Research Article
Austin J Clin Immunol. 2014;1(4): 1017.
Molecular Network of NLRP3 Inflammasome Activation-Responsive Genes in a Human Monocyte Cell Line
Natsuki Kawana, Yoji Yamamoto, Yoshihiro Kino and Jun-ichi Satoh*
Department of Bioinformatics and Molecular Neuropathology, Meiji Pharmaceutical University, Japan
*Corresponding author: Jun-ichi Satoh, Department of Bioinformatics and Molecular Neuropathology, Meiji Pharmaceutical University, 2-522-1 Noshio, Kiyose, Tokyo 204-8588, Japan
Received: May 05, 2014; Accepted: June 11, 2014; Published: June 13, 2014
Abstract
Background: Inflammasome, activated by pathogen-derived and host-derived danger signals, constitutes a multimolecular signaling complex that serves as a platform for Caspase-1 (CASP1) activation and interleukin-1β (IL-1β) maturation. The activation of NLRP3 inflammasome requires two-step signals. The first ““priming” signal enhances gene expression of inflammasome components. The second “activation” signal promotes the assembly of inflammasome components. Deregulated activation of NLRP3 inflammasome contributes to the pathological processes of Alzheimer’s disease (AD), and Multiple Sclerosis (MS). However, at present, the precise mechanism regulating NLRP3 inflammasome activation and deactivation remains largely unknown.
Methods: By genome-wide gene expression profiling, we studied the molecular network of NLRP3 inflammasome activation-responsive genes in a human monocyte cell line THP-1 sequentially given two-step signals.
Results: We identified the set of 83 NLRP3 inflammasome activation-responsive genes. Among them, we found the NR4A nuclear receptor family NR4A1, NR4A2, and NR4A3, the EGR family EGR1, EGR2, and EGR3, the IκB family NFKBIZ, NFKBID, and NFKBIA as a key group of the genes that possibly constitute a negative feedback loop for shutting down inflammation following NLRP3 inflammasome activation. By molecular network analysis, we identified a complex network of NLRP3 inflammasome activation-responsive genes involved in cellular development and death, and immune and inflammatory responses, where transcription factors AP-1, NR4A, and EGR serve as a hub.
Conclusion: NLRP3 inflammasome activation-responsive genes constitute the molecular network composed of a set of negative feedback regulators for prompt resolution of inflammation.
Keywords: Mast cells; Autoimmune diseases
Introduction
Inflammasome serves as a multi molecular signaling complex involved in activation of Caspase-1 (CASP1) and maturation of interleukin-1β (IL-1β) and IL-18 [1,2]. A wide variety of exogenous and endogenous stimuli, characterized by microbe-derived Pathogen- Associated Molecular Patterns (PAMPs) and host- or environment-derived Danger-Associated Molecular Patterns (DAMPs), are recognized by an intracellular sensor called the NOD-like Receptors (NLRs), resulting in rapid induction of inflammasome formation by ordered assembly of self-oligomerizing components.
Among various classes of inflammasome, the nucleotide-binding oligomerization domain, leucine rich repeat and pyrin domain containing 3 (NLRP3) inflammasome has been most intensively studied. It is composed of NLRP3, the adaptor molecule named apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC), and the precursor form of the cysteine protease pro-CASP1 [1,2]. NLRP3 contains a central nucleotide-binding and oligomerization (NACHT) domain essential for activation of the signaling complex via ATP-dependent oligomerization, flanked by a C-terminal Leucine-Rich Repeat (LRR) pivotal for ligand sensing and auto regulation and a N-terminal pyrin (PYD) domain involved in a homotypic protein-protein interaction between NLRP3 and ASC. The molecular interaction of NLRP3 with ASC recruits pro-CASP1 by a homotypic interaction of Caspase Activation and Recruitment (CARD) domains between ASC and pro-CASP1. Subsequently, the proximity-induced pro-CASP1 oligomerization causes autocatalytic activation of CASP1, resulting in processing of pro-IL-1β or pro- IL-18 into biologically active IL-1β and IL-18. Both of them act as a central regulator for induction of cytokines and chemokines that amplify inflammation by recruiting immune effector cells.
