Research Article
Austin J Clin Immunol. 2014;1(3): 1015.
Molecular Identification of an MHC Class Ib (H2-Q9) Restricted T Cell Receptor Specific for a Mouse Polyomavirus Peptide VP2.139
Zemin Zhou1, Xiao He1* and Peter E Jensen1,2
1Department of Pathology, University of Utah, USA
2ARUP Laboratories, University of Utah, USA
*Corresponding author: Xiao He, Department of Pathology, University of Utah, 15 North Medical Drive East, Suite 1200, Salt Lake City, UT 84112-5650, USA
Received: February 22, 2014; Accepted: April 29, 2014; Published: May 01, 2014
Abstract
The murine major his to compatibility complex (MHC) class Ib region encodes more than 20proteins with diverse biological functions, and many have not been characterized. Evidence emerging in recent years indicates that MHC class Ib–restricted CD8 T cells can play an important role in antiviral immunity. We molecularly cloned an α⁄β T cell receptor (TCR) from a T cell hydridoma cell line, that is restricted by an MHC class Ib molecule H2–Q9, and specifically recognizes the VP2.139 peptide epitope from mouse polyoma virus (PyV). We further demonstrated that the cloned TCR, using TCRα10 and TCRβ4 genes, is functional and mediates the development of Q9⁄VP2.139–restricted CD8 T cells in vivo. The cloning of a H2–Q9–restricted, PyV–specific α⁄β TCR provides a useful tool for future study of the potential role of MHC class Ib restricted CD8 T cells in antiviral immunity.
Keywords: MHC Class Ib; Qa–2; H2–Q9; CD8+ T cell; TCR; Polyomavirus
Introduction
Major his to compatibility complex (MHC) class I proteins play an important role in adaptive immunity [1,2]. They provide a mechanism of immune surveillance by presenting intracellular antigens from either self or pathogenicorigin to CD8 T cells, natural killer (NK) cells and⁄or natural killer T (NKT) cells [1,3–5]. The functions of the highly polymorphic classical MHC class I (MHC class Ia) molecules, including H2–K, –D and –L in mouse and HLA–A, –B and –C in human, are well established [2,6]; however, most of the non–classical MHC class I (MHC class Ib) molecules, with limited polymorphism, are not well studied [6,7]. The Qa–2 antigen (encoded by the H2–Q6, 7, 8, 9) is one of the best characterized class Ib molecules in mouse (possibly homologous to HLA–G in human). H2–Q9 encodes two typesof Qa–2 isoforms, membrane–bound and soluble, through alternative splicing of transcripts [8]. Multiple biological functions of Q9, as well as that of HLA–G in human, have been documented, such as participating in preimplantation embryo development [9,10], tumor immunity [11,12], and antiviral immunity [13]. However, the selection of CD8+ T cells by Q9 and the characteristics of Q9–restricted CD8+ T cells are poor understood.
Cytotoxic CD8+ T cells directly eradicate or control intracellular pathogens by killing infected cells or by releasing cytokines to inhibit further spread of infection [14]. The selection and development of antiviral CD8+ T cells by class Ia molecules has been well documented [1,2]. However, the role of class Ib–restricted antiviral CD8+ T cells needs further investigation. The potential mechanismsofcontrol of viral infection by class Ib–restricted CD8+ T cells areunclear and the development pathways for this subset of CD8+ T cells arepoorly defined. Recently, CD8+ T cells have been identified that can recognize viral epitopes presented by MHC class Ib molecules [15–17], and there is evidence that class Ib–restricted CD8+ T cells contribute to the control or eradication of virus infection [13,18,19].
Polyomaviruses are double–stranded DNA viruses, persistently infecting humans or animals, and potentially inducing tumors in immune compromised individuals [20]. Since the 1970s, at least 6 polyomaviruses, including BK virus, JC virus, KI polyoma virus, WU polyoma virus, Merkel cell polyomavirus and trichodysplasiaspinulosa virus, have been identified in humans, [20–24]. Mouse polyomavirus (PyV) was the first identified polyomavirus [20,25] and it can induce tumors in immune compromised mouse strains [25,26]. T antigens of PyV can induce class Ia–restricted CD8+ T cell responses [27,28]. Recently, Swanson et al. successfully identified MHC class Ib, Q9– restricted CD8+ T cells that responded to a PyV–derived epitopeVP2.139 [13], which provided the first solid evidence demonstrating that class Ib–restricted CD8+ T cells might play a role in controlling persistent viral infection.
In the current study, we molecularly identified a T cell receptor (TCR) from Q9⁄VP2.139–specific CD8+ T cells and confirmed its function in transducedcells as well as in retrogenic mice. To our knowledge, this was the first identified MHC class Ib–restricted TCR that responds to viral infection.
Materials and Methods
Mice
C57BL⁄6J and Rag1–⁄– mice were purchased from the Jackson Laboratory. OT–I TCR transgenic mice, with specificity to OVA357– 364⁄Kb(OVA–I⁄Kb), have been previously described [29]. Kb–⁄–Db–⁄– CIITA–⁄– mice were generated in our laboratory as previously described [30]. Eight– to 12–week–old mice were used in the experiments. Mice were bred and housed in a specific pathogen–free facility at the University of Utah and were handled according to the Institutional Animal Care and Use Committee policies.
