
Research Article
Austin Chromatogr. 2021; 7(1): 1053.
Influence of Hydrogen Ion Concentrations on the Protein Mobility in Native-PAGE Buffers
Ganesh A, Hilda K, Bhuvaragavan S, Mullainadhan P and Janarthanan S*
Department of Zoology, University of Madras, Guindy Campus, India
*Corresponding author:S Janarthanan, Professor and Head, Department of Zoology, University of Madras, Guindy Campus, India
Received: January 05, 2021; Accepted: January 29, 2021; Published: February 05, 2021
Abstract
The study demonstrates the effect of hydrogen ion concentrations (pH) of Tank Buffer (TB) and resolving gel buffer (RGB) in a native-PAGE system on protein samples. A pH range of 7.8 to 9.3 for RGB and 8.3 to 9.3 for TB were used. Proteins in the samples under native-PAGE at varying pH of RGB such as 7.8, 8.3 and 8.8 with pH of TB, 8.3 and 8.8 were resolved well. The total running time for the samples to reach the end of the dye front ranged between 2.30 h to 4.30 h at the above pH combinations. It was observed as 3.30 h as total running time under normal pH conditions (RGB, pH 8.8; TB, pH 8.3). At the same time, buffers at higher pH or the combination of extreme pH (7.8 vs. 9.3) in the buffers were not favored good protein mobility/resolution, and bands were diffused. Longer running time was observed in various combinations of pH of RGB and pH of TB with 18.00 h being a longest with a pH of 8.3 and 9.3 for RGB and TB, respectively. This indicated the importance of pH of electrophoresis buffers for ionization of various proteins for better separation.
Keywords: Native-PAGE; pH; Protein Mobility; Resolving Gel Buffer; Tank Buffer
Abbreviations
TB: Tank Buffer; RGB: Resolving Gel Buffer; PAGE: Polyacrylamide Gel Electrophoresis; pIs: Isoelectric Points; BSA: Bovine Serum Albumin; RBC: Red Blood Cells; PTU: Phenylthiourea
Introduction
Acrylamide gel electrophoresis is a standard technique used to separate, analyse and purify molecules such as proteins based on their net charge and size [1-3]. Separation is influenced by the type of proteins, pH of the buffers used, electrical field strength and temperature [4,5]. The pH of the electrophoresis buffer required to be in the range over which proteins of interest are stable or at which they retain their biological activity [6]. The pH should be appropriately chosen with respect to the isoelectric points (pIs) of proteins. The pH of the resolving gel buffer must be adequately distant away from the pIs of the proteins and so they carry enough net charge to travel through the gel in a reasonable time period [7]. The resolution and stability of proteins are often decided by the choice of pH [8]. A protein possesses charge as a result of the constituents of its acidic (glutamic and aspartic acids) and basic (lysine and arginine) amino acids. However, it is only the net charge on the molecule that is significant in deciding its movement on a matrix. Since each molecule possesses unique charge and mass, it migrates to a specific position within the electric field in a given duration of time [9]. Therefore, if a mixture of proteins is subjected to electrophoresis, each of the protein would be expected to concentrate into a tight migrating band at a unique position in the electric field on a matrix [10]. Different protein molecules in the sample move through the matrix at various velocities under the influence of an applied electrical current [11]. Polyacrylamide is the most common matrix used for separating proteins [1,12]. Although studies on standardization of PAGE according to the different model proteins in regard to various pH have reported, there is no information pertaining to the effect of changes of hydrogen ion concentration in resolving gel buffer which is crucial for separation of biological macromolecules [10,13,14]. In that context, this study is aimed to optimize a polyacrylamide gel to separate native proteins with good resolution by varying the pH of buffers used in both electrophoresis and resolving gel.
Materials and Methods
Protein Samples
Lysozyme and Bovine Serum Albumin (BSA) fraction-V were purchased from Hi-Media, haemoglobin from ox red blood cells (RBC) and haemolymph of Bombyx mori (fifth instar larva) were used for this study. The average molecular weight of lysozyme is 14.6 kDa and its pI value is 11.0. The molecular weight of BSA is 66.3 kDa and pI value is 4.9. The protein concentration of samples was measured using bovine serum albumin as standard by following Lowry et al. [15]. Alsever’s solution was prepared by following Garvey et al. [16] for maintaining ox RBC.
Preparation of Protein Samples
Lysozyme muramidase from chicken egg white
Lysozyme (5 mg lyophilized powder) was weighed and dissolved in 1 ml of double distilled water (stock). From stock, 5 μl (25 μg) was taken and mixed with 10 μl of sample buffer for electrophoresis.
