
Research Article
Austin J Cancer Clin Res 2015;2(7): 1059.
Residual Tumor Characteristics Following Neoadjuvant Chemotherapy in Breast Cancer
Mani CS¹*, Chander V², Kurian A³, Subramani A³, Mallikarjun VS4, Mahalingam S5 and Ramakrishnan V6
¹Cancer Research and Relief Trust, India
²Department of Pathology, Saveetha Medical College, India
³Department of Pathology, Apollo Specialty Hospitals, India
4Department of Pathology, SRM Medical College, India
5Department of Biotechnology, IIT-Madras, India
6Madras Medical College, Indi
*Corresponding author: Mani CS, Consultant Surgical Oncologist, Cancer Research and Relief Trust, 39, TTk 1st cross street, Alwarpet, Chennai 600018, India
Received: June 16, 2015; Accepted: August 09, 2015; Published: August 12, 2015
Abstract
Neoadjuvant chemotherapy for breast cancer leaves behind a residual tumor that is substantially different in its characteristics from the original tumor. The histological changes and receptor changes have been systematically evaluated in large studies. However, the nucleolar morphology of the residual cancer has not been well characterized. This study evaluated the nucleolar morphology in the residual cancer using haematoxylin eosin for evaluation of nucleolar number and size in relation to mitotic index, mAgNOR assessment, and an immunohistochemistry panel consisting of Ki-67, p53, CCND1, E-Cad, ER, PR, Her2Neu, CD3, CD8 and CD68. Another sample of patients were studied by exome analysis of a 344 candidate cancer gene panel. The results were collated to infer the nature of the residual tumor. There was an increase in nucleolar number and size following neoadjuvant chemotherapy. The mitotic rate, and positive staining for Ki-67 and CCND1 were reduced in the residual tumor. 344 candidate cancer gene panel exome analyses showed residual tumor with fewer mutations and the more common known driver mutation in breast cancer such as CREBP, ERBB2, PARP, TOP2A, P53 etc. COL1A1 was the only gene mutation demonstrated in the residual tumor that was not found in the untreated primary surgical group.
Keywords: Breast cancer; Nucleoli; mAgNOR; Exome analysis; Neoadjuvant chemotherapy; Residual tumor
Abbreviations
mAgNOR: Mean AgNOR; NAC: Neoadjuvant Chemotherapy; NSABP: B18 National Surgical Adjuvant Breast and Bowel Project; QC: Quality Control; ER: Estrogen Receptor; PR: Progesterone Receptor; FISH: Fluorescent in Situ Hybridization; RD: Residual Disease; IHC: Immunohistochemistry
Introduction
Response to neoadjuvant chemotherapy (NAC) in breast cancer has a wide spectrum. It ranges from complete pathological response to no response, or progression of disease. Chemotherapy results in histologic changes that have been well studied but its effect on nucleolar morphology has received little attention.
It appears that tumor cells remaining after NAC contain a cancer cell population that is intrinsically resistant to chemotherapy. They represent clones that are capable of further growth and cell division and ultimately result in distant metastases. They are unlikely to respond to further chemotherapy especially to the agents directed against the pathways that were previously targeted. It is postulated that these residual cells could be a resistant clone or acquire features of cancer stem cells.
Neoadjuvant chemotherapy is the standard of care in locally advanced breast cancer [1]. It is also being increasingly used in early stage breast cancers that have poor prognosis such as triple negative breast cancer and the Her2 Neu positive cancers [2,3]. Neo adjuvant chemotherapy offers the advantage of early institution of systemic treatment in order to sterilize potential micro metastases before local therapy such as surgery or radiation is performed. It provides an idea of in vivo chemo sensitivity of the tumor. It also improves the chances of breast conservation surgery in a substantial number of patients. The NSABP- B18 trial that addressed the issue of neoadjuvant treatment is the largest randomized controlled trial comparing neoadjuvant chemotherapy to adjuvant chemotherapy and its 9 years follow up study showed no difference in the overall and disease specific survival of patients between the 2 arms [4].
