolerant rice cultivars through different approaches such as conventional breeding and marker assisted breeding. However, further development is needed to fulfill the increasing demand of the staple crop, rice.
Acknowledgment
AW, PK and RSP wish to thank Guru Gobind Singh Indraprastha University, New Delhi, India for providing financial and infrastructural support.
References
- Mohammadi-Nejad G, Singh RK, Arzani A, RezaieAM, Sabouri H, Gregorio GB. Evaluation of salinity tolerance in rice genotypes. Int J Plant Prod. 2010; 4: 199-208.
- Sabouri H, Sabouri A. New evidence of QTLs attributed to salinity tolerance in rice. Afr J Biotechnol. 2008; 7: 4376-4383.
- Islam MR, Salam MA, Hassan L, Collard BCY, Singh RK, Gregorio GB. QTL mapping for salinity tolerance at seedling stage in rice. Emir J Food Agric. 2011; 23: 137-146.
- Ganie SA, Borgohain MJ, Kritika K, Talukdar A, Pani DR, Mondal TK. Assessment of genetic diversity of Saltol QTL among the rice (Oryza sativa L.) genotypes. Physiol Mol Biol Plants. 2016.
- Nazar R, Iqbal N, Masood A, Syeed S, Khan NA. Understanding the significance of sulfur in improving salinity tolerance in plants. Environ Exp Bot. 2011; 70: 80-87.
- Huyen LTN, Cuc LM, Ham LH, Khanh TD. Introgression the Saltol QTL into Q5BD, the elite variety of Vietnam using marker assisted selection (MAS). Am J Biosci. 2013; 1:80-84.
- Li JY, Wang J, Zeigler RS. The 3,000 rice genomes project: new opportunities and challenges for future rice research. Gigascience. 2014; 3: 8.
- Mass EV, Hoffman GJ. Crop salt tolerance-Current assessment. J Irrig Drainage Div ASCE. 1977.
- Lafitte HR, Ismail A, Bennett J. Abiotic stress tolerance in rice for Asia: progress and the future. Fischer T, Turner N, Angus J, McIntyre L, Robertson M, Borrell A, et al., editors. In: New directions for a diverse planet: Proceedings of the 4th International Crop Science Congress. Brisbane, Australia. 2004.
- Flowers TJ, Yeo AR. Variability in the resistance of sodium chloride salinity within rice (Oryza sativa L.) varieties. New Phytol. 1981; 88: 363-373.
- Lutts S, Kinet JM, Bouharmont J. Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J Exp Bot. 1995; 46: 1843-1852.
- Zeng L, Shannon MC. Salinity effects on the seedling growth and yield components of rice. Crop Sci. 2000; 40: 996-1003.
- Ismail AM, Heuer S, Thomson MJ, Wissuwa M. Genetic and genomic approaches to develop rice germplasm for problem soils. Plant Mol Biol. 2007; 65: 547-570.
- Reddy MA, Francies RM, Rasool SN, Reddy VRP. Breeding for tolerance to stress triggered by salinity in rice. Int J Appl Biol Pharm Tech. 2014; 5: 166-176.
- Thomson MJ, de Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, et al., Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice. 2010; 3: 148-160.
- Gregorio GB, Senadhira D, Mendoza RD. Screening rice for salinity tolerance. IRRI Discussion Paper Series Number 22. International Rice Research Institute, Manila, Philippines. 1997; 1-30.
- Chattopadhyay K, Nath D, Mohanta RL, Bhattacharyya S, Marndi BC, Nayak AK, et al. Diversity and validation of microsatellite markers in 'Saltol' QTL region in contrasting rice genotypes for salt tolerance at the early vegetative stage. Aust J Crop Sci. 2014; 8: 356-362.
- Kumari S, Sabharwal VP, Kushwaha HR, Sopory SK, Singla-Pareek SL, Pareek A. Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct Integr Genomics.2009; 9: 109-123.
- Soda N, Kushwaha HR, Soni P, Singla-Pareek SL, Pareek A. A suite of new genes defining salinity stress tolerance in seedlings of contrasting rice genotypes. Funct Integr Genomics. 2013; 13: 351-365.
- Ali S, Gautam RK, Mahajan R, Krishnamurthy SL, Sharma SK, Singh RK, et al. Stress indices and selectable traits in Saltol QTL introgressed rice genotypes for reproductive stage tolerance to sodicity and salinity stresses. Field Crops Res. 2013; 154: 65-73.
- Zhang GY, Guo Y, Chen SL, Chen SY. RFLP tagging of a salt tolerance gene in rice. Plant Sci. 1995; 110: 227-234.
- Lin HX, Yangihara S, Zhuang JY, Senboku T, Zheng KL, Yashima S, et al. Mapping of QTL for salt tolerance in rice (Oryza sativa L.) via molecular markers. Chin Rice Res News Lett. 1997; 5: 1-2.