Bovine Serum Albumin (BSA) fraction-V
BSA (1 mg) was weighed and dissolved in 1 ml of double distilled water (stock). From this 3 μl (3 μg) was taken and mixed with 10 μl of sample buffer for electrophoresis.
Haemoglobin from Ox RBC
Ox RBC was collected from the Corporation Slaughter House, Perambur, Chennai. It was collected in Alsever’s solution. The RBC was washed thrice with 0.9% physiological saline by centrifugation at 500×g for 10 min. From the suspension, 10 μl of RBC was taken and diluted to 2 fold with 25 mM NaCl. This solution was incubated for 30 min at Room Temperature (RT), centrifuged at 10,000 rpm for 15 min. From this 2 μl (28μg) of supernatant was directly loaded on the gel.
Haemolymph
A healthy fifth instar silkworm (Bombyx mori) larva was wiped on the ventral portion with ethanol. With the help of sterile scissors, the foreleg was cut and the oozed out haemolymph was collected in vial containing Phenylthiourea (PTU) to prevent melanisation. It was centrifuged at 10,000 rpm for 10 min. The supernatant (1 μl equivalent to 18 μg of protein) was directly loaded on the gel.
Analysis of protein samples in Native-PAGE
The protein samples were analyzed using native PAGE [17]. The gel plates were assembled according to the manufacturer’s instruction and the volume of the gel mould was determined (170 x 150 x 1.5 mm). In a beaker, the resolving gel mixture for 8% (2.70 ml of 30% acrylamide stock + 2.50 ml of 1.5 M Tris HCl (pH 7.8, 8.3, 8.8 and 9.3), + 4.60 ml of double distilled water + 100 μl of 10% ammonium per sulphate + 6 μl of TEMED) was prepared. The components were mixed thoroughly and without delay the resolving gel mixture was poured into the glass mould and overlaid with distilled water carefully. After polymerization (approximately 20 minutes), the overlay was removed. 5% stacking gel was prepared by mixing 0.67 ml of 30% acrylamide stock, 0.50 ml of 0.5 M Tris HCl, pH 6.8, 2.70 ml of distilled water, 40 μl of 10% ammonium per sulphate and 4 μl of TEMED. These components were mixed and poured over the resolving gel. Immediately a comb was inserted and the gel was allowed to polymerize (approximately 15 min). Once it got polymerized, comb was removed and lanes were washed with distilled water. The gel mould was placed in an electrophoresis apparatus and completely filled with electrophoresis (tank) buffer (pH 8.3, 8.8 and 9.3).
Using a gel loading tips, lysozyme (25 μg) was mixed with 10 μl of sample buffer and loaded in the I lane. Ox haemoglobin (20 μg) was mixed with 10 μl of sample buffer and loaded in the II lane. BSA (3 μg) was mixed with 10 μl of sample buffer and loaded in the III lane. Haemolymph (18 μg) was mixed with 10 μl of sample buffer and loaded in the IV lane.
Electrophoresis at different combinations of pH in electrophoresis (tank) and resolving gel buffers.
In one set of experiment, the pH of the resolving gel was kept constant at 7.8 and the pH of the electrophoresis buffer was changed to 8.3, 8.8 and 9.3. In the second experiment, the pH of the resolving gel was kept at 8.3 and the pH of the electrophoresis buffer was changed to 8.3, 8.8 and 9.3. In the third experiment, the pH of the resolving gel was kept at 8.8 and the pH of the electrophoresis buffer was changed to 8.3, 8.8 and 9.3. In the fourth set, the pH of the resolving gel was kept at 9.3 and the pH of the electrophoresis buffer was changed to 8.3, 8.8 and 9.3.
The apparatus was plugged to the power supply unit taking care of anode and cathode wire plugged properly. Initially the voltage was set at 60 for stacking gel and later 120V for resolving gel. The power supply was turned off, once the bromophenol blue reached the bottom of the resolving gel. The gel mould was removed apart using a spatula. The orientation of the gel was marked. The gel was stained with staining solution. Each box was labelled for identification. After 4 hours, each gel was washed with distilled water and destaining solution was poured. Destaining solution was changed until the background was cleared. It was stored in 7% acetic acid. The bands were visualized using an illuminator and then photographed.
Results
Silkworm haemolymph sample, hemoglobin from ox RBC and two commercially purchased proteins such as lysozyme and BSA were used in the study to show the influence of pH in the buffer systems used in native-PAGE. The results obtained were presented in the order of constant pH of resolving gel buffer with differing pH of tank buffers. At a resolving gel buffer of pH 8.8, three different pH of electrophoresis or tank buffers (pH: 8.3, 8.8 and 9.3) were used to separate various samples in the acrylamide gel. The native gel system comprised of resolving gel pH 8.8 with a combination of tank buffer pH 8.3 separated lysozyme, haemoglobin, BSA and the silkworm haemolypmph proteins better than other pH variants of tank buffers (i.e., pH 8.8 & 9.3) (Figure 1A). The increasing pH of tank buffers made proteins to diffuse over the gel and separate poorly in native- PAGE (Figures 1B and 1C).
