
Research Article
Austin Chromatogr. 2015;2(1): 1027.
Construction, Purification and Characterization of Novel, Fluorescent RuvA Chimeras
Hui Yin Tan1,2, Syafiq Abdul Wahab1,2, Jiun Xiang Seet3 and Piero R Bianco1,2,3*
1Department of Microbiology and Immunology, University at Buffalo, USA
2Center of Single Molecule Biophysics, University of Buffalo, USA
3Department of Biochemistry, University at Buffalo, USA
*Corresponding author: Bianco PR, Center for Single Molecule Biophysics, Department of Microbiology and Immunology, University at Buffalo, Buffalo, NY 14214, USA
Received: January 12, 2015; Accepted: February 17, 2015; Published: February 26, 2015
Abstract
The Escherichia coli RuvA and RuvB proteins play important roles in the late stages of recombinational DNA repair and genetic recombination. RuvB is a DNA-stimulated helicase, whose activity is controlled by the homotetrameric RuvA protein. In order to facilitate the studies of the interaction and the role of these proteins both in vivo and in vitro, we constructed a series of novel, RuvAautofluorescent protein fusions. The fusions were then expressed with a “wild type” RuvA using a dual plasmid expression system. The resulting heterogeneous populations of chimeras are readily separated using chromatography. The purified chimeras contain one to four fluorescent tagged subunits and maintain full functionality with RuvB in the presence of Holliday junction substrates.
Introduction
Genetic recombination is essential for maintaining genomic integrity and generating genetic diversity in living organisms. In Escherichia coli, this multi-enzyme process requires the close interplay between several crucial enzymes. Included in this list (not all inclusive) are the recombinase RecA, the helicase-nuclease RecBCD, the Single-Stranded DNA Binding protein (SSB) and the hetero-oligomeric, resolvase complex, RuvABC. Even though the recombinase RecA is capable of catalyzing unidirectional branch migration in vitro, there are enzymes which drive this reaction, possibly more efficiently. One such enzyme is the well-studied RuvAB branch migration complex which binds to the central recombination intermediate, the Holliday Junction (HJ) and catalyzes its migration during the late stages of genetic recombination and recombinational DNA repair [1-5]. While RuvAB is responsible for branch migration of HJs, the central, four-stranded recombination intermediates, RuvC is responsible for HJ cleavage.
The RuvAB complex is composed of two non-identical subunits encoded by the ruvA and ruvB genes [6,7]. The active branch migration complex shown in Figure 1A, consists of at least a symmetric tetramer of RuvA protein (monomer mass, 22 kDa) which binds one face of the Holliday junction and two homohexameric rings of RuvB (monomer mass, 37 kDa) which function as chemo mechanical motors to drive branch migration [1,4,8,9]. The resolution complex forms when a RuvC dimer (monomer mass 19kDa) responsible for HJ cleavage at the crossover point, associates with RuvAB [10,11]. Branch migration and junction cleavage require the coordinated actions of all three proteins [4].
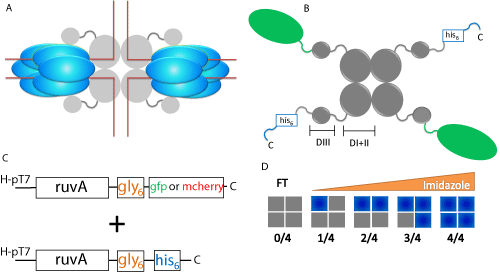
Figure 1: Scheme for dual-plasmid expression and purification of RuvA
chimeras. (A) Schematic diagram of homotetrameric RuvA and hexameric
RuvB assembled on a Holliday junction (red lines). Grey spheres represent
RuvA and blue ovals represent RuvB. (B) Schematic of the RuvA tetramer.
Grey spheres represent RuvA, with domains I & II and domain III being
highlighted by the brackets. The C-terminal histidine tag is indicated in blue
and the fluorescent protein is represented by green ovals. C, C-terminal; D
I+II, domains I & II; D III, domain III. (C) Dual plasmids express the RuvAgly6
6
-fluorescent fusion protein and RuvA-gly6
6
-histidine tag. The designation
H-pT7 (pEAW106 and pET28a+ based) refers to a high copy number plasmid
where the expression of RuvA is under the control of T7 promoters and is
induced by IPTG. (D) Purification scheme for RuvA chimeras. Following
cell lysis, the heterogeneous population of RuvA tetramers is subjected to
nickel column chromatography and can be separated by elution with a linear
imidazole gradient. Grey boxes represent non-histidine tagged RuvA and
blue boxes represent histidine tagged RuvA. FT, flow through.
Branch migration by RuvAB occurs in the 5’ -3' direction and requires a screw motion and lateral pulling or pumping of double stranded DNA (dsDNA), which passes through the center of the RuvB hexamers, and over the surface of the RuvA tetramer, which uses four acidic pins (residues Glu55 and Asp56) to direct the path of each DNA strand through the complex [1,12-15]. Two models have been proposed to describe how RuvB facilitates the screw motion. The first involves a static RuvA - RuvB interaction, with a subset of RuvB monomers within each hexamer participating in passage of DNA and ATPase activity in cyclical fashion around the interior of each hexamer [8]. The second proposes a rotation of RuvB hexamers around the dsDNA, relative to the RuvA complex [14]. Rotation is brought about by ATP hydrolysis and is driven by interactions of RuvB monomers with the DNA and RuvA. In this model, RuvB is proposed to function as a rotating DNA motor, analogous to the F1-ATPase, another AAA+ motor [16]. However, an elegant single molecule study demonstrates that the first model is correct and that it is the DNA that rotates, and not RuvB during branch migration [17].