- Gong JM, He P, Qian Q, Shen LS, Zhu LH, Chen SY. Identification of salt-tolerance QTL in rice (Oryza sativa L.). Chin Sci Bull. 1999; 44: 68-71.
- Prasad SR, Bagali PG, Shailaja H, Shashidhar HE, Hittalmani S. Molecular mapping of quantitative trait loci associated with seedling tolerance to salt stress in rice (Oryza sativa L.). Curr Sci. 2000; 78: 162-164.
- Bonilla P, Dvorak J, Mackill D, Deal K, Gregorio G. RFLP and SSL mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philippine J Agric Sci. 2002; 85: 68-76.
- Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, et al. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet. 2004; 108: 253-260.
- Ammar MHM, Pandit A, Singh RK, Sameena S, Chauhan MS, Singh AK, et al. Mapping of QTLs controlling Na+, K+ and Cl- ion concentrations in salt tolerant indica rice variety CSR27. J Plant Biochem Biotechnol. 2009; 18: 139-150.
- Gregorio GB, Senadhira D, Mendoza RD, Manigbas NL, Roxas JP, Guerta CQ. Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res. 2002; 76: 91-101.
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, et al. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005; 139: 822-835.
- Kim SH, Bhat PR, Cui X, Walia H, Xu J, Wanamaker S, et al. Detection and validation of single feature polymorphisms using RNA expression data from a rice genome array. BMC Plant Biol. 2009; 9: 65.
- Lakra N, Nutan KK, Das P, Anwar K, Singla-Pareek SL, Pareek A. A nuclear-localized histone-gene binding protein from rice (OsHBP1b) functions in salinity and drought stress tolerance by maintaining chlorophyll content and improving the antioxidant machinery. J Plant Physiol. 2015; 176: 36-46.
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009; 23: 1805-1817.
- Liu C, Mao B, Ou S, Wang W, Liu L, Wu Y, et al. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol. 2014; 84: 19-36.
- Hasan MM, Rafii MY, Ismail MR, Mahmood M, Rahim HA, Alam MA, et al. Marker-assisted backcrossing: a useful method for rice improvement. Biotechnol Biotechnol Equip. 2015; 29: 237-254.
- Singh AK, Gopalakrishnan S, Singh VP, Prabhu KV, Mohapatra T, Singh NK, et al. Marker assisted selection: a paradigm shift in Basmati breeding. Indian J Genet Plant Breed. 2011; 71: 120-128.
- Luu TNH, Luu MC, Abdelbagi MI, Le HH. Introgression the salinity tolerance QTLs Saltol into AS996, the elite rice variety of Vietnam. Am J Plant Sci. 2012; 3: 981-987.
- Linh le H, Linh TH, Xuan TD, Ham le H, Ismail AM, Khanh TD. Molecular Breeding to Improve Salt Tolerance of Rice (Oryza sativa L.) in the Red River Delta of Vietnam. Int J Plant Genomics. 2012; 2012: 949038.
- Vu HTT, Le DD, Ismail AM, Le HH. Marker-assisted backcrossing (MABC) for improved salinity tolerance in rice (Oryza sativa L.) to cope with climate change in Vietnam. Aust J Crop Sci. 2012; 6: 1649-1654.
- Hoque ABMZ, Haque MA, Sarker MRA, Rahman MA. Marker-assisted introgression of Saltol locus into genetic background of BRRI Dhan-49. Int J Biosci. 2015; 6: 71-80.
- Usatov AV, Alabushev AV, Kostylev PI, Azarin KV, Makarenko MS, Usatova OA. Introgression the Saltol QTL into the Elite Rice Variety of Russia by Marker-Assisted Selection. Am J Agric Biol Sci. 2015; 10: 165-169.
- Lee SY, Ahn JH, Cha YS, Yun DW, Lee MC, Ko JC, et al. Mapping of quantitative trait loci for salt tolerance at the seedling stage in rice. Mol Cells. 2006; 21: 192-196.
- Lee SY, Ahn JH, Cha YS, Yun DW, Lee MC, Ko JC, et al., Mapping QTLs related to salinity tolerance of rice at the young seedling stage. Plant Breed. 2007; 126: 43-46.
- Ghomi K, Rabiei B, Sabouri H, Sabouri A. Mapping QTLs for traits related to salinity tolerance at seedling stage of rice (Oryza sativa L.): an agrigenomics study of an Iranian rice population. OMICS. 2013; 17: 242-251.
- Bimpong IK, Manneh B, El-Namaky R, Diaw F, Amoah NKA, Sanneh B, et al., Mapping QTLs related to salt tolerance in rice at the young seedling stage using 384-plex single nucleotide polymorphism SNP, marker sets. Mol Plant Breed. 2014; 5: 47-63.