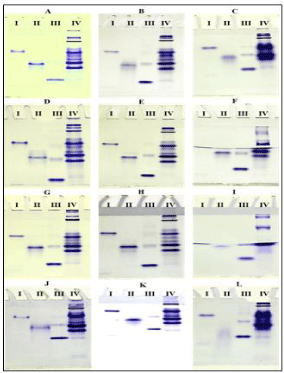
Figure 1: Protein profile of various samples in Native-PAGE (RGB –
Resolving Gel Buffer; TB – Tank Buffer). (A) RGB - pH 8.8, TB – pH 8.3 (B)
RGB - pH 8.8, TB – pH 8.8 (C) RGB - pH 8.8, TB – pH 9.3 (D) RGB - pH 7.8,
TB – pH 8.3 (E) RGB - pH 7.8, TB – pH 8.8 (F) RGB - pH 7.8, TB – pH 9.3 (G)
RGB - pH 8.3, TB – pH 8.3 (H) RGB - pH 8.3, TB – pH 8.8 (I) RGB - pH 8.3,
TB – pH 9.3 (J) RGB - pH 9.3, TB – pH 8.3 (K) RGB - pH 9.3, TB – pH 8.8 (L)
RGB - pH 9.3, TB – pH 9.3. I. Lysozyme (14.6 kDa, pI 11); II. Haemoglobin
(64 kDa, pI 7.3); III. BSA (66.3 kDa, pI 4.9); IV. Silkworm haemolymph.
Different proteins in the haemolymph and other standard proteins were resolved much better with combination of resolving gel (pH 7.8) and both pH of tank buffers (i.e., 8.3 & 8.8). Individual protein molecules were also dense and sharply packed in the gel (Figures 1D and 1E). But at the same pH of resolving gel, the pH 9.3 of tank buffer showed diffusion, damage of lanes and disappearance of some protein molecules (Figure 1F). At pH 8.3 of resolving gel buffer and tank buffer, there was incomplete transfer of haemolymph proteins from the stacking gel (Figure. 1G). But, electrophoresis of various silkworm haemolymph proteins in the case of resolving gel buffer at pH 8.3 and tank buffer pH 8.8 showed a better resolution with dense packing of individual protein fractions (Figure 1H). Native-PAGE with a combination of resolving gel buffer and tank buffer at pH 8.3 and 9.3, respectively showed diffusion and disappearance of proteins (Figure 1I). At a high pH of 9.3 in resolving gel buffer, the native-PAGE failed to show a good profile of various proteins in all the three different pH variants of tank buffer 8.3, 8.8 and 9.3 (Figures 1J, 1K and 1L).
Discussion
In the present study, the influence of hydrogen ion concentrations (pH) of tank buffer and resolving gel buffer on vertical gel electrophoresis system was demonstrated. Proteins are biomolecules that migrate in the electric field on the basis of their net charge, mass and size. This principle of gel electrophoresis sets an interesting hypothesis that the net charge is greatly influenced by the pH of the buffers used in the electrophoresis system [9,18]. The state of ionization of the proteins is also dependent on the group of amino acids they possess, which is either acidic or basic or neutral. The ionization grossly differs at a particular pH [13,19]. Under standard electrophoresis conditions, the pH of the resolving gel buffer would be 8.8, and the electrophoresis or tank buffer would be 8.3. This condition is usually an ideal condition for most of the proteins, because at this pH, 95% of the ionization is achieved. In the present study, BSA and lysozyme whose molecular weight 66.3 kDa and 14.6 kDa, respectively were used for electrophoresis with different combinations of pH of both resolving gel and tank buffers. BSA being high molecular weight than lysozyme, BSA should electrophorese at the top in gel than lysozyme. This was not observed in Figure. 1A because lysozyme gets ionized faster when compared to BSA. Likewise, when the pH of the tank buffer was increased to 8.8, there was partial diffusion of haemoglobin, and when the pH was increased to 9.3, there was a complete diffusion or either denaturation of the haemoglobin. This might be due to the slow migration of haemoglobin. The resolution of proteins in haemolymph of silkworm was not clear at higher pH of tank buffer. At a state of resolving gel buffer pH 8.3 and the tank buffer pH 8.8, high resolution was observed in all the samples that were loaded in native-PAGE and there was no lagging of protein in stacking gel. But, when the pH of the tank buffer was reduced to 8.3, there was some lagging of proteins observed in silkworm haemolymph proteins. This might be due to the improper ionization of proteins in the stacking gel region. At the same time, when the pH of the tank buffer was increased to 9.3, resolution of proteins in resolving gel was too slow and it electrophoresed improperly. This is because the trailing ions (glycine) are present at very low concentration. Correspondingly, the same results were obtained when the pH of the resolving gel buffer was lowered to pH 7.8 and tank buffer pH was set to 9.3. Subsequently, when the pH of the tank buffer was set to 8.8 and 8.3, clear resolutions of protein profiles were observed. There was no lagging of proteins in stacking gel.