For branch migration to occur, RuvAB must assemble on the Holliday junction. Here, RuvA (a stable tetramer [18] binds to a dimer of RuvB and the complex binds to the HJ in a reaction that requires only Mg2+ ions [7,19-21]. Thereafter, the remaining 10 monomers of RuvB bind to complete the formation of the diametrically opposed hexameric rings sandwiching the RuvA-HJ complex (Figure 1A). ATP hydrolysis-dependent branch migration then ensues.
RuvA plays an essential role in branch migration by RuvAB [22]. It functions to change the configuration of a Holliday junction to an open-square structure that is energetically more favorable for branch migration. It targets RuvB to the junction and stimulates its DNA helicase activity, and finally, it facilitates binding of RuvC leading to resolution. Structural and biochemical studies have shown that RuvA consists of three domains. Domains I and II constitute the core of the protein which is capable of tetramer formation and HJ binding [12,23,24]. Domain III, which is flexible, interacts with RuvB and modulates its ATPase and consequently its branch migration activity as well [25,26]. Because the ATP binding sites in each of the subunits of RuvB are nonequivalent, ATP hydrolysis moves in cyclic fashion around the hexameric ring [27]. The rate of passage of the cyclical motion of ATP hydrolysis is thought to be regulated by domain III of RuvA [25]. These cyclical passages of ATP hydrolysis around the hexameric RuvB rings may be directly responsible for the screw motion producing branch migration.
To provide further insight into the biochemical mechanism of branch migration and Holliday junction resolution, fluorescent tagging of the individual components of the resolvase are required. Tagging of RuvA with fluorophores would assist in real-time visualization in vivo and on single molecules of DNA, and in in vitro FRET assays. Due to its central role in the function of RuvABC, the attachment of fluorophores to RuvA must be carefully done to ensure a fully active protein. In this study, we describe our rationale for C-terminal tagging of RuvA. Details of the tagging of RuvB and RuvC will be published elsewhere. Then, we demonstrate that the resulting tetramers are fully active in stimulating the ATPase activity of RuvB in the presence of Holliday junction substrates in vitro.
Material and Methods
Materials
Phosphoenolpyruvicacids (PEP), Nicotinamide Adenine Dinucleotide (NADH), Molecular Weight Marker Kit, Pyruvate Kinase (PK) and Lactate Dehydrogenase (LDH) were from Sigma. Gel Filtration Calibration Kit, Adenosine Triphosphate (ATP), NAP-25 columns, Q-Sepharose Fast Flow, HiPrep 16/10 Heparin FF column, Superose 6 10/300 GL and the HisTrap FF crude columns were from GE Healthcare Biosciences. Dithiothreitol (DTT) was from Acros Organics. Bovine Serum Albumin (BSA) was purchased from New England Biolabs. In-fusion HD Cloning Kit was from Clontech Laboratories’, Inc. Oligonucleotides used to construct model fork substrates were purchased from Integrated DNA Technologies (IDT).
Reagents
All solutions were prepared using Barnstead Nanopure water and filtered by 0.2μm membrane. Stock solutions of PEP were prepared in 0.5 M Tris-OAc (pH 7.5). ATP was dissolved as a concentrated stock in 0.5 M Tris-HCl (pH 7.5), with the concentration determined spectrophotometrically using an extinction coefficient of 1.54 x 105 M-1 cm-1. NADH was dissolved in 10 mM Tris-OAc (pH 7.5), concentration determined using an extinction coefficient of 6.25 x 103 M-1 cm-1, and stored in small aliquots at -80°C. DTT was dissolved as a 1M stock in nanopure water and stored at -80°C. All reaction buffers described below were assembled at 10 times reaction concentration and stored in 1mL aliquots at -80 °C.
Cloning
Plasmid pEAW106-RuvA was a gift from Dr. Mike Cox (University of Wisconsin, Madison, WI). Full length RuvA and domain I & II of RuvA were amplified from this plasmid via PCR and cloned into pET28a(+)-gly6 at the NcoI and HindIII sites using In-fusion Cloning. Resulting clones (pET28a(+)-RuvA-gly6-his6& pET28a(+)-RuvA-domain I & II-gly6-his6) were confirmed using NdeI restriction enzyme mapping and sequencing.
Plasmid pEAW106-RuvA-gly6-his6 was made by ligating electroelution purified pieces of pET28a (+)-RuvA-gly6-his6 and pEAW106- RuvA digested using DraIII. Clones were confirmed with an NgoMIV digest.
The GFP and mcherry genes were amplified using PCR with different primer sets that either place ruvA in frame or out of frame with the C-terminal histidine tag of pET28a(+)-RuvA-gly6-his6. PCR fragments were cloned into pET28a (+)-RuvA-gly6-his6 at the XhoI site using In-fusion Cloning. Resulting clones (pET28a(+)-RuvAgly6-GFP, pET28a(+)-RuvA-gly6-GFP-his6, pET28a(+)-RuvA-gly6- mcherry & pET28a(+)-RuvA-gly6-mcherry-his6) were confirmed using XhoI restriction enzyme mapping.
Tuner (DE3) ΔruvAcells were made by P1 transduction of MG1655ΔruvA60::Tn 10 (Gift from Dr. Robert Llyod) into Tuner (DE3) cells.