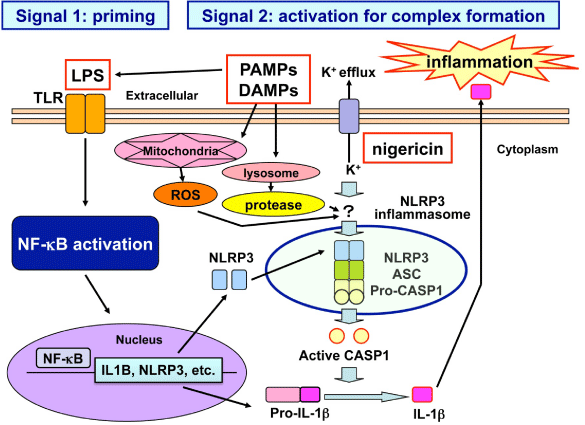
The activation of NLRP3 inflammasome requires two-step signals (Figure 1) [3,4]. The first “priming” signal termed Signal 1, such as microbe-derived Lipo Poly Saccharide (LPS), enhances gene expression of inflammasome components and target proteins via activation of transcription factor nuclear factor-kappa B (NF- κB). The second “activation” signal termed Signal 2 promotes the organized assembly of inflammasome components. The second signal involves three major mechanisms, such as generation of Reactive Oxygen Species (ROS), lysosomal protease leakage, and the potassium efflux [1,2]. Mitochondria often serve as the principal source of ROS. Blockade of mitophagy induces accumulation of ROS-generating mitochondria that activates NLRP3 inflammasome [5]. Furthermore, oxidized mitochondrial DNA directly activates NLRP3 inflammasome following induction of apoptosis [6]. By serving as an inducer of two-step signals, a diverse range of danger signals armed with PAMPs, such as Listeria monocytogenes, Candida albicans, and influenza A virus and those with DAMPs, such as amyloid-β (Aβ), uric acid and cholesterol crystals, asbestos, silica, alum, hyaluronan, and Adenosine 5’-Triphosphate (ATP), promptly activate the NLRP3 inflammasome [7,8].
Figure 1: Two-step signals for NLRP3 inflammasome activation. Activation of NLRP3 inflammasome, composed of NLRP3, ASC, and pro-CASP1, is tightly regulated by two-step signals. The first “priming” signal, such as LPS, enhances the expression of inflammasome components and target proteins via activation of transcription factor NF-κB. The second “activation” signal promotes the assembly of inflammasome components. The second signal involves three major mechanisms, including generation of ROS, lysosomal damage, and the potassium efflux. Abbreviations: LPS, lipopolysaccharide; TLR, toll-like receptor; PAMPs, pathogen-associated molecular patterns; DAMPs, danger-associated molecular patterns; ROS, reactive oxygen species.
Deregulated activation of NLRP3 inflammasome contributes to the pathological processes of various diseases, such as type 2 diabetes, Alzheimer’s disease (AD), and Multiple Sclerosis (MS) [9-11]. Lack of NLRP3 inflammasome components skews micro glial cells to an anti-inflammatory M2 phenotype with an enhanced capacity of amyloid-β (Aβ) clearance in a mouse model of AD [10]. Nlrp3-knockout mice showed reduced severity of Experimental Autoimmune Encephalomyelitis (EAE), a mouse model of MS, characterized by substantial attenuation of inflammation, demyelination and astrogliosis [12]. In active inflammatory demyelinating lesions of MS, reactive astrocytes and perivascular macrophages expressed all three components of NLRP3 inflammasome, such as NLRP3, ASC, and CASP1, along with IL-1β, suggesting that biochemical agents and monoclonal antibodies designed to block specifically NLRP3 inflammasome activation might be highly effective in treatment of active MS [11]. However, at present, the precise mechanism regulating NLRP3 inflammasome activation and deactivation remains largely unknown. In the present study, by genome-wide gene expression profiling, we attempts to clarify the comprehensive molecular network of NLRP3 inflammasome activation-responsive genes in a human monocyte cell line given consecutively two-step signals.
Materials and Methods
NLRP3 inflammasome activation
A human monocyte cell line THP-1 was obtained from RIKEN Cell Bank (Saitama, Japan). The cells were maintained in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS), 55 μM 2-mercaptoethanol, 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (feeding medium). To load the Signal 1, the cells were incubated for 3 hours with or without 0.2 μg/ml lipo polysaccharide (LPS; Sigma, St. Louis, MO, USA). To load the Signal 2, they were washed twice by Phosphate-Buffered Saline (PBS) and incubated further for 0.5 or 2 hours with 10 μM nigericin sodium salt (Wako Pure Chemical, Osaka, Japan) dissolved in ethanol or the equal v/v% concentration of ethanol (vehicle). Then, protein extract of the cells was processed for western blot analysis with a rabbit antibody against the C-terminal peptide of the human CASP1 p10 protein (sc-515, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and a rabbit antibody against the peptide mapping at amino acid residues of 117-269 of the human IL-1β protein (sc-7884, Santa Cruz Biotechnology).