Cell lines and culture conditions
The Q9⁄VP2.139–specific C1–21 T cell clone and T cell hybridoma lines C3K and 3H6, and the recipient linesZ6.21–3B2.E7 and BWZwere provided by Dr. Aron Lukacher (Emory University).The recipient cell lines58α–⁄β– and 5KC⁄CD4were provided by Dr. Piotr Kraj (Georgia Regents University) and maintained as previously described [31]. 3H6 T cell hybridoma was further sorted into 3H6 (+) and (−), based on the presence or absence of surface expression of TCR Vα2. 3H6 (–) cells, which did not produce IL–2 upon stimulation of VP2.139 peptides were used as recipient cells in retroviral transductionexperiments. The retroviral packaging cell line Plat–E was described previously [32]. The C1–21 T cell clone was maintained in IMDM with 10% FBS (Hybrid), 10 U⁄ml of mouse IL–2, 10 uM of VP2.139 peptides and irradiated Kb–⁄–Db–⁄–CIITA–⁄– mouse splenocytes as antigen presenting cells (APCs).C3K and 3H6 T cell hybridomas, recipient cell line sand Plat–E packaging cell lineswere maintained in DMEM with 10% FBS, supplemented with 100 U⁄ml of penicillin, 100 mg⁄ml of streptomycin, 292 mg⁄ml of L–glutamine, 100 mM of nonessential amino acids, 1 mM of sodium pyruvate, and 55 uMof 2–ME (Invitrogen),as complete DMEM as previously described [18]. Cells were maintained in a humidified 37oC incubator supplied with 5% CO2.
Generation of soluble H2–Kb, H2–Q9 proteins and Q9⁄VP2.139 tetramer
The peptides L19 (ILMEHIHKL), VP2.139 (HALNVVHDW), OVA–I (SIINFEKL) were synthesized, and substitutedL19–H5C, VP2.139–V5C and OVA–I–N4C peptides were synthesized and biotinylatedat the core facility ofUniversity of Utah. Soluble H2– Kb and H2–Q9 proteins were purified from E. coli (BL21)–derived proteins and refolded with the appropriated peptides in a 1:1 molar ratioto produceQ9⁄L19, Q9⁄VP2.139 or Kb⁄OVA–I monomers, using ourpreviously describedmethod [33]. To generate Q9⁄VP2 tetramers, refolded Q9⁄VP2.139 monomer was biotinylated and tetramerized with streptavidin–APC (Invitrogen) in a 4:1 molar ratio as previously described [13,34]. Peptide dissociation rates and MHC–peptide complex half–liveswere measured as previously described [33,35]. Briefly, Q9⁄L19, Q9⁄VP2.139 or Kb⁄OVA–I complex (50 nM) was incubated with biotin–L19, biotin–VP2.139 or biotin–OVA–I (1 μM) peptide, respectively, in PBS with 0.01% Nonidet P–40 at RT for overnight. Then, unlabeled L19, VP2.139 or OVA–I peptide (200 μM), respectively, was added and incubated at 37°C for the indicated lengths of time. The remained biotin–peptide that bound to the MHC class I molecules was measured by Eu–ELISAusing an anti–β2m capture mAb [35].
Abs and flow cytometry
Fluorophore–conjugated mAbs to mouse CD4 (H129.19), CD8 (RPA–T8), CD62L (MEL–14), Vα2 (B20.1), Vα3.2 (B21.14), Vα8.3(RR3–16), Vα11(RR8–1), isotype–matched control mAbs,and a mouse Vβ TCR Screening Panel (557004) were purchased from BD Pharmingen. Cell surface staining and Q9⁄VP2 tetramer staining were performed according to the standard procedures. Stained cells were analyzed on a FACS can flow cytometer (BD Bioscience) and data were analyzed using Flow Jo 8.4 software.
Cloning of the TCR α chain and β chain gene and generation of expression constructs
Total mRNA of C3K hybridoma cells was extracted using the RN easy Kit (Qiagen) and the cDNA was reversely transcribed, with the Super ScriptTM III Reverse Transcriptase Kit (Invitrogen) following the manufacture’s instructions, for further cloning.The TCRExpress™ mouse TCR Vα screening kit was purchased from BioMed Immunotech. TCR variable region prediction and open reading frame analysis were performed using the IMGT website for mouse TCR analysis (http:⁄⁄www.imgt.org⁄IMGT_vquest⁄), as well as the sequences previously published [36–38]. The primers used for cloning of the full length TCRα10 and TCRβ4 cloning were listed as below:
TCRα10 forward (TCRVα10f–F): 5’–GTGCGGCCGCGTTT A A A C A T G A A G A G G C T G C T G T G C T C T C T G C – 3 ’ , TCRα10 reverse (TCRCαf–R): 5’–TTCCTGCAGGGTT TA AACTCAACTGGACCACAGCCTCAG–3’; TCRβ4 forward (TCRVb4f–F): 5’– GCTTAATTAAGTTTAAACATGGGCTCCA TTTTCCTCAGTTG–3’, TCRβ4 reverse (TCRCb1fR–R): 5’– ATGGCGCGCCGTTTAAACTCATGAATTCTTTCTTTTGA CCATAGCC–3’. To co–express TCRα10 and TCRβ4, we fused full–length TCRα10 (excluding stop codon) and TCRβ4 genes with T2A (GAGGGCAGAGGAAGTCTTCTAACATGCGGTG ACGTGGAGGAGAATCCCGGCCCT) sequence [39], using primers TRA10–2A–R (5’–CTCCTCCACGTCACCGCATGTT AGAAGAC T T C C T C TGC C C T CGT TAACAC TGGAC CACAGCCTCAGCG–3’) and 2A–TRB4 –F (5’–CTTCTAAC ATGCGGTGACGTGGAGGAGAATCCCGGCCCTATGG GCTCCATTTTCCTCAGTTGC–3’), paired with TCRVα10f–F or TCRCβ1fR–R, respectively, in two rounds of PCR amplification,to generate a TCRα10–T2A–β4 (C3K) construct. All of these PCR products were amplified with the Platinum® Taq DNA Polymerase High FidelityPCR Kit (Invitrogen) following the manufacture’sinstruction and cloned into the pCR2.1 vector using the TOPO® TA CloningKit (Invitrogen).The DNA sequences were determined by sequencing(the Gen Bank accession number: KJ755332). Then, the TCRα10–T2A–β4 construct in the pCR2.1 vector were digested with PmeI (Roche) and further cloned into a retroviral vector MigR1 as previously described [40,41]. The OT–I TCR construct TCRα2–T2A–β5.2 in MigR1,used as a positive control in generation of the retrogenicmice, was a gift from Dr. KateVignali (St. Jude Children’s Research Hospital).