Proteins
wtRuvA and wtRuvB proteins were purified as described previously [28]. The concentration of RuvA was determined using an extinction coefficient of 5,550 M-1 cm-1 [27]. For the RuvB purification, the DEAE BiogelA column was replaced by a 100mL Q-Sepharose column that was equilibrated with TEGD buffer (20mM Tris-acetate, 1mM EDTA, 10% (v/v) Glycerol, and 1mM DTT) and the protein was eluted with a 1 L linear gradient from 0 to 500mM potassium acetate. The concentration of RuvB protein was determined using an extinction coefficient of 16,400 M-1 cm-1 [27].
For dual expression of wtRuvA and RuvA-gly6-GFP/mcherry with either one histidine-tagged, plasmids were transformed into Tuner (λDE3) ΔlacZYΔruvA cells and expression of protein was verified by SDS-PAGE and/or with measurement of fluorescence (GFP excitation at 488nm, emission at 514nm; mcherry excitation at 588nm, emission at 611nm). Large cultures were grown to an OD600 ~ 0.4, induced with IPTG and grown until the culture reached stationary phase. Cells were harvested and lysed, and the resulting lysates were subjected to nickel column chromatography [29]. All different RuvA tetramers with histidine tags were purified using only the nickel column while wild type RuvA/RuvA-gly6-GFP-his6 and RuvA-domain I&II-gly6- his6were subjected to heparin column chromatography as well. The HiPrep 16/10 Heparin FF column was equilibrated with Buffer R (20mM Tris-HCl, pH7.5, 1mM EDTA, 1mM DTT, 150mMKCl and 10% glycerol). Proteins were eluted off the column with a gradient of 150mM–1M KCl. All purified proteins were dialyzed against Buffer R with 150mM NaCl instead of KCl and then against Storage buffer (20mM Tris-HCl, pH7.5, 1mM EDTA, 1mM DTT, 150mM NaCl and 20% glycerol) and stored in aliquots at -80°C.
The extinction coefficients used to determine the concentration of RuvA tetramers are as followed: wild type RuvA and RuvA-gly6- his6, e=5,960M-1cm-1; RuvA-gly6-GFP and RuvA-gly6-GFP-his6, Σ=27975M-1cm-1; RuvA-gly6-mcherry and RuvA-gly6-mcherry-his6, e=40,340M-1cm-1. All extinction coefficients were calculated based on amino acid sequences of subunit. The ratio of either wild-type, histidine tagged and/or fluorophore fusion protein in the RuvA tetramer determines the sum of extinction coefficient used to calculate the concentration of RuvA tetramer [30].
DNA cofactors
M13 mp18 ssDNA was prepared as described [31]. The concentration of DNA was determined spectrophotometrically using an extinction coefficient of 8, 780 M-1cm-1 (nucleotides). Purified DNA was stored in small aliquots at -80°C.
The sequences of oligonucleotides used to construct model fork substrates were adapted from those used previously [32,33] and contain a mobile homologous core flanked by heterologous sequences. Oligonucleotides were purified using denaturing polyacrylamide gels. This was followed by gel filtration using NAP-25 columns and ethanol precipitation. The concentration of each oligonucleotide was determined spectrophotometrically using the extinction coefficient provided by IDT.
The Holliday Junction contained oligonucleotides PB170, 173, 345 and 346 and was prepared by annealing four oligonucleotides: PB170 (5’–CTAGAGACGCTGCCGAATTCTGGCTTGGATCTGATGCT GTCTAGAGGCCTCCACTATGAAATCGCTGCA–3’), PB173 (5’– CCGGGCTGCAGAGCTCATAGATCGATAGTCTCTAGACAGC ATCAGATCCAAGCCAGAATTCGGCAGCGTCT–3’), PB345 (5’–GCGATTTCATAGTGGAGGCCTCTAGACAGCACG CCGTTGAATGGGCGGATGCTAATTACTATCTC–3’) and PB346 5’–GAGATAGTAATTAGCATCCGCCCATTCAACG G C G T G C T G T C T A G A G A C T A T C G A T C T A TGAGCTCTGCAGC–3’). Purified oligonucleotides (1 – 10 μM molecules each in different annealing experiments) were annealed in a total volume of 50 μl containing 10 mM Tris-HCl (pH 7.5) or 10mM Tris-OAc (pH 7.5), 100mM NaCl and 10 mM MgOAc. Annealing reaction involved incubation of the DNA-buffer mixture in thin-walled PCR tubes at 100°C for 5 min, followed by an overnight cooling step to room temperature. Junctions were added directly to ATPase assays without further purification. The extent of annealing was verified by non-denaturing PAGE using 5’-end labeled oligonucleotides annealed under identical conditions (data not shown). Typically, >95% of the DNA present was found to be in the annealed substrate (data not shown).
ATP hydrolysis assay
The hydrolysis of ATP was monitored using a coupled spectrophotometric assay carried at 37°C as described previously [31,32]. The standard reaction buffer for RuvAB contained 20 mM Tris-OAc (pH 7.5), 2 mM DTT, 0.3 mM NADH, 7.5 mM PEP, 20 U/mL PK, 20 U/mL LDH, 100 μg/mL BSA, 1 mM ATP, and 10 mM MgOAc. Assays were performed in a reaction volume of 150μl, and were initiated by the addition of enzyme following a 2 minute preincubation at 37°C of all other components. For assays with M13 ssDNA, 10μM DNA is present and the concentration of RuvB was held constant at 1 μM and concentration of RuvA varied. Both proteins were mixed together on ice prior to addition to assays [28]. In contrast, in assays with Holliday junctions, addition of preformed RuvAB complexes did not result in ATPase activity ([33] and data not shown). To observe activity, proteins were added sequentially to reaction mixes containing 100nM DNA and 1mM ATP as follows: RuvA first (400 nM monomer, final), followed by 5 minute incubation; then varying amount of RuvB was added to initiate reactions. The rate of ATP hydrolysis was calculated by multiplying the slope of a tangent drawn to linear portions of time courses by 159. In a typical reaction, close to 200 data points were used to draw a linear fit to the data to calculate reaction rates. Curve fitting was done using Prism v 5.04 (GraphPad Software, Inc.). Typically, 2 to 4 assays were done on separate days for each reaction condition to obtain reaction rates.