Neoadjuvant treatment is therefore started after obtaining a core needle biopsy that determines the histology, grade and the immunohistochemical markers predictive of treatment options such as hormonal receptors and Her2Neu expression. The tissue obtained in a core needle biopsy is limited but adequate to obtain the necessary information. Concerns about tumor heterogeneity and lack of adequate tissue to evaluate novel prognostic markers remain with such an approach. NAC alters the tumor in remarkable ways [5]. It fragments the tumor into islands of necrosis and fibrosis with residual viable tumor in between. These residual tumor cells may differ substantially from the original tumors in many respects. The histology changes to more of a lobular type, histiocytoid cellular changes are noted, and expression of hormonal receptors are altered significantly, while Her2Neu receptor expression remains constant. Molecular profiling and whole genome sequencing of the residual tumors have been studied to gain insight into chemo resistance and the outcome of these patients. But less is known on how the nucleolus responds to neoadjuvant treatment and contributes to the architectural and molecular changes of the residual tumor.
The purpose of the current study was to systematically evaluate the changes in nucleolar morphology due to neoadjuvant chemotherapy after NAC. We also wanted to understand whether nucleolar morphology and function in the residual cancer cells after chemotherapy contribute to either the resistance or stemness.
Methods
Consecutive breast cancer patients who consented to this study at the Cancer Research and Relief Trust between 2013 and 2014 were recruited to this study after obtaining approval from the ethics committee and institutional review board. Concept study of 11 patients with infiltrating ductal carcinoma who had received NAC with anthracycline or taxol containing regimens were taken to study the nucleolar morphology before and after chemotherapy on routine H&E staining (Sample A).
An additional cohort of 46 patients, 7of whom had been treated with NAC was taken up for mAgNOR scoring and detailed studies with an immunohistochemistry (IHC) panel containing CCND1, p53, Bcl2, Ki-67, E-Cad, ER, PR, Her2Neu CD3, CD8 and CD68 (Sample B).
The staining protocol consisted of numerous steps in which reagents were incubated for pre-determined times at specific temperatures. The paraffin embedded tissues that were used for the original hematoxylin and eosin stained sections were used for IHC. Sections were cut at 3 micron thickness. A standard IHC technique was performed using VENTANA BENCHMARK XT.
Tissues obtained from a third set of 11 patients with residual tumor who had received NAC were analyzed by exome sequencing with a 344 gene panel (Sample C). The exome analysis procedure consisted of DNA/RNA extraction, QC, DNA library preparation, enrichment capture of comprehensive candidate cancer genes, cluster amplification, and running of sample on Illumina HiSeq platform, generating quality report of raw data and advanced data analysis.
Genomic DNA was isolated to meet quality specifications. The genomic DNA was sheared and used to perform enrichment using the probes in the cancer gene panel. Cancer gene panel targets about 344 genes with an approximate target region of 7MB. Captured fragments were adapted to produce libraries that were sequenced on Illumina HiSeq 2000 to generate paired end 2 x 100bp sequence reads to produce 100 x coverage. The generated sequence data were analyzed after quality control for variant calling and annotation. Bioanalyzer plots used at Bioanalyzer plots were used at every step to assess library size and qPCR was used for measuring the quantity of the library before sequencing.
The results were computed and analyzed using SPSS2.
Results
The number of nucleoli per cell was counted in tissue obtained from patients in Sample A. The mean number of nucleoli per cell was distinctly higher in the post treatment residual viable cells withmacroenlargement of the nucleoli (Figures 1, 2, 3).