Retroviral transfection and retrogenic mouse generation
Retroviral supernatants were generated by transfection of Plat–E cells with MigR1–TCRα10–T2A–β4 (C3K) or MigR1–TCRα2– T2A–β5.2 (OT–I) as previously described [32,41,42]. The C3K and OT–I retrogenic mice were generated as previously described [43]. Briefly, 5x107bone marrow cells were harvested from 6–week–old Rag1–⁄–mouseand cultured in 15 ml of complete DMEM with 20% FBS, supplemented with IL3 (20 ng⁄ml), IL–6 (50 ng⁄ml) and SCF (50 ng⁄ml) (R&D systems, Inc) for 48 hours., The infected bone marrow cells were re–infected by adding 5 ml of fresh C3K or OT–I retroviral supernatants produced from Plat–E transfectants. Four days post–transduction, semi–lethally irradiated (450 rad) Rag1–⁄– recipient micewerereconstituted with the transduced bone marrow cells, following the previously described protocol [43]. Five–weeks post reconstitution, peripheral bloodsamples werecollected and analyzed for GFP expression and cell surface staining. Six weeks postreconstitution, retrogenic mice were sacrificed for analysis.
T cell activation assay and measurement of IL–2 production
To stimulateC1–21 T cells, 3H6 and C3K T cell hybridoma cells, or the TCR transductant, 2x105 cells (per well in a 96–well plate), with 2x105of irradiated Kb–⁄–Db–⁄–CIITA–⁄–splenocytes, which only expresses MHC Ib [30], as APCs, and 0, 1, or 10 uM of VP2.139,OVA–I or L19 peptides, were added. For retrogenic mice T cell activation assay, 4x105 cellsper well of C3K retrogenic or OT–ITCR transgenic mouse splenocytes were cultured with 0, 0.5, 1.0, 2.0, or 4.0 uM of VP2.139,OVA–I or L19 peptides. Eighteenhourslater, the supernatants were harvested and measured for mouse IL–2 production by the standard IL–2 ELISA procedure (eBioscience).
Results
Characterization ofQ9⁄VP2.139complex stability andT cell specificity
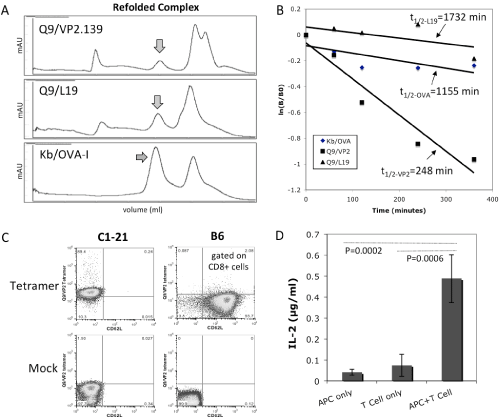
It has been shown previously that VP2.139 peptide bound to Q9 canstimulate specific CD8+ T cells during mouse polyomavirus infection [13]. In order to further characterize the VP2.139 peptide binding capacity with Q9 and to generate a Q9⁄VP2.139 tetramer reagent for staining of T cells, soluble recombinant Q9 protein (α1 toα3 domains without the transmembrane and cytoplasmic domains) was expressed in E.coli and the inclusion body was purifiedand refolded with VP2.139 peptide or L19 peptide(as acontrol), a known Q9 bound endogenous peptide derived from mouse 60S ribosomal protein [44,45]. In addition, soluble Kb [33]was prepared using the same method (Figure 1A). Interestingly, the Q9⁄VP2.139 complex has relatively lower complex stability with a half–life of 248 min, compared with the complex of Q9⁄L19 or Kb⁄OVA–I with half–life’s of 1732 min or 1155 min, respectively (Figure 1B), which is consistent with previously observations [46].
Figure 1: Generation of Q9⁄VP2.139 peptide complexes and characterization of C3K, a VP2.139 peptide specific CD8 T cell hybridoma line. (A) Refolding of soluble Q9 with a polyomavirus–derived VP2.139 peptide, or L19 peptide, and of Kb with OVA–I peptide as a positive control. The arrows indicated the refolded MHC–I⁄peptide complexes. (B) Comparison of peptide dissociation rates. The arrows show the calculated half–lives (t1⁄2) of biotin–labeled peptides dissociated from the MHC–I⁄peptide complexes. (C) Cell surface staining of Q9⁄VP2.139–specific CD8 T cell clone by Q9⁄VP2 tetramers. Non–biotinylated Q9⁄VP2.139 monomers mixed with SA–APC, and CD62L mAb matched isotype control, were used for the mock staining. (D) T cell activation assay by measuring the secreted IL–2 cytokine for C1–21–derived C3K T cell hybridomacells stimulated with 2x105 cells per well, using Kb–⁄–Db–⁄–CIITA–⁄–splenocytes as APCs and 10 uM of VP2.139 peptides.