Gel Filtration
250μl of individual Molecular Weight Markers and protein samples were applied separately to the Superose 6 10/300 GL equilibrated with buffer containing 20mM Tris-HCl, pH8.0, 1mM EDTA and 100mM NaCl. Molecular weight markers were prepared in the concentration recommended by manufacturer. Concentration of protein samples were: wild type RuvA, 91.1μM (0.543 mg/ml); RuvA-gly6-6, 103.5μM (0.617 mg/ml); 3 RuvA-gly6-GFP6/1 RuvAgly6-66, 4.98μM (0.448 mg/ml); 2 RuvA-gly6-GFP6/2 RuvA-gly6-his6, 4.79μM (1.004 mg/ml).The column was run using Bio-Rad Biologic Duoflow Chromatography system. A calibration curve was generated using Kav of molecular weight markers that were chromatographed in the same buffer: Ferritin (440,000Da), Catalase (232,000Da), Aldolase (158,000Da), Alcohol dehydrogenase (150,000Da), Conalbumin (75,000Da), Ovalbumin (44,000Da), Carbonic anhydrase (29,000Da) and Ribonuclease A (13,700Da). Elution volume was determined by measuring the volume of the eluent from the point of injection to the center of elution peak. The void volume (7.2ml) of this column was estimated to be 30% of the column volume (24ml). The Kav value of each molecular weight standards was calculated and a calibration curve was graphed with a plot of Kav vs log of molecular weight. The molecular weight of each protein sample was determined by extrapolating the Kav from the calibration curve.
Results and Discussion
Design of chimeric and fluorescent RuvA proteins
RuvA is purified as a stable tetramer [34]. Each monomer within the tetramer consists of three domains [23,26]. The first two, domains I and II, span the N-terminal, one third of the protein and comprise the tetramerization and DNA binding domains. Domains I and II are visible in RuvA crystal structures and electron microscopy images [8,35]. The C-terminal domain III which is flexible and not visible in the structures or electron microscopy images, extends away from the core of the protein, and is essential for binding to RuvB, to stimulation of its ATPase activity and to branch migration [25,26]. As the N-terminus of each RuvA monomer is buried within the tetramer core, facile histidine and fluorescent protein tagging must be done at the accessible C-terminus (Figure 1B) [23,26]. We reasoned that adding tags at this position would have minimal effects on tetramer formation and DNA binding. To further position tag away from the RuvA core and in an attempt to minimize RuvB-binding defects, an additional six glycine residues were added between the C-terminus and the tags. These fusion protein designs aim to minimize the effect of tagging on RuvA and to maintain its functionality.
The scheme for production of active and fluorescent, chimeric RuvA proteins is to utilize a dual, high copy number plasmid expression system that has been successfully used for the E.coli singlestranded DNA binding protein [29,36]. The first plasmid expresses a C-terminal fluorescent RuvA fusion and the other plasmid expresses C-terminal histidine tagged RuvA (Figure 1C). An alternative design utilizing wild type RuvA and RuvA-gly6-GFP/mcherry-his6 has also been employed producing similar results as described in subsequent sections.
Over expressing both types of protein in the cell ensures all possible ratios of “wild type” RuvA to RuvA fluorescent protein fusions can be obtained (Figure 1D). Bl21 cells deleted for lacY and ruvA were used for the over expression of the ruvA genes encoded by these plasmids. These “Tuner” strains allow for carefully controlled levels of expression via IPTG titration as a result of the mutation in the lac permease and ensure that the only RuvA present in the cells is plasmid encoded.
When the ruvA genes encoded by these plasmids are over expressed in the same cell, a heterogeneous population of RuvA tetramers is created, which can be separated into distinct species using nickel column chromatography by eluting with linear imidazole gradient as shown previously for SSB [29,36]. This strategy makes sense as tetramers with a single histidine tag elute at lower imidazole concentration while tetramers with two, three and four tags are eluted at increasingly higher imidazole concentration (Figure 1D).
Purification of chimeric and fluorescent RuvA proteins
Following cell lysis, the cleared cell lysates were subjected to nickel column chromatography to separate individual RuvA chimeras. Homotetramers that do not have a histidine tag do not bind to the resin and are eluted in the flow through, identical to what we observed for SSB (Figure 1D and [29,36]. The remaining tetramers bind to the nickel column with different affinities based on the number of histidine tagged subunits present. By applying a linear imidazole gradient to the column, tetramers with increasing number of histidine tags are eluted with increasingly higher imidazole concentration (Figure 2A). Fractions are pooled with caution according to SDS-PAGE results as there are overlapping regions containing mixed ratios of chimeras. Pooled RuvA chimeras with contaminating species were further purified using heparin columns (data not shown).