Microarray analysis
Total cellular RNA was isolated by using the TRIZOL plus RNA Purification kit (Invitrogen). The quality of total RNA was evaluated on Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Three hundred ng of total RNA was processed for cDNA synthesis, fragmentation, and terminal labeling with the GeneChip Whole Transcript Sense Target Labeling and Control Reagents (Affymetrix, Santa Clara, CA, USA). Then, the labeled cRNA was processed for hybridization at 45°C for 17 hours with Human Gene 1.0 ST Array (28,869 genes; Affymetrix). The arrays were washed in the Gene Chip Fluidic Station 450 (Affymetrix), and scanned by the Gene Chip Scanner 3000 7G (Affymetrix). The raw data were expressed as CEL files and normalized by the Robust Multi Array average (RMA) method with the Expression Console software (Affymetrix).
Quantitative reverse transcription (RT)-polymerase chain reaction (qPCR) analysis
DNase-treated total RNA isolated from THP-1 cells was processed for cDNA synthesis using oligo (dT) [12-18] primers and Super Script II reverse transcriptase (Invitrogen). Then, cDNA was amplified by PCR in Light Cycler ST300 (Roche Diagnostics, Tokyo, Japan) using SYBR Green I and a panel of sense and antisense primer sets following: 5’ccagcactgccaaactggactact3’ and 5’ acagctcagcaaagccagggatct3’ for an 162 bp product of nuclear receptor subfamily 4, group A, member 1 (NR4A1); 5’ccaaagccgaccaagacctgcttt3’ and 5’ctgtgcaagaccaccccattgcaa3’ for an 124 bp product of nuclear receptor subfamily 4, group A, member 2 (NR4A2); 5’gagggctgcaagggctttttcaag3’ and 5’ gagggctgagaaggttcctgttgt3’ for a 242 bp product of nuclear receptor subfamily 4, group A, member 3 (NR4A3); and 5’ccatgttcgtcatgggtgtgaacca3’ and 5’gccagtagaggcagggatgatgttc3’ for a 251 bp product of the glyceraldehyde-3-phosphate dehydrogenase (G3PDH) gene that serves as an endogenous control. The expression levels of target genes were standardized against the levels of G3PDH detected in the corresponding cDNA samples. All the assays were performed in triplicate.
Molecular network analysis
To identify biologically relevant molecular networks, we imported corresponding Entrez Gene IDs into Ingenuity Pathways Analysis (IPA) (Ingenuity Systems, Redwood City, CA, USA), KeyMolnet (Institute of Medicinal Molecular Design, Tokyo, Japan), or Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) 9.1. STRING is an open-access database, while IPA and KeyMolnet are commercial resources.
STRING is a database that contains known and predicted, physiological and functional protein-protein interactions composed of 5,214,234 proteins from 1133 organisms [13]. STRING integrates the information from numerous resources, including experimental repositories, computational prediction methods, and public text collections. By uploading the list of UniProt IDs or Gene Symbols, STRING illustrates the union of all possible association networks.
IPA is a knowledgebase that contains approximately 3,000,000 biological and chemical interactions and functional annotations with definite scientific evidence. By uploading the list of Gene IDs and expression values, the network-generation algorithm identifies focused genes integrated in a global molecular network. IPA calculates the score p-value that reflects the statistical significance of association between the genes and the networks by the Fisher’s exact test.
KeyMolnet contains knowledge-based contents on 164,000 relationships among human genes and proteins, small molecules, diseases, pathways and drugs [14]. They include the core contents collected from selected review articles with the highest reliability. By importing the list of Gene ID and expression values, KeyMolnet automatically provides corresponding molecules as nodes on the network. The neighboring network-search algorithm selected one or more molecules as starting points to generate the network of all kinds of molecular interactions around starting molecules, including direct activation/inactivation, transcriptional activation/repression, and the complex formation within one path from starting points. The generated network was compared side by side with 501 human canonical pathways of the KeyMolnet library. The algorithm counting the number of overlapping molecular relations between the extracted network and the canonical pathway makes it possible to identify the canonical pathway showing the most significant contribution to the extracted network.