The C3K T cell hybridoma used for TCR cloning was derived from a previously reported Q9⁄VP2.139–specific CD8+ T cell clone, C1–21[13], which was also confirmed for its TCR binding specificity using our Q9⁄VP2.139 tetramer in staining (Figure 1C). When stimulated with the VP2.139 peptide, the C3K T cell hybridoma cells were activated to produce IL–2 cytokine (Figure 1D), indicating that the TCR used in C3K was the same as its parental CTL clone C1–21. Thus the C3K T cell hybridoma line was used to clone the α and β chains of its TCR.
Cloning of the α and β chains of TCR from C3K T cell hybridoma
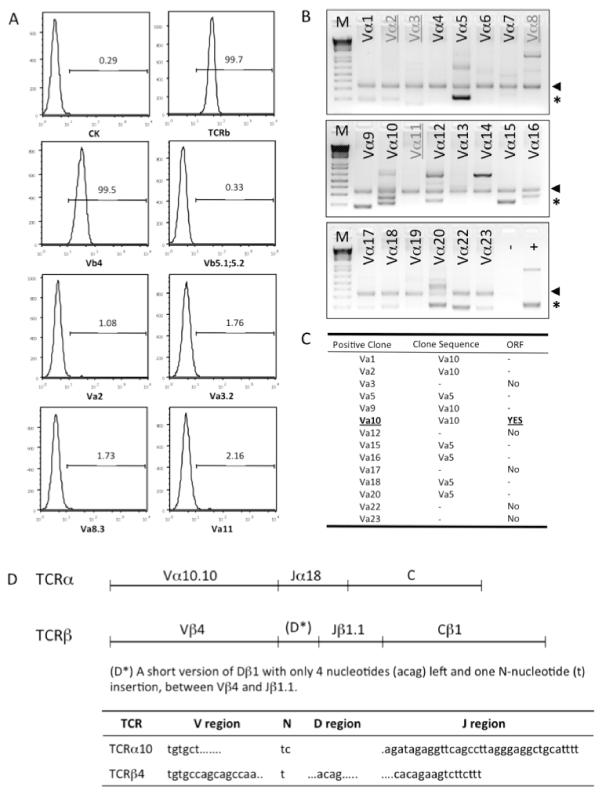
To identify the α and β chains of TCR used by C3K T cell hybridoma, C3K cells were firstly surface stained for the TCR Vβusage by usinga mouse Vβ TCR Screening Panel. As shown in Figure 2A, only anti–TCR Vβ4 mAβ positively stained the C3K cells, among all of the 17 anti–TCR Vβ mAβs included in the Screening Panel, but none of the commercially available anti–TCR Vα antibodies, V&aVα2, Vα3.2, Vα8.3 ανδ Vα11,stainedthe C3K cells. In order to ermine the TCRα chain usage of C3K, we next screened the TCR Vα region by PCR using a TCR Express™ mouse TCR Vα screening kit. Initially severalVα regions were positivelyamplified (Figure 2B). After sequencing the PCR products; we found that many of the PCR products were from non–specific amplification due to the sequence similarity, such as Vα1, Vα2 and Vα9. Vα5 was another transcript amplified in several different reactions (Vα5, Vα15, Vα16, Vα18 and Vα20), but there was no open reading frame (ORF) predicted at the translation level (Figure 2C). Therefore, we concludedthat TCRα10 and TCRβ4 were used in C3K.
Figure 2: Cloning of T cell receptor from C3K T cell hybridoma. (A) Cell surface staining to screen usage of TCRα and TCRβ in C3K T cell hybridoma. (B) PCR screening of the variable region(s) of TCRα chain expressed in C3K T cell hybridoma. The arrows show the internal control designed in the TCR PCR screening kit (BioMed Immunotech). The asterisks indicate the positive PCR products. All of the positive bands were purified and cloned into a TA cloning vector and the sequence was determined by sequencing. The signs of (−) and (+) indicate the negative and positive PCR controls. (C) Identification of Vα10 as the variable region used in C3K T cell hybridoma by sequencing analysis and open reading frame (ORF) prediction. Some of the amplified bands were proved to be non-specific PCR amplification products, confirmed by sequencing (−) and without ORF (no). (D) The detailed usage of V, D, J and C regions in full length of TCRα and TCRβ chains cloned from C3K T cell hybridoma, showed in upper panel. The lower panel shows the information of P– and N–nucleotides and the V and J regions coding the CDR3 region of the TCRα10 and TCR β4 chains.
Next, the full–length genes of both TCRα10 and TCRβ4 were cloned from C3K cDNA. Sequence analysis confirmed that the variable region of C3K TCRα contained Vα10.10 and Jα18, and that of C3K TCRβcontained Vβ4, Dβ1 and Jβ1.1 with the constant region of Cβ1. Interestingly, only a very short D region of Dβ1 (acag) with one N–nucleoside (t) insertion was detected between Vβ4 and Jβ1.1. Both TCRα10 and TCRβ4 from C3K showed open reading frames inthe correct frame (Figure 2D), with a 14aa of coding complementarity determining region 3 (CDR3) sequence, CASDRGSALGRLHF, for TCRα10, and a 12aa of CDR3 sequence, CASSQYSTEVFF, for TCRβ4, respectively.