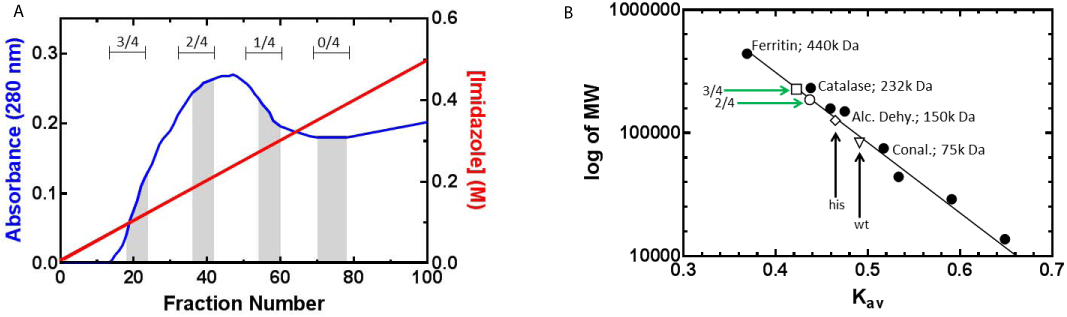
Figure 2: 2: Different composition of RuvA chimeras are purified by nickel
column chromatography. (A) Elution profile of nickel column from expression
of dual-plasmids: RuvA-gly6-his6 and RuvA-gly6-GFP. Proteins were eluted
using a linear imidazole gradient (30 to 500 mM). The blue curve indicates
absorbance at 280 nm (or protein) and the red line indicates the imidazole
concentration. The ratios of autofluorescent tagged to histidine tagged protein
are indicated above of each highlighted grey area. Homotetrameric RuvAgly
6-his6 elutes last (0/4). (B) Molecular weights of wild type and chimeric
RuvA proteins determined by gel filtration chromatography. Molecular weight
standards (•) used were: Ferritin (440,000 Da), Catalase (232,000 Da),
Aldolase (158,000 Da), Alcohol dehydrogenase (150,000 Da), Conalbumin
(75,000 Da), Ovalbumin (44,000 Da), Carbonic anhydrase (29,000 Da)
and Ribonuclease A (13,700 Da). The Kav value of each molecular weight
standard was determined and the calibration curve was generated as a plot
of Kav vs log of molecular weight. The molecular weights of protein samples
were determined by extrapolating their Kav values from the calibration curve.
Protein samples include: wild type (wt) RuvA (∇); 0/4, RuvA-gly6-his6 (his) (◊);
2/4, RuvA-gly6-GFP / 2RuvA-gly6-his6 (Ο); and 3/4, RuvA-gly6-GFP / 1RuvAgly
6-his6 (⟩)
Tagging of RuvA results in tetrameric chimeras with different predicted molecular weight as, calculated from the protein sequence. To further confirm the ratio of fusion protein in the tetramers and the homogeneity of population as evaluated by SDS-PAGE, protein samples were subjected to gel filtration chromatography. A calibration graph consisting of Kav of standards versus their respective molecular weights was generated (Figure 2B). Then separate chromatographic runs of wild type and homotetrameric RuvA-gly6-his6 were carried out. The wild type eluted from the column with a Kav value that is consistent with its molecular weight of 88,344 Da. In contrast, the histidine tagged RuvA homotetramer elutes earlier than expected, with an observed molecular weight 126,377 Daltons as derived from the Kav. This is 1.3 fold higher than the expected molecular weight of 95,448 Da. At present, we do not know the reason for this, but we speculate that the histidine tag may alter the conformation of the protein so it elutes larger than expected molecular weight.
Next, the RuvA chimeras were subjected to gel filtration and the resulting Kav values were utilized to determine their molecular weights. The different RuvA chimeras eluted off the gel filtration column in the order consistent with the molecular weights calculated from the amino acid sequences. That is, a tetramer with 3 GFP tags eluted first, followed by a tetramer with 2 tags (Figure 2B). However, as for the homotetrameric RuvA-gly6-his6 protein, we observed the same disparity in molecular weights. That is, the observed molecular weights of chimeras containing one or more histidine tags are greater by a factor of 1.3. The molecular weights determined by their Kav values are 186, 209 Da for 2 GFP-tagged RuvA, and 227,335 Da for a RuvA tetramer containing 3 GFP tags.
RuvA chimeras are partially active in the presence of M13 ssDNA
RuvA facilitates RuvB binding to DNA substrates and additionally stimulates the ATPase activity of RuvB in the presence of DNA [37]. Previous work has shown that the domain III of RuvA, which is located at the C-terminus, is responsible for binding to RuvB [26]. In this study, we created chimeras of RuvA fused with fluorescent protein and/or a histidine tag at the C-terminus of the subunit. These additional residues could disrupt RuvA-RuvB binding, resulting in inactive complexes.
To determine whether the additional residues perturb the interaction of RuvA with RuvB, we examined the rate of ATP hydrolysis by RuvB in the presence and absence of M13 single-
stranded DNA. The positive control was wild type RuvA and the negative control was a mutant protein designated RuvA ΔDIII that contains only domains I and II. In the absence of DNA, RuvB exhibited ATPase activity [27]. This is not stimulated by either wild type or any of the various chimeric RuvA tetramers or RuvA ΔDIII (Figure 3A). In contrast, wild type and homotetrameric RuvA-gly6-his6 proteins stimulated the ssDNA-dependent ATPase activity of RuvB three-fold relative to the DNA-independent rate. This stimulation is not simply due to RuvB acting alone, as shown in the RuvB-ssDNA control reaction. As expected, the RuvA ΔDIII protein did not simulate the ATPase activity of RuvB in the presence of ssDNA (Figure 3A). Next, the chimeric RuvA proteins were tested and the results showed that they stimulated RuvAB ATPase activity 1.5- to 2-fold relative to the DNA-independent rate.