Finally, when the pH of the resolving gel buffer and the tank buffer were raised to 9.3, there was poor resolution of proteins and this could be because of high proportion of chlorine and tris ions when compared to the glycine ions and hence ineffective ionization. When the pH of the tank buffer was 8.8 and 8.3, there was good resolution in the protein profile. Throughout the study the tertiary structure of BSA remained unaltered; this is because BSA is shown to keep its structure unaltered at wide range of pH varying from 4.0 to 9.0, even without undergoing any significant conformational change [20]. It is clear from the study that hydrogen ion concentrations (pH) play a very distinct role in migration of proteins. Our observations clearly showed that pH 9.3 of tank buffer was not suitable for the samples used in the present study. Likewise, buffers with low pH i.e., less than 8.0 made the irregular pattern of protein profile in the native-PAGE. The pH range of 8.3 to 8.8 in buffers showed clear protein profile in PAGE.
References
- Westermeier R. Vertical PAGE. Electrophoresis in Practice. Weinheim, Germany: Wiley-VCH. 2001; 201.
- Nelson DL, Lehninger AL, Cox MM. Lehninger Principles of Biochemistry. New York: Macmillan, Freeman Company. 2004; 72.
- Dunn MJ. Gel Electrophoresis of Proteins. Techno House, Redcliffe way, Bristol, England: IOP Publishing Ltd. 2014.
- Swank RT, Munkres KD. Molecular weight analysis of oligopeptides by electrophoresis in polyacrylamide gel with sodium dodecyl sulfate. Anal Biochem. 1971; 39: 462-477.
- Chiari M, Righetti PG. New types of separation matrices for electrophoresis. Electrophoresis. 1995; 16: 1815-1829.
- Shoji M, Kato M, Hashizume S. Electrophoretic recovery of proteins from polyacrylamide gel. J Chromatogr A. 1995; 698: 145-162.
- Pomastowski P, Buszewski B. Two-dimensional gel electrophoresis in the light of new developments. Trac-Trend Anal Chem. 2014; 53: 167-177.
- Yamaoka T. Pore gradient gel electrophoresis: theory, practice, and applications. Anal Chim Acta. 1998; 372: 91-98.
- Shaw KL, Grimsley GR, Yakovlev GI, Makarov AA, Pace CN. The effect of net charge on the solubility, activity, and stability of ribonuclease Sa. Protein Sci. 2001; 10: 1206-1215.
- McLellan T. Electrophoresis buffers for polyacrylamide gels at various pH. Anal Biochem. 1982; 126: 94-99.
- Cooper TG. The Tools of Biochemistry. New Delhi, India: Wiley India Pvt. Ltd. 1977; 194.
- Westermeier RM, Naven T, Hopker HR. Proteomics in Practice. 2nd edn. Weinheim, Germany: Wiley-VCH. 2008.
- Geisthardt D, Kruppa J. Polyacrylamide gel electrophoresis: reaction of acrylamide at alkaline pH with buffer components and proteins. Anal Biochem. 1987; 160: 184-191.
- Hachmann JP, Amshey JW. Models of protein modification in Tris-glycine and neutral pH Bis-Tris gels during electrophoresis: Effect of gel pH. Anal Biochem. 2005; 342: 237-245.
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with Folin phenol reagent. J Biol Chem. 1951; 193: 265-275.
- Garvey JS, Cremer NE, Sussdorf DH. Methods in Immunology. 3rd edn. Massachusetts: Benjamin WA. Inc. Reading. 1978; 545.
- Maurer HR. Disc Electrophoresis and Related Techniques of Polyacrylamide Gel Electrophoresis. New York: Walter de Gruyter. 1972.
- Zewert TE, Harrington MG. Cross-linked poly (N-acetylethylenimine) as an isoelectric focusing matrix. Electrophoresis. 1999; 20: 1339-1348.
- Righetti PG. Macroporous gels: facts and misfacts. J Chromatogr A. 1995; 698: 3-17.
- Barbosa LR, Ortore MG, Spinozzi F, Mariani P, Bernstorff S, Itri R. The importance of protein-protein interactions on the pH-induced conformational changes of bovine serum albumin: a small-angle X-ray scattering study. Biophys J. 2010; 98: 147-157.