Verification of the function of cloned TCRα10 and TCRβ4 incell lines
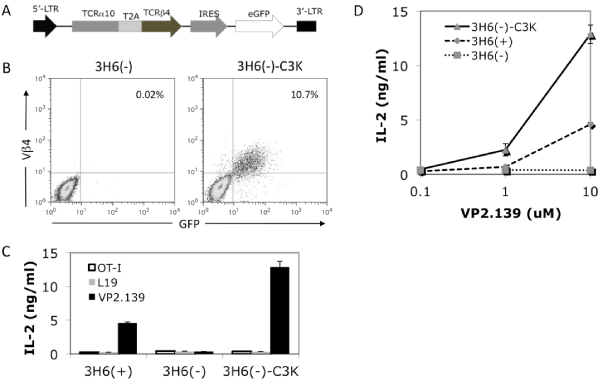
In general, a given T cell expresses only one TCR α chain and β chain pair. However, a single T cell can express more than one α chain [47–49]. To test whether the cloned C3K TCR α and βchains indeed pair with each other and are functional, we tried to express the construct of TCRα10–T2A–β4–MigR1, which also contained an eGFP reporter driven by an IRES (Figure 3A), in several cell lines, including 58α–⁄β–, 5KC⁄CD4, Z6.21–3B2.E7 and BWZ. Although pairing and surface expression of the TCR was observed in several of these cell lines, responsiveness to the VP2.139 peptide was not conferred (data not shown).
Figure 3: Expression and function of the cloned TCR in a T cell hybridoma line. (A) The diagram shows the construct used in retroviral transfection of 3H6(–) to express TCRα10 and TCRβ4 fused by T2A sequence with eGFP as a reporter driven by IRES. (B) Cell surface staining of TCR Vβ4 expression on transduced 3H6(–) cells. 3H6(–)–C3K represent the 3H6(–) cells transfected with TCRα10–β4 genes cloned from C3K T cell hybridoma. (C) IL–2 production of untransfected or transfected 3H6(–) and 3H6(+) cells stimulated with different peptides at 10 uM, and (D) dose responses of IL–2 production of untransfected or transfected 3H6(–) and 3H6(+) cells upon stimulation with 0 uM, 1 uM or 10 uM. Both (C) and (D) use Kb–⁄–Db–⁄–CIITA–⁄– splenocytes as APCs. The recipient cell line, 3H6(–), was isolated from another non–clonal VP2.139 peptide responsible T cell hybridoma 3H6, which has different TCR usage (data not shown) from the C3K T cell hybridoma. The population of Vα2 positive, 3H6(+), respond to VP2.139 stimulation, but not the population of Vα2 negative, 3H6(–).
The non–clonal T cell hybridoma 3H6 is derived from another Q9⁄VP2.139–responsive T cell clone, C3–8 [13], but this hybridoma actually includes at least two populations, denoted as 3H6 (–) and 3H6 (+), sorted by the expression of TCR Vα2 (data not shown). The 3H6(–)cells did not express Vβ4 nor respond to VP2.139 peptide stimulation (Figure 3B, left panel, and Figure 3C).When 3H6(–) cells were retrovirally transduced with the same TCRα10–T2A– β4–MigR1 construct, as shown in Figure 3B, the cloned TCR could be expressed on the cell surface, as stained by anti–TCR Vβ4mAb. Upon stimulation by VP2.139 peptides, the transduced 3H6 (–) cells specifically produced IL–2 in a dose dependent manner. No IL–2 was produced when stimulated with either L19 or OVA–I (OT–I TCR specific) control peptides (Figures 3C and 3D). We also separately expressed the cloned TCRα10 or β4 single chain in 3H6 (–) cells. As expected, none of the cells transfected with single chain of α10 or β4 can be stimulated with VP2.139 peptides, while the cells sequentially transfected with both TCRα10 and β4 (in separate constructs) can be stimulated (data not shown). These results indicated that the TCRα10 and TCRβ4 genes cloned from C3K T cell hybridoma were functional, conferring specificity for the VP2.139 peptide. However, the possibility remained that an unknown factor expressed in 3H6 (–) cells was required for the ability to respond to Q9⁄VP2.139.
Functional CD8+ T cell development mediated by the cloned TCRa10 and TCRb4 in vivo
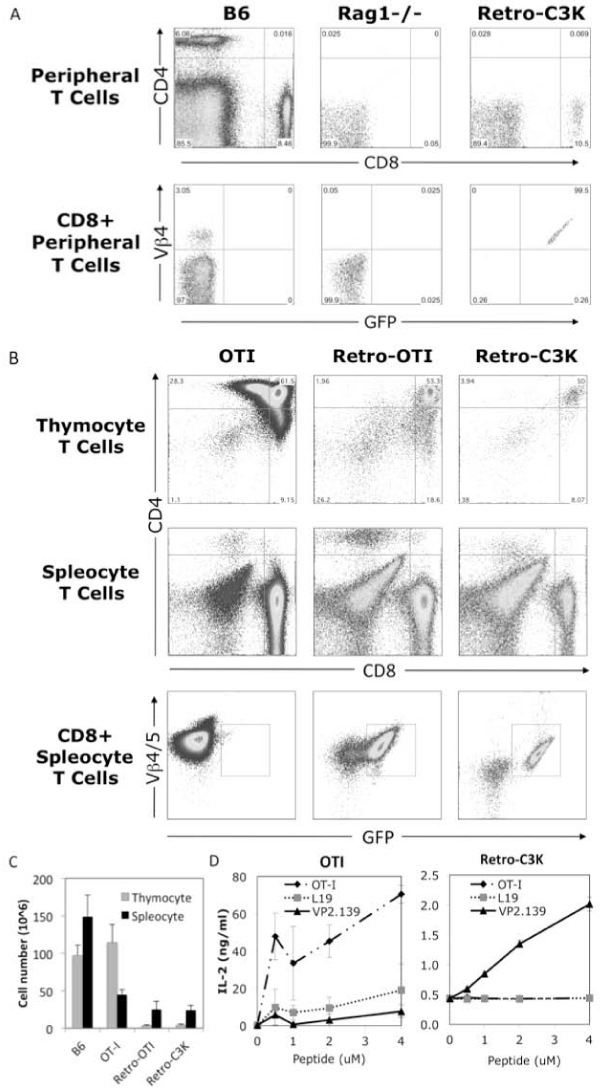
To further examine the function of the cloned C3K TCRα10 and β4genes, we generated retrogenic mice expressing the TCRα10– T2A–β4–MigR1 construct to test whether this TCR can specifically support the thymic selection and development ofCD8+ T cell, and mediate survival and function of T cells in the periphery. To generate C3Kretrogenic mice, semi–lethally irradiated Rag1–⁄– mice were reconstituted with retroviral transfected Rag1–⁄– bone marrow expressing the C3K TCR or the OT–I TCR(as a control).The C3K TCR specifically mediate the development of CD8+ T cells in recipient Rag1–⁄– mice and all of theCD8+ T cells from C3K retroviral mice were TCR Vβ4 and GFP double–positive, as shown by staining of peripheral blood from C3K retrogenic mice at 5 weeks post bone marrow reconstitution (Figure 4A), representing a population of CD8+ T cells thatdeveloped from the bone marrow T cell progenitors expressing the transfected C3K TCR. Further comparison of thymocytes and splenocytes in retrogenic mice showed that both the C3K TCR, as well as the control OT–1 TCR, rescued CD8+ T cell development in Rag1–⁄– mice (Figure 4B). The total thymocyte and splenocyte numbers were comparable between C3K and OT–I retrogenic mice, even though the numbers were lower when compared with wild type B6 or OT–I TCR transgenic mice (Figure 4C). Most importantly, the splenocytes from the C3K retrogenic mice responded specifically to stimulation of VP2.139 peptide to produce IL–2, but not to stimulation by control L19 or OT–I peptides. By contrast, the OT–I retrogenicsplenocytes were stimulated by the OT–I peptide, not the L19 or VP2.139 (Figure 4D). These results clearly demonstrated that the cloned C3K TCR geneswereable to support selection of CD8+ T cells in vivo, and that our cloned TCR genes conferspecificity to the VP2.139 peptide.