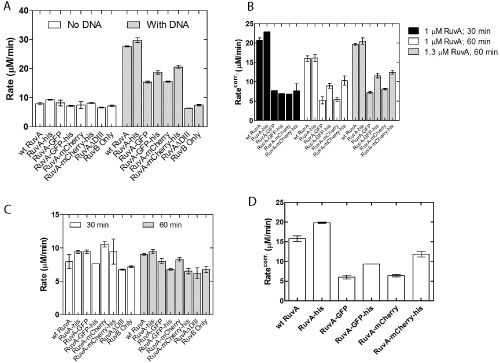
Figure 3: RuvA chimeras stimulate the DNA-dependent ATPase activity
of RuvB. (A) Comparison of wild type and chimeric RuvA proteins. (B)
Pre-incubation time and RuvA concentration lead to modest RuvB rate
enhancements. (C) Rate enhancements are not the result of effects on the
DNA-independent ATPase activity of RuvB. (D) The chimeras are not as
effective as wild type or RuvA-gly6-his6 in stimulating the ATPase of RuvB.
The rates shown in (A) and (D) have been corrected for the DNA-independent
ATPase rate of RuvAB. ATPase assays were performed as described in
Experimental Procedures and were initiated by the addition of preformed
RuvAB complexes. The concentration of RuvB was held constant at 1 μM in
each assay, and RuvA was present at the indicated concentrations (A and B)
and at 1.3 uM in panels C and D.. All protein concentrations were reported in
terms of monomers. Error bars represent the mean with standard deviation of
at least two separate experiments. Corr., RuvB ATP hydrolysis rate corrected
for the DNA-independent rate. The column labeled RuvA-GFP is 2 RuvAgly
6-GFP / 2 RuvA-gly6-his6; RuvA-GFP-his, 2 RuvA-gly6-GFP-his6 / 2 RuvA;
RuvA-mcherry, 2 RuvA-gly6-mcherry / 2 RuvA-gly6-his6; RuvA-mcherry-his, 2
RuvA-gly6-mcherry-his6 / 2 RuvA.
In these reactions, RuvA and RuvB were pre-incubated on ice for 30 minutes prior to initiating reactions [28,32,33]. The inability of the chimeras to fully stimulate RuvB to the levels observed for wild type or the homotetrameric RuvA-his protein could be due to a failure to properly form RuvAB complexes. To test whether this was occurring, the incubation time for complex formation was extended to 60 minutes. This resulted in only a small increase for the RuvAgly 6-GFP-his6 and RuvA-gly6-mcherry-his6 proteins (Figure 3B,C). This is not due to an increase in the DNA-independent activity as no enhancement was seen in these reactions (Figure 3C). Finally, the concentration of RuvA was increased to 1.3 μM and the preincubation time of 60 minutes used. No further stimulation was observed (Figure 3B). Collectively, these results suggested that in the presence of M13 ssDNA, the addition of the auto fluorescent protein to the C-terminus of RuvA perturbs the RuvA-RuvB interaction (Figure 3D). This perturbation is likely at the RuvAB complex assembly step and results in chimeras that are only 38 to 50% as active as wild type.
RuvA chimeras are fully active in the presence of a Holliday junction
The failure of the RuvA chimeras to fully stimulate the ATPase activity of RuvB in the presence of ssDNA could be due to these novel RuvA proteins existing in equilibrium between inactive and active configurations. As RuvAB is well known as a branch migration complex catalyzing the movement of Holliday junctions, we surmised that perhaps this DNA substrate could shift the RuvA equilibrium into the fully active configuration, resulting in maximal stimulation of the ATPase activity of RuvB. To test this, assays were done using a model Holliday Junction that has been extensively used to characterize the activity of the wild type RuvAB complex [32,33].
In contrast to ATPase assays in the presence of M13 ssDNA, no reaction is observed in the presence of Holliday Junction reactions initiated with preformed RuvAB complexes [32,33] and data not shown). Therefore, reactions were assembled sequentially: RuvA was added to pre-warmed reaction mixes containing DNA, to allow binding to occur. Two minutes later, RuvB was added to initiate the reaction. The results show that as expected, wild type and homotetrameric RuvA-gly6-his6 stimulated the ATPase activity of RuvB and the RuvA ΔDIII mutant did not (Figure 4A).