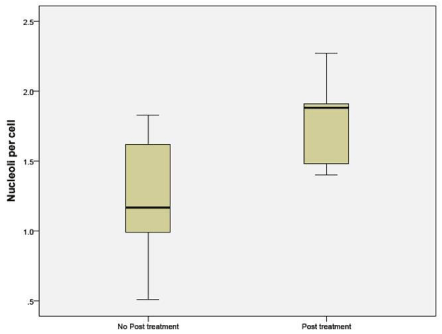
Figure 1: Number of nucleoli per cell in the untreated and NAC groups.
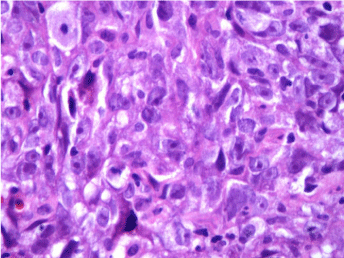
Figure 2: H & E staining of Grade 3 infiltrating duct carcinoma –Untreated.
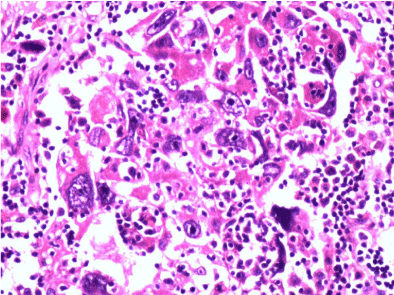
Figure 3: H & E staining of infiltrating duct carcinoma- NAC group.
mAgNOR was compared between tissue obtained from patients who had received NAC and untreated patients. The mAgNOR in untreated patients was some what higher in the NAC group, but did not reach statistical significance (p 0.33) and showed a trend towards macroenlargement (Figure 4).
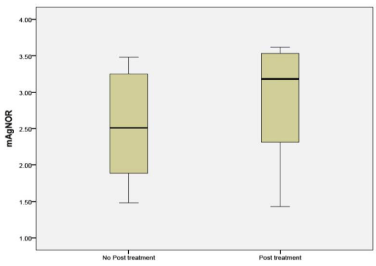
Figure 4: mAgNOR counts in untreated and NAC groups.
Mitosis per high power field in untreated patients was significantly higher than in the NAC group (p 0.042) (Figure 5).
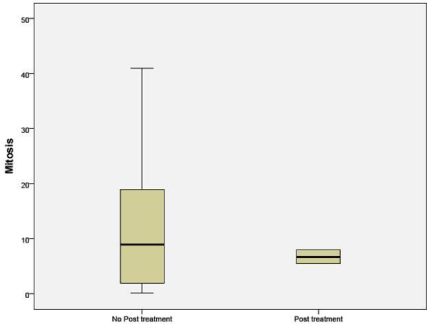
Figure 5: Number of mitosis per hpf in untreated and NAC groups.
Immunohistochemistry for Ki-67 was significantly different between untreated (51.36%) and NAC groups (29.62%) (p 0.05). The percentages indicate the number of cells that stained positive for Ki- 67 (Figure 6).
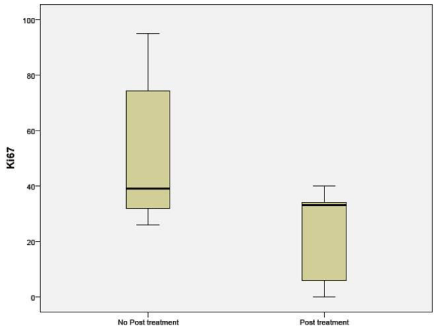
Figure 6: Percentage of cells expressing Ki-67 in untreated and NAC groups.
CCND1 when compared between the NAC (41%) and untreated groups (63%) showed a significant difference (p 0.036) (Figure 7).
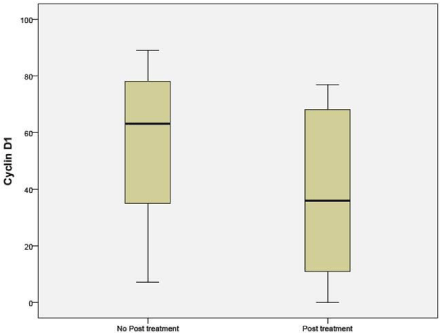
Figure 7: Percentage of cells expressing Cyclin D1 in untreated and NAC
groups.