Firure 1: Generation of retrogenic mice expressing the cloned C3K TCR genes. (A) Surface staining of Vβ4 positive CD8+ T cells in peripheral blood of retrogenic mice in 5 weeks post bone marrow reconstitution. (B) Surface staining of Vß4 positive CD8+ T cells in thymus and spleens of C3K TCR retrogenic, OT–I TCR retrogenic and OT–I transgenic mice (as controls). (C) Total cell number of thymocyte and splenocyte in age–matched wild type B6, OT–I transgenic mice, OT–I and C3K TCR retrogenic mice. The data represents at least 3 mice in each group. (D) IL–2 production from the splenocytes of OT–I transgenic or C3K TCR retrogenic mice in vitro stimulated with 0, 0.5, 2 or 4 uM of OT–I, L19 or VP2.139 peptides.
Discussion
In this study, we molecularly identified a Q9⁄VP2.139–specific TCR and confirmed its function to respond to mouse PyV derived peptide stimulation. We also showedevidences that this TCR can be expressed in vivoand mediate specific CD8 T cell development. Thus we not only generated a useful tool in study of class Ib–restricted CD8 T cells in antiviral immunity, but also provide a unique system (MHC class Ib⁄peptide⁄TCR)to be used in the future study, such as the generation of an MHC class Ib restricted TCR transgenic mouse, only a few of which have been generated so far, and co–crystallization of MHC class Ib MHC⁄peptide⁄TCR for structural analysis.
Intriguingly, we notice a shorter half–life and relative instabilityof the Q9⁄VP2.139 complex, compared with a class Ia complex (Figure 1B) and Q9 bound to the L19 peptide, a high affinity endogenous selfpeptide [44,50]. This observation provides a newpiece of evidence consistent with the previous hypothesisthatthe dependence of CD28 signaling and continuous CD4 T cell help to expand Q9–restricted CD8 T cells is due to the weak interaction among MHC, peptide and TCR [46]. However, further evidence isrequired from a direct measurement of interaction among Q9, VP2.139 peptide and the cloned C3K TCR. It is plausible that the relative instability of Q9 and VP2.139 peptide might also weaken the interaction between pMHC ligand and TCR, such that it cannot fully stimulate the Q9⁄VP2.139– specific CD8 T cells. Consistent with this,these CD8 T cells express low level of inhibitory PD–1but comparable level of activating CD28 [46,51].
It was reported that upon VP2.139 peptide stimulation, the Q9⁄ VP2.139–specific CD8 T cells exhibited functional impairment in IFNγproduction [13], in contrast to robust IFNγ production often observed with class Ia–restricted CD8 T cells in antiviral immunity [52,53].We found that C3K T cells produce significant amount of IL–2 when stimulated with VP2.139 peptides (Figure 1D and Figure 4C). IL–2 is necessary for the growth, differentiation and survival of CD4 and CD8 T cells, especially memory T cell development [54– 56]. The observation of impaired production of IFNγ but maintaining IL–2 productionhas several implications for the potential role of class Ia–restricted CD8 T cells: (1) the intrinsic potency to develop into memory type of CD8 T cell; (2) help in promoting survival and memory development in other CD4 and CD8 T cells; (3) stimulationto prolongthe life of effector CD4 helper T cells, and especially promotion of effector MHC class Ia–restricted CD8 T cells with a capacityto control the persistent viral infection. Consistent with these proposed implications, it has been reported [57,13] that Q9⁄VP2.139–specific CD8 T cells are long lived, and express CD122, a component in CD122–dependent signaling critically involved in CD8 T cell immunity and memory [56,58]. However, further work is required to test these possibilities. Generation of transgenic mice expressing the C3K TCR should provide an opportunity to characterize the developmental pathway of Q9–restricted T cells, the requirements for positive and negative thymic selection, the requirements for expression of Q9 in T cell survival and homing, and the potential for Q9–restricted T cells to mediate viral clearance and establish immunological memory. Transgenic C3K T cells have the potential to provide a valuable tool to explore the potential of MHC class Ib–restricted T cells to participate in the anti–viral immune response.