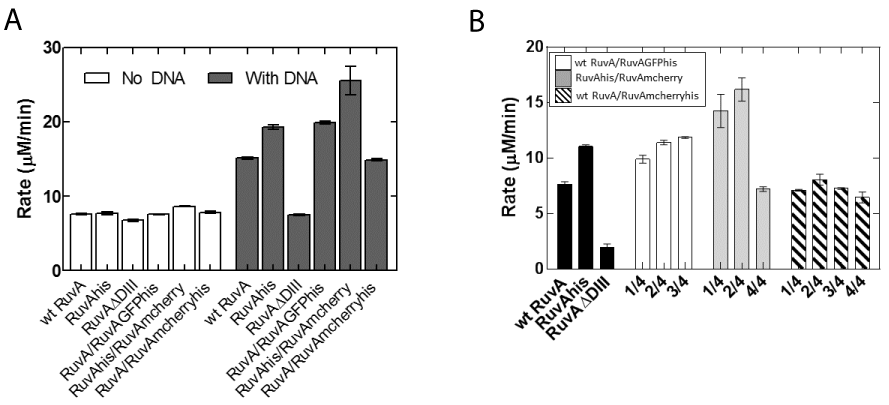
Figure 4: RuvA chimeras stimulate RuvB ATPase activity in the presence
of Holliday junctions. (A) Stimulation of RuvB ATPase activity requires
the Holliday junction. Reactions contained 1200 nM RuvB. (B) Different
chimeras stimulate the ATPase activity of RuvB in the presence of 3600
nM RuvB. ATPase assays were performed as described in Experimental
Procedures. 100 nM Holliday junction was incubated with 400nM RuvA and
assays were initiated by the addition of RuvB. All protein concentrations
are reported in terms of monomers. Error bars represent the mean with
standard deviation of at least two separate experiments. Rate in panel B is
the RuvB ATP hydrolysis rate corrected for the DNA-independent rate. In
Panel (A), the column labeled RuvA/RuvAGFPhis is 2 RuvA / 2 RuvA-gly6-
GFP-his6; RuvAhis/RuvAmcherry, 2 RuvA-gly6-his6 /2 RuvA-gly6-mcherry;
RuvA/RuvAmcherryhis, 2 RuvA/2 RuvA-gly6-mcherry-his6. In Panel (B), 1/4
represents 1 fluorescent tagged subunit; 2/4, 2 fluoresecent tagged subunits;
3/4, 3 fluorescent tagged subunits; 4/4, 4 fluorescent subunits per tetramer.
In contrast to reactions with M13 ssDNA, RuvA chimeras were able to fully stimulate the ATPase activity of RuvB. When expressed as a fraction of wild type in the presence of M13 ssDNA, the chimeras are now 75-140% as active. Similarly, when the rate of ATP hydrolysis for each of the chimeras is expressed as a percent of wild type in the presence of Holliday junctions, these proteins are 100-180% as active. No further increase was observed when the RuvB concentration was increased 3-fold (Figure 4B). These results are consistent with our model that in solution, the chimeras (and possibly wild type as well) exist in an equilibrium between active and inactive states. In the inactive state, they cannot associate with RuvB properly. When the chimeras are bound to a Holliday Junction, the protein is now in the active configuration, resulting in full stimulation of the ATPase activity of RuvB. Therefore, the chimeras are fully active. Furthermore, our design adding a six glycine linker to position the auto fluorescent protein away from the residues required for RuvB binding functions as anticipated. In addition, the identity of auto fluorescent protein did affect activity as the GFP fusions were more active than mcherry ones. Additional experiments are in progress to understand this, but we suggest that this is likely due to preparation to preparation variation. Finally, the presence of one, two or three autofluorescent proteins in a RuvA tetramer resulted in comparable stimulation in the levels of RuvB ATPase activity.
Conclusion
The design of the RuvA fusion genes in this study and the dual plasmid system produces fluorescent-tagged protein that retains full activity in the presence of Holliday Junctions. We propose that in solution, RuvA exists in equilibrium between an inactive and active state. This equilibrium is shifted towards the active state by Holliday junction binding.
Homogeneous preparations of the different composition RuvA chimeras can be easily obtained by sequential nickel column chromatography and gel filtration. The RuvA chimeras containing one to four autofluorescent proteins are as active as the wild type, thus the autofluorescent proteins either do not make inhibitory contacts with RuvB, or are free to rotate due to the incorporation of the glycine linkers.
Acknowledgement
Work in the Bianco laboratory is supported by NIH Grant GM100156 to PRB.
References
- Rafferty JB, Sedelnikova SE, Hargreaves D, Artymiuk PJ, Baker PJ, Sharples GJ, et al. Crystal structure of DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science. 1996; 274: 415-421.
- West SC. Formation, translocation and resolution of Holliday junctions during homologous genetic recombination. Philosophical Transactions of the Royal Society of London - Series B: Biological Sciences. 1995; 347: 21-25.
- Eggleston AK, Mitchell AH, West SC. In vitro reconstitution of the late steps of genetic recombination in E coli Cell. 1997; 89: 607-617.
- West SC. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997; 31: 213-244.
- Muller B, West SC. Processing of Holliday junctions by the Escherichia coli RuvA, RuvB, RuvC and RecG proteins. Experientia. 1994; 50: 216-222.
- Sharples GJ, Benson FE, Illing GT, Lloyd RG. Molecular and functional analysis of the ruv region of Escherichia coli K-12 reveals three genes involved in DNA repair and recombination. Mol Gen Genet. 1990; 221: 219-226.
- Shiba T, Iwasaki H, Nakata A, Shinagawa H. Escherichia coli RuvA and RuvB proteins involved in recombination repair: physical properties and interactions with DNA. Mol Gen Genet. 1993; 237: 395-399.
- Yu X, West SC, Egelman EH. Structure and subunit composition of the RuvAB-Holliday junction complex. J Mol Biol. 1997; 266: 217-222.
- Hargreaves D, Rice DW, Sedelnikova SE, Artymiuk PJ, Lloyd RG, Rafferty JB. Crystal structure of E.coli RuvA with bound DNA Holliday junction at 6 A resolution. Nat Struct Biol. 1998; 5: 441-446.
- Connolly B, Parsons CA, Benson FE, Dunderdale HJ, Sharples GJ, Lloyd RG, et al. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc Natl Acad Sci USA. 1991; 88: 6063-6067.
- Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991; 10: 4381-4389.
- Ariyoshi M, Nishino T, Iwasaki H, Shinagawa H, Morikawa K. Crystal structure of the holliday junction DNA in complex with a single RuvA tetramer. Proc Natl Acad Sci USA. 2000; 97: 8257-8262.