Results
NLRP3 inflammasome activation in THP-1 cells following introduction of two-step signals
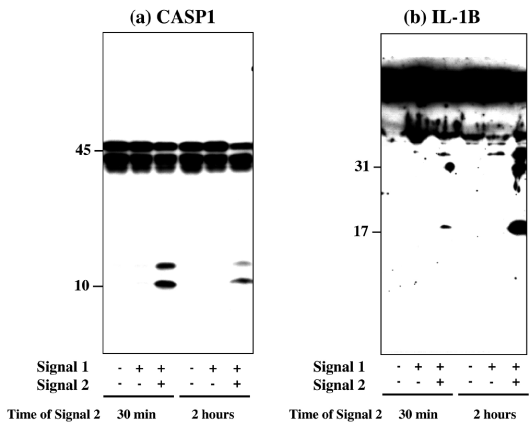
First, by western blot analysis, we studied NLRP3 inflammasome activation in THP-1 treated initially with exposure to 0.2 μg/ml LPS for 3 hours (Signal 1), followed by exposure to 10 μM nigericin for 30 min or 2 hours (Signal 2). The consecutive load of Signal 1 and Signal 2 markedly activated NLRP3 inflammasome in THP-1 cells, as indicated by production of cleaved products of CASP1 (Figure 2, panel a) and IL-1β (Figure 2, panel b). In contrast, the introduction of Signal 1 alone was not enough to activate NLRP3 inflammasome in THP-1 cells (Figure 2, panels a and b).
Figure 2: Two-step signals activate NLRP3 inflammasome in THP-1 cells. NLRP3 inflammasome activation was determined by western blot in THP-1 cells following exposure to 0.2 μg/ml LPS for 3 hours (Signal 1), followed by exposure to 10 μM nigericin for 30 min or for 2 hours (Signal 2). The panels (a, b) indicate western blot of (a) CASP1 in the cellular protein extract, and (b) IL-1β in the culture supernatant.
Gene expression profile during NLRP3 inflammasome activation
Next, we studied the genome-wide gene expression profile of THP-1 cells pretreated with 0.2 μg/ml LPS for 3 hours (Signal 1), washed by PBS, and exposed to 10 μM nigericin or vehicle for 2 hours (Signal 2). Then, total RNA was immediately processed for gene expression profiling on a Human Gene 1.0 ST Array. To identify NLRP3 inflammasome activation-responsive genes, we extracted the set of 83 annotated and protein-coding genes that satisfied fold change (FC) in Signal 1 (the presence of LPS versus the absence of LPS) smaller than 2-fold and FC in Signal 2 (the presence of nigericin versus the absence of nigericin) greater than 2-fold (Table 1). This gene enrichment procedure minimized the genes that were activated simply by exposure to LPS alone but not directly related to NLRP3 inflammasome activation.
Rank
FC Related to Signal 1
FC Related to Signal 2
Entrez Gene ID
Gene Symbol
Gene Name
1
1.06819645
18.61247501
8013
NR4A3
nuclear receptor subfamily 4, group A, member 3
2
1.942378012
12.91651537
6348
CCL3
chemokine (C-C motif) ligand 3
3
1.63109973
11.69111
414062
CCL3L3
chemokine (C-C motif) ligand 3-like 3
4
1.100615838
11.24166642
9308
CD83
CD83 molecule
5
1.819566773
10.85127008
3576
IL8
interleukin 8
6
1.292541852
7.633454043
1960
EGR3
early growth response 3
7
0.948867136
6.576691539
4929
NR4A2
nuclear receptor subfamily 4, group A, member 2
8
1.116320272
5.51767318
3164
NR4A1
nuclear receptor subfamily 4, group A, member 1
9
1.842348508
5.271896351
64332
NFKBIZ
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
10
1.268131184
4.992502002
643616
MOP-1
MOP-1
11
1.222058201
4.99018398
1959
EGR2
early growth response 2
12
1.716614387
4.456895103
5734
PTGER4
prostaglandin E receptor 4 (subtype EP4)
13
1.067764134
4.401932449
10746
MAP3K2
mitogen-activated protein kinase kinase kinase 2
14
1.076240121
4.353030131
2920
CXCL2
chemokine (C-X-C motif) ligand 2
15
1.443866138
4.329651804
6364
CCL20
chemokine (C-C motif) ligand 20
16
1.506881527
4.037790353
5743
PTGS2
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
17
1.143021068
3.908082725
153020
RASGEF1B
RasGEF domain family, member 1B
18
1.00701348
3.793627448
1958
EGR1
early growth response 1
19
1.188818931
3.318906546
23645
PPP1R15A