In the NAC group, ER status in the exome sequencing (sample 3) group showed low positive (<30%) in 5 cases and high positivity in 5 cases.
There was no difference in the expression of CD3, CD8 and CD68 between the groups.
Whole genome sequencing for exome analysis of 344 genes wasperformed on a sample of tumors that had been exposed to NAC. The results are as follows:
Mutations were identified in 33/55 samples obtained from the untreated group and in 4/11 patients in the NAC group (Table 1) 29 of the 33 patients in the untreated group had two or more mutations (Table 2). From the exome analyses of the 344 cancer gene panel, COL1A1 was the unique gene mutated in the NAC group while the other genes CREBBP, ERBB2, FN1, GNAS, NSD1, PARP1, TCF3, TOP2A and TPR were found with equal frequency in both groups.
Mutation absent
Mutation present
Total
Primary surgery
22
33
55
Neoadjuvant treatment
7
4
11
Total
29
37
66
Table 1: Distribution of mutations in samples from the untreated and NAC groups.
0
1
2
3
4 or more
Primary surgery
22
4
6
4
19
Neoadjuvant treatment
7
0
2
1
1
Table 2: Number of mutations per patient in untreated and NAC groups.
Discussion
Thenucleolar size and number following neoadjuvant treatmentshoweda trend towards increased number and macroenlargement. In NAC (post treatment), comparable or enlarged nucleoli did not appear to be related to enhanced ribosome biogenesis alone, as theresidual tumors were quiescent with lesser number of mitosis and a lower Ki-67value. Based on these novel observations, the following interesting questions can be posed. Does chemoresistance require enlargement of nucleoli? Alternatively, does the enlarged nucleolus signify a non ribosomal role in the primary tumor that continues in the residual tumor as a cause of chemoresistance? Do they mark for a cancer stem cell nature? Could it be the acquired stemness that provides the mechanism for chemoresistance? [6].
Mitosis and Ki67 were decreased in this subset suggesting that they did not have high proliferative potential in spite of having slightly higher mAgNOR and an increased number and enlargement of nucleoli. Hence, it suggests if they are cells with stem cell like properties rather than resistant clones of chemotherapy [7,8].
Mitotic activity index as a surrogate marker for the ribosomal function of nucleoli was considered. It was observed while untreated cases had a wide range of mitotic figures; the residual tumor from the NAC group had only a narrow range and fewer of mitotic figures [9,10].
Nucleoli per cell counted by H&E stain in sample A, showed increased number of nucleoli in the post treatment residual tumor. It ranged from no change to doubling in number. The same findings were duplicated in the sample B (Figures 1, 2, 3). mAgNOR values were at 2.5 for those without treatment and at 3.2 in those with neoadjuvant treatment (Figure 4).
Number of mitosis in the residual tumor were significantly less than the primary surgical group (Figure 5). The ER and PR expression in the post treatment scenario changed but did not reach statistical significance. ER status may be altered following a course of NAC but is usually concordant with pre-treatment status. The literature contains only few studies of pre and post NAC comparison of biomarker status [11,12]. For ER and PR status, studies have reported changes in around 10% of cases but some studies document changes in up to a third of cases; most commonly of a change in the ER status from positive to negative. Her2 status appears to remain more stable throughout NAC. Concordance between pre and post NAC Her2 status ranges from 65% to 100%. FISH testing appears more stable than immunohistochemical testing for Her2, with concordance of 87% and higher having been reported.
Immunohistochemistry (IHC) of the proliferation marker Ki67 in the post-NAC residual disease (RD) was computed in this sample of patients. Ki67 levels being low post treatment in this study along with prominent or increased number of nucleoli per cell suggests that chemotherapy leaves behind the residual cells that have lower proliferative potential.