Acknowledgement
We thank Dr. Aron Lukacher for providing C1–21 T cell clone, and C3K and 3H6, T cell hybridomas, Dr. Piotr Kraj for providing reagents and suggestions on TCR cloning and expression. Dr. Hai–tao He for providing Plat–E packaging cells.
References
- Jensen PE. Recent advances in antigen processing and presentation. Nat Immunol. 2007; 8: 1041-1048.
- Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011; 11: 823-836.
- Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011; 331: 44-49.
- Berzins SP, Smyth MJ, Baxter AG. Presumed guilty: natural killer T cell defects and human disease. Nat Rev Immunol. 2011; 11: 131-142.
- Salio M, Silk JD, Yvonne Jones E, Cerundolo V. Biology of CD1- and MR1-Restricted T Cells. Annu Rev Immunol. 2014; 32: 323-366.
- Adams EJ, Luoma AM. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu Rev Immunol. 2013; 31: 529-561.
- Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005; 5: 459-471.
- Tabaczewski P, Shirwan H, Lewis K, Stroynowski I. Alternative splicing of class Ib major histocompatibility complex transcripts in vivo leads to the expression of soluble Qa-2 molecules in murine blood. Proc Natl Acad Sci U S A. 1994; 91: 1883-1887.
- Comiskey M, Domino KE, Warner CM. HLA-G is found in lipid rafts and can act as a signaling molecule. Hum Immunol. 2007; 68: 1-11.
- Sun LL, Wang AM, Haines CJ, Han Y, Yao YQ. Down-regulation of HLA-G Attenuates Cleavage Rate in Human Triploid Embryos. J Reprod Infertil. 2011; 12: 215-220.
- Chiang EY, Stroynowski I. Protective immunity against disparate tumors is mediated by a nonpolymorphic MHC class I molecule. J Immunol. 2005; 174: 5367-5374.
- Gomes AQ, Correia DV, Silva-Santos B. Non-classical major histocompatibility complex proteins as determinants of tumour immunosurveillance. EMBO Rep. 2007; 8: 1024-1030.
- Swanson PA 2nd, Pack CD, Hadley A, Wang CR, Stroynowski I, Jensen PE, et al. An MHC class Ib-restricted CD8 T cell response confers antiviral immunity. J Exp Med. 2008; 205: 1647-1657.
- Yewdell JW, Haeryfar SM. Understanding presentation of viral antigens to CD8+ T cells in vivo: the key to rational vaccine design. Annu Rev Immunol. 2005; 23: 651-682.
- Milligan GN, Flaherty L, Braciale VL, Braciale TJ. Nonconventional (TL-encoded) major histocompatibility complex molecules present processed viral antigen to cytotoxic T lymphocytes. J Exp Med. 1991; 174: 133-138.
- Byers DE, Fischer Lindahl K. H2-M3 presents a nonformylated viral epitope to CTLs generated in vitro. J Immunol. 1998; 161: 90-96.
- Pietra G, Romagnani C, Mazzarino P, Falco M, Millo E, Moretta A, et al. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc Natl Acad Sci U S A. 2003; 100: 10896-10901.
- Chen L, Jay DC, Fairbanks JD, He X, Jensen PE. An MHC class Ib-restricted CD8+ T cell response to lymphocytic choriomeningitis virus. J Immunol. 2011; 187: 6463-6472.
- Braaten DC, McClellan JS, Messaoudi I, Tibbetts SA, McClellan KB, Nikolich-Zugich J, et al. Effective control of chronic gamma-herpesvirus infection by unconventional MHC Class Ia-independent CD8 T cells. PLoS Pathog. 2006; 2: e37.
- Pinto M, Dobson S. BK and JC virus: a review. J Infect. 2014; 68 Suppl 1: S2-8.
- Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, et al. Identification of a third human polyomavirus. J Virol. 2007; 81: 4130-4136.
- Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, et al. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007; 3: e64.
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008; 319: 1096-1100.
- van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010; 6: e1001024.
- GROSS L. A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice. Proc Soc Exp Biol Med. 1953; 83: 414-421.
- Freund R, Dubensky T, Bronson R, Sotnikov A, Carroll J, Benjamin T. Polyoma tumorigenesis in mice: evidence for dominant resistance and dominant susceptibility genes of the host. Virology. 1992; 191: 724-731.
- Kemball CC, Lee ED, Vezys V, Pearson TC, Larsen CP, Lukacher AE. Late priming and variability of epitope-specific CD8+ T cell responses during a persistent virus infection. J Immunol. 2005; 174: 7950-7960.
- Wilson CS, Moser JM, Altman JD, Jensen PE, Lukacher AE. Cross-recognition of two middle T protein epitopes by immunodominant polyoma virus-specific CTL. J Immunol. 1999; 162: 3933-3941.
- Goldrath AW, Bevan MJ. Low-affinity ligands for the TCR drive proliferation of mature CD8+ T cells in lymphopenic hosts. Immunity. 1999; 11: 183-190.
- Jay DC, Reed-Loisel LM, Jensen PE. Polyclonal MHC Ib-restricted CD8+ T cells undergo homeostatic expansion in the absence of conventional MHC-restricted T cells. J Immunol. 2008; 180: 2805-2814.