- Ingleston SM, Sharples GJ, Lloyd RG. The acidic pin of RuvA modulates Holliday junction binding and processing by the RuvABC resolvasome. EMBO J. 2000; 19: 6266-6274.
- Putnam CD, Clancy SB, Tsuruta H, Gonzalez S, Wetmur JG, Tainer JA. Structure and mechanism of the RuvB Holliday junction branch migration motor. J Mol Biol. 2001; 311: 297-310.
- Stasiak A, Tsaneva IR, West SC, Benson CJ, Yu X, Egelman EH. The Escherichia coli RuvB branch migration protein forms double hexameric rings around DNA. Proc Natl Acad Sci USA. 1994; 91: 7618-7622.
- Kinosita K, Yasuda R, Noji H, Ishiwata S, Yoshida M. F1-ATPase: a rotary motor made of a single molecule. Cell. 1998; 93: 21-24.
- Han Y, Tani T, Hayashi M, Hishida T, Iwasaki H, Shinagawa H, et al. Direct observation of DNA rotation during branch migration of Holliday junction DNA by Escherichia coli RuvA-RuvB protein complex. Proc Natl Acad Sci USA. 2006; 103: 11544-11548.
- Tsaneva IR, Illing G, Lloyd RG, West SC. Purification and properties of the RuvA and RuvB proteins of Escherichia coli. Mol Gen Genet. 1992; 235: 1-10.
- Hiom K, West SC. Characterisation of RuvAB-Holliday junction complexes by glycerol gradient sedimentation. Nucleic Acids Res. 1995; 23: 3621-3626.
- Müller B, Tsaneva I, West S. Branch migration of Holliday junctions promoted by the Escherichia coli RuvA and RuvB proteins. I. Comparison of RuvAB- and RuvB-mediated reactions. J Biol Chem. 1993; 268: 17179-17184.
- Müller B, Tsaneva I, West S. Branch migration of Holliday junctions promoted by the Escherichia coli RuvA and RuvB proteins. II. Interaction of RuvB with DNA. J Biol Chem. 1993; 268: 17185-17189.
- Cassuto E, Sequret S, Hejna J, Morel P. Opposite effects of UvrD in RecA-promoted homologous pairing and strand transfer. J Cell Biol. 1992; 34.
- Nishino T, Ariyoshi M, Iwasaki H, Shinagawa H, Morikawa K. Functional analyses of the domain structure in the Holliday junction binding protein RuvA. Structure. 1998; 6: 11-21.
- Yamada K, Kunishima N, Mayanagi K, Ohnishi T, Nishino T, Iwasaki H, et al. Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8. Proc Natl Acad Sci USA. 2001; 98: 1442-1447.
- Nishino T, Iwasaki H, Kataoka M, Ariyoshi M, Fujita T, Shinagawa H, et al. Modulation of RuvB function by the mobile domain III of the Holliday junction recognition protein RuvA. J Mol Biol. 2000; 298: 407-416.
- Yamada K, Miyata T, Tsuchiya D, Oyama T, Fujiwara Y, Ohnishi T, et al. Crystal structure of the RuvA-RuvB complex: a structural basis for the Holliday junction migrating motor machinery. Mol Cell. 2002; 10: 671-681.
- Marrione PE, Cox MM. RuvB protein-mediated ATP hydrolysis: functional asymmetry in the RuvB hexamer. Biochemistry. 1995; 34: 9809-9818.
- Marrione PE, Cox MM. Allosteric effects of RuvA protein, ATP, and DNA on RuvB protein-mediated ATP hydrolysis. Biochemistry. 1996; 35: 11228-11238.
- Liu J, Choi M, Stanenas AG, Byrd AK, Raney KD, Cohan C, et al. Novel, fluorescent, SSB protein chimeras with broad utility. Protein Sci. 2011; 20: 1005-1020.
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989; 182: 319-326.
- Slocum SL, Buss JA, Kimura Y, Bianco PR. Characterization of the ATPase activity of the Escherichia coli RecG protein reveals that the preferred cofactor is negatively supercoiled DNA. J Mol Biol. 2007; 367: 647-664.
- Buss JA, Kimura Y, Bianco PR. RecG interacts directly with SSB: implications for stalled replication fork regression. Nucleic Acids Res. 2008; 36: 7029-7042.
- Abd Wahab S, Choi M, Bianco PR. Characterization of the ATPase activity of RecG and RuvAB proteins on model fork structures reveals insight into stalled DNA replication fork repair. J Biol Chem. 2013; 288: 26397-26409.
- Tsaneva IR, Illing G, Lloyd RG, West SC. Purification and properties of the RuvA and RuvB proteins of Escherichia coli. Mol Gen Genet. 1992; 235: 1-10.
- Sedelnikova SE, Rafferty JB, Hargreaves D, Mahdi AA, Lloyd RG, Rice DW. Crystallization of E. coli RuvA gives insights into the symmetry of a Holliday junction/protein complex. Acta Crystallogr D Biol Crystallogr. 1997; 53: 122-124.
- Bianco PR, Stanenas AJ, Liu J, Cohan CS. Fluorescent single-stranded DNA-binding proteins enable in vitro and in vivo studies. Methods Mol Biol. 2012; 922: 235-244.
- Mitchell AH, West SC. Role of RuvA in branch migration reactions catalyzed by the RuvA and RuvB proteins of Escherichia coli. J Biol Chem. 1996; 271: 19497-19502.