Petric M et al. showed Post treatment Ki67 correlate with patient outcome [13]. Maur M et al. showed A Ki-67 =15% and nodal positivity after treatment was predictive of inferior disease-free survival [14,15]. Other studies have yielded conflicting results. There is some literature regarding the prognostic ability of Ki67 after NAC (that included all subtypes of breast cancer i.e. HER2-enriched, luminal A, luminal B and basal-like), others have recently shown that these subtypes differ vastly in their post-NAC Ki67 scores, confounding the prognostic utility of Ki67 [16,17].
Furthermore, Ki67 scoring is difficult to standardize among clinical laboratories and many studies have defined different ‘cutoffs’ for patient stratification, ranging from 14-50% [18]. Finally, the Ki67 scoring of the post-NAC residual tumor does not help in clinical decision making as there is no identifiable pathogenic driver of the tumor at that stage and, as such, a tailoring of drug treatment decision cannot be rationally done.
Penault-Llorca F et al. also demonstrated, significant variation before and after neo adjuvant chemotherapy was only for Cyclin D1 and mitotic index. These results are consistent with our study [19].
One of the most commonly altered genes in oncology is p53 and the status of this tumor suppressor gene has been shown to play a pivotal role in the response to a large number of anticancer drugs. Previous studies suggested that breast cancers with TP53 mutations might be either resistant or sensitive to anticancer drugs. However, the issue could not be resolved, because most of the available clinical reports involved small sample sizes, and the results were therefore unable to determine the value of TP53 status for predicting the response to chemotherapy. Additionally, IHC, which lacks sensitivity and specificity, or various DNA sequencing techniques, some of which also lack sensitivity, were the main techniques used in these studies. Meta-analyses of p53 status on tumors prior to treatment have shown it can help predict outcomes. However, it is not predictive of the choice of treatment [20,21]. In triple negative breast cancers too p53 predicts for complete pathological response following neoadjuvant chemotherapy [22]. Nucleolar involvement in regulation p53 forms an important consituent in driving tumors as well as defining responses. Thus its central role in the assessment post NAC.
In Penault-Llorca F et al. demonstrated significant variation before and after neo adjuvant chemotherapy was noticed only for cyclin D1 and mitotic index. These results are the consistent with our study [23].
Mutation analyses in the post treatment cases using the targeted exome sequencing of 344 genes in 11 cases showed no mutations in 7 cases in the residual tumor. Of the 4 cases with mutations identified in the residual tumor, the number of mutations were fewer than 4 in 3 cases. The predominant mutation involved p53 in 2 cases and other genes include ERRB2, CREBBP, HDAC1, TRAPP, PARP1 and GNAS. Our study differed from the others in that it did not show as many mutations. The mutational profiles were also different. This could be attributed to our small sample size. In a study done by Balko JM et al. on molecular profiling of triple negative breast cancers after NAC, the identified alterations were categorized into several key pathway or functional groups: cell cycle alterations (amplifications in CDK4, CDK6, CCND1-3, CCNE1 or AURKA and loss of CDKN2A, CDKN2B or RB1); PI3K/mTOR alterations (amplifications of AKT1- 3, PIK3CA, RAPTOR, RICTOR, loss or mutation of PTEN, truncations or nonsense mutations in TSC1, amplifications or mutations in PIK3CA or PIK3R1); growth factor receptor (GFR) amplifications (EGFR, MET, KIT, FGFR1-2 and 4 or IGF1R); Ras/MAPK alterations (amplifications/gains of KRAS, BRAF, RAF1, or truncations of NF1); or DNA repair alterations (truncations, loss or mutations of BRCA1, BRCA2, or mutations in ATM). They highlighted similarity of their series to basal-like primary breast cancers in the TCGA with respect to potentially targetable alterations. These included PTEN alterations (PI3K and AKT inhibitors), amplifications of JAK2 (ruxolinib or tofacitinib), CDK6, CCND1, CCND2, CCND3 (CDK4/6 inhibitors) and IGF1R (dalotuzumab). Importantly, several patients’ tumors showed an enrichment of AK family CNAs and cyclin D family CNAs after NAC, suggesting an association of these alterations with resistance to chemotherapy [24].