- Kraj P, Pacholczyk R, Ignatowicz L. Alpha beta TCRs differ in the degree of their specificity for the positively selecting MHC/peptide ligand. J Immunol. 2001; 166: 2251-2259.
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000; 7: 1063-1066.
- Kambayashi T, Kraft-Leavy JR, Dauner JG, Sullivan BA, Laur O, Jensen PE. The nonclassical MHC class I molecule Qa-1 forms unstable peptide complexes. J Immunol. 2004; 172: 1661-1669.
- Bieganowska K, Höllsberg P, Buckle GJ, Lim DG, Greten TF, Schneck J, et al. Direct analysis of viral-specific CD8+ T cells with soluble HLA-A2/Tax11-19 tetramer complexes in patients with human T cell lymphotropic virus-associated myelopathy. J Immunol. 1999; 162: 1765-1771.
- Kraft JR, Vance RE, Pohl J, Martin AM, Raulet DH, Jensen PE. Analysis of Qa-1(b) peptide binding specificity and the capacity of CD94/NKG2A to discriminate between Qa-1-peptide complexes. J Exp Med. 2000; 192: 613-624.
- Clark SP, Arden B, Kabelitz D, Mak TW. Comparison of human and mouse T-cell receptor variable gene segment subfamilies. Immunogenetics. 1995; 42: 531-540.
- Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995; 42: 501-530.
- Casanova JL, Romero P, Widmann C, Kourilsky P, Maryanski JL. T cell receptor genes in a series of class I major histocompatibility complex-restricted cytotoxic T lymphocyte clones specific for a Plasmodium berghei nonapeptide: implications for T cell allelic exclusion and antigen-specific repertoire. J Exp Med. 1991; 174: 1371-1383.
- Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, et al. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. J Gen Virol. 2001; 82: 1013-1025.
- Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998; 92: 3780-3792.
- Zhou Z, Callaway KA, Weber DA, Jensen PE. Cutting edge: HLA-DM functions through a mechanism that does not require specific conserved hydrogen bonds in class II MHC-peptide complexes. J Immunol. 2009; 183: 4187-4191.
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993; 90: 8392-8396.
- Holst J, Szymczak-Workman AL, Vignali KM, Burton AR, Workman CJ, Vignali DA. Generation of T-cell receptor retrogenic mice. Nat Protoc. 2006; 1: 406-417.
- Joyce S, Tabaczewski P, Angeletti RH, Nathenson SG, Stroynowski I. A nonpolymorphic major histocompatibility complex class Ib molecule binds a large array of diverse self-peptides. J Exp Med. 1994; 179: 579-588.
- He X, Tabaczewski P, Ho J, Stroynowski I, Garcia KC. Promiscuous antigen presentation by the nonclassical MHC Ib Qa-2 is enabled by a shallow, hydrophobic groove and self-stabilized peptide conformation. Structure. 2001; 9: 1213-1224.
- Hofstetter AR, Ford ML, Sullivan LC, Wilson JJ, Hadley A, Brooks AG, et al. MHC class Ib-restricted CD8 T cells differ in dependence on CD4 T cell help and CD28 costimulation over the course of mouse polyomavirus infection. J Immunol. 2012; 188: 3071-3079.
- Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Expression of two T cell receptor alpha chains: dual receptor T cells. Science. 1993; 262: 422-424.
- Wang F, Huang CY, Kanagawa O. Rapid deletion of rearranged T cell antigen receptor (TCR) Valpha-Jalpha segment by secondary rearrangement in the thymus: role of continuous rearrangement of TCR alpha chain gene and positive selection in the T cell repertoire formation. Proc Natl Acad Sci USA. 1998; 95: 11834-11839.
- Polic B, Kunkel D, Scheffold A, Rajewsky K. How alpha beta T cells deal with induced TCR alpha ablation. Proc Natl Acad Sci U S A. 2001; 98: 8744-8749.
- Tabaczewski P, Chiang E, Henson M, Stroynowski I. Alternative peptide binding motifs of Qa-2 class Ib molecules define rules for binding of self and nonself peptides. J Immunol. 1997; 159: 2771-2781.
- Hofstetter AR, Evavold BD, Lukacher AE. Peptide immunization elicits polyomavirus-specific MHC class ib-restricted CD8 T cells in MHC class ia allogeneic mice. Viral Immunol. 2013; 26: 109-113.
- Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997; 15: 749-795.
- Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA, Pantaleo G. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol Rev. 2006; 211: 236-254.
- Dooms H, Wolslegel K, Lin P, Abbas AK. Interleukin-2 enhances CD4+ T cell memory by promoting the generation of IL-7R alpha-expressing cells. J Exp Med. 2007; 204: 547-557.
- Mitchell DM, Ravkov EV, Williams MA. Distinct roles for IL-2 and IL-15 in the differentiation and survival of CD8+ effector and memory T cells. J Immunol. 2010; 184: 6719-6730.
- Castro I, Yu A, Dee MJ, Malek TR. The basis of distinctive IL-2- and IL-15-dependent signaling: weak CD122-dependent signaling favors CD8+ T central-memory cell survival but not T effector-memory cell development. J Immunol. 2011; 187: 5170-5182.
- Swanson PA 2nd, Hofstetter AR, Wilson JJ, Lukacher AE. Cutting edge: shift in antigen dependence by an antiviral MHC class Ib-restricted CD8 T cell response during persistent viral infection. J Immunol. 2009; 182: 5198-5202.
- Lazarevic V, Glimcher LH, Lord GM. T-bet: a bridge between innate and adaptive immunity. Nat Rev Immunol. 2013; 13: 777-789.