Our sample contained only 2 cases with triple negative cancers, 4 cases each was hormone receptor positive and Her 2 Neu positive. Of the targetable mutations PARP1, HDAC1 were identified.
The significance of macronucleoli and increased nucleoli per cell count in the residual tumor after NAC is unclear. If it represents a stress response to the chemotherapeutic agents they signify genotoxic stress with DNA repair mechanisms. As majority of such residual tumors did not have any representative mutation in the 344 gene exome sequencing panel, it is still an open question if there are other mechanisms to the increased and enlarged nucleoli in these cells. Perhaps they are to do with cancer stem cells?
Conclusions and Limitations
The role of nucleoli in a residual tumor following NAC is not easy to understand. Many chemotherapeutic drugs used in treating breast cancer act directly or indirectly through the various functions of nucleoli. Through its ribosomal manufacturing role the RNA pol 1 and its regulators can be affected by chemotherapy. Or through its non-ribosomal function of sequestering many proteins it may affect cell cycle regulation, senescence and apoptosis. This concept study clearly established that residual cancer after NAC has comparable or increased number and size of nucleoli. Mitosis and Ki67 are significantly reduced. Cyclin D1 is also reduced significantly. Exome analyses of 344 candidate cancer gene panel showed fewer tumors with mutations and none more unique except COL1A. They suggest there is more to these cells than chemo resistance. This study is limited by its smaller sample size. The markers for apoptosis and telomerase could not be completed due to technical difficulty. Further studies incorporating the above panel and whole genome sequencing of residual tumor may resolve these issues.
References
- Simmons CE, Hogeveen S, Leonard R, Rajmohan Y, Han D, Wong A, et al. A Canadian national expert consensus on neoadjuvant therapy for breast cancer: linking practice to evidence and beyond. Curr Oncol. 2015; 22: S43-53.
- Rastogi P, Anderson SJ, Bear HD, Geyer CE, Kahlenberg MS, Robidoux A, et al. Preoperative chemotherapy: updates of National Surgical Adjuvant Breast and Bowel Project Protocols B-18 and B-27. J Clin Oncol. 2008; 26: 778-785.
- Mamounas EP, Anderson SJ, Dignam JJ, Bear HD, Julian TB, Geyer CE Jr, et al. Predictors of locoregional recurrence after neoadjuvant chemotherapy: results from combined analysis of National Surgical Adjuvant Breast and Bowel Project B-18 and B-27. J Clin Oncol. 2012; 30: 3960-3966.
- Wolmark N, Wang J, Mamounas E, Bryant J, Fisher B. Preoperative chemotherapy in patients with operable breast cancer: nine-year results from National Surgical Adjuvant Breast and Bowel Project B-18. J Natl Cancer Inst Monogr. 2001; 96-102.
- Kurosumi M. Significance and problems in evaluations of pathological responses to neoadjuvant therapy for breast cancer. Breast Cancer. 2006; 13: 254-259.
- Kapinas K, Grandy R, Ghule P, Medina R, Becker K, Pardee A, et al. The abbreviated pluripotent cell cycle. J Cell Physiol. 2013; 228: 9-20.
- Moore N, Lyle S. Quiescent, slow-cycling stem cell populations in cancer: a review of the evidence and discussion of significance. J Oncol. 2011; 2011.
- Bomken S, Fiser K, Heidenreich O, Vormoor J. Understanding the cancer stem cell. Br J Cancer. 2010; 103: 439-445.
- Yigit N, Gunal A, Kucukodaci Z, Karslioglu Y, Onguru O, Ozcan A. Are we counting mitoses correctly? Ann Diagn Pathol. 2013; 17: 536-539.
- Bertucci F, Finetti P, Roche H, Le Doussal JM, Marisa L, Martin AL, et al. Comparison of the prognostic value of genomic grade index, Ki67 expression and mitotic activity index in early node-positive breast cancer patients. Ann Oncol. 2013; 24: 625-632.
- El-Sayed MI, Maximous DW, Zakhary MM, Mikhail NN. Biological markers and response to neoadjuvant taxane-based chemotherapy in patients with locally advanced breast cancer. ISRN Oncol. 2012; 2012: 245891.
- Chuah BY, Putti T, Salto-Tellez M, Charlton A, Iau P, Buhari SA, et al. Serial changes in the expression of breast cancer-related proteins in response to neoadjuvant chemotherapy. Ann Oncol. 2011; 22: 1748-1754.
- Petric M, Martinez S, Acevedo F, Oddo D, Artigas R, Camus M, et al. Correlation between Ki67 and Histological Grade in Breast Cancer Patients Treated with Preoperative Chemotherapy. Asian Pacific Journal of Cancer Prevention. 2015; 15: 10277-10280.
- Guarneri V, Broglio K, Kau SW, Cristofanilli M, Buzdar AU, Valero V, et al. Prognostic value of pathologic complete response after primary chemotherapy in relation to hormone receptor status and other factors. J Clin Oncol. 2006; 24: 1037-1044.
- Maur M, Guarneri V, Frassoldati A, Conte PF. Primary systemic therapy in operable breast cancer: clinical data and biological fall-out. Ann Oncol. 2006; 17 Suppl 5: v158-164.
- Caldarella A, Crocetti E, Paci E. Ki67 in breast cancer: a useful prognostic marker? Ann Oncol. 2014; 25: 542.
- Denkert C, Loibl S, Müller BM, Eidtmann H, Schmitt WD, Eiermann W, et al. Ki67 levels as predictive and prognostic parameters in pretherapeutic breast cancer core biopsies: a translational investigation in the neoadjuvant GeparTrio trial. Ann Oncol. 2013; 24: 2786-2793.
- Sahebjam S, Aloyz R, Pilavdzic D, Brisson ML, Ferrario C, Bouganim N, et al. Ki 67 is a major, but not the sole determinant of Oncotype Dx recurrence score. Br J Cancer. 2011; 105: 1342-1345.
- Polley MY, Leung SC, McShane LM, Gao D, Hugh JC, Mastropasqua MG, et al. An international Ki67 reproducibility study. J Natl Cancer Inst. 2013; 105: 1897-1906.
- Bonnefoi H, Piccart M, Bogaerts J, Mauriac L, Fumoleau P, Brain E, et al. TP53 status for prediction of sensitivity to taxane versus non-taxane neoadjuvant chemotherapy in breast cancer (EORTC 10994/BIG 1-00): a randomised phase 3 trial. Lancet Oncol. 2011; 12: 527-539.
- Chen MB, Zhu YQ, Xu JY, Wang LQ, Liu CY, Ji ZY, et al. Value of TP53 status for predicting response to neoadjuvant chemotherapy in breast cancer: a meta-analysis. PLoS One. 2012; 7: e39655.
- Kim T, Han W, Kim MK, Lee JW, Kim J, Ahn SK, et al. Predictive Significance of p53, Ki-67, and Bcl-2 Expression for Pathologic Complete Response after Neoadjuvant Chemotherapy for Triple-Negative Breast Cancer. J Breast Cancer. 2015; 18: 16-21.
- Penault-Llorca F, Abrial C, Raoelfils I, Chollet P, Cayre A, Mouret-Reynier MA, et al. Changes and predictive and prognostic value of the mitotic index, Ki-67, cyclin D1, and cyclo-oxygenase-2 in 710 operable breast cancer patients treated with neoadjuvant chemotherapy. Oncologist. 2008; 13: 1235-1245.
- Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014; 4: 232-245.