protein phosphatase 1, regulatory (inhibitor) subunit 15A
20
0.978133301
3.154899408
65125
WNK1
WNK lysine deficient protein kinase 1
21
1.116953399
3.113268501
84807
NFKBID
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta
22
1.431860551
3.025219884
51561
IL23A
interleukin 23, alpha subunit p19
23
0.654486344
2.985745104
645188
LOC645188
hypothetical LOC645188
24
1.082721348
2.867304268
1843
DUSP1
dual specificity phosphatase 1
25
1.877501415
2.813972064
8870
IER3
immediate early response 3
26
1.458901009
2.788511085
9021
SOCS3
suppressor of cytokine signaling 3
27
0.930381294
2.730662487
728715
LOC728715
ovostatin homolog 2-like
28
1.251031395
2.703465614
2353
FOS
v-fos FBJ murine osteosarcoma viral oncogene homolog
29
1.994627015
2.654181457
27289
RND1
Rho family GTPase 1
30
0.877732964
2.64583117
23499
MACF1
microtubule-actin crosslinking factor 1
Table 1 part 1 of 3: The set of 83 up-regulated genes in THP-1 monocytes following activation of NLRP3 inflammasome.
31
1.18363314
2.591793912
7538
ZFP36
zinc finger protein 36, C3H type, homolog (mouse)
32
0.768263434
2.584281103
79101
TAF1D
TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa
33
1.895682029
2.568793654
90668
LRRC16B
leucine rich repeat containing 16B
34
0.916615124
2.536018037
259296
TAS2R50
taste receptor, type 2, member 50
35
0.895110685
2.535538194
728741
LOC728741
hypothetical LOC728741
36
0.870604266
2.532650507
84319
CMSS1
cms1 ribosomal small subunit homolog (yeast)
37
0.474895831
2.525788794
4072
EPCAM
epithelial cell adhesion molecule
38
1.667878267
2.514873802
1326
MAP3K8
mitogen-activated protein kinase kinase kinase 8
39
1.107775084
2.496005315
8744
TNFSF9
tumor necrosis factor (ligand) superfamily, member 9
40
1.024389944
2.491488658
4616
GADD45B
growth arrest and DNA-damage-inducible, beta
41
0.97810347
2.470592388
2354
FOSB
FBJ murine osteosarcoma viral oncogene homolog B
42
1.017380957
2.461870724
643036
SLED1
RTFV9368
43
1.017380957
2.377675786
2152
F3
coagulation factor III (thromboplastin, tissue factor)
44
1.038770533
2.373054125
1973
EIF4A1
eukaryotic translation initiation factor 4A, isoform 1
45
1.596962012
2.3683134
4792
NFKBIA
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
46
0.872659044
2.354224669
1736
DKC1
dyskeratosis congenita 1, dyskerin
47
1.254570022
2.347010028
50515
CHST11
carbohydrate (chondroitin 4) sulfotransferase 11
48
0.818985035
2.34454831
50840
TAS2R14
taste receptor, type 2, member 14
49
0.649089802
2.278082518
85028
SNHG12
small nucleolar RNA host gene 12 (non-protein coding)
50
0.978928228
2.273044623
2889
RAPGEF1
Rap guanine nucleotide exchange factor (GEF) 1
51
0.689249392
2.247537218
55795
PCID2
PCI domain containing 2
52
0.827575589
2.246739728
54765
TRIM44
tripartite motif-containing 44
53
1.067300921
2.243145194
1263
PLK3
polo-like kinase 3 (Drosophila)
54
0.767788042
2.229552244
337867
UBAC2
UBA domain containing 2
55
1.306111439
2.229215371
3759
KCNJ2
potassium inwardly-rectifying channel, subfamily J, member 2
56
1.925222241
2.191743556
80149
ZC3H12A
zinc finger CCCH-type containing 12A
57
0.882964289
2.185060168
58155
PTBP2
polypyrimidine tract binding protein 2
58
1.545906426
2.181251323
56895
AGPAT4
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
59
1.05509141
2.155321381
10896
OCLM
oculomedin
60
1.05361515
2.15489714
9659
PDE4DIP
phosphodiesterase 4D interacting protein
Table 1 part 2 of 3: The set of 83 up-regulated genes in THP-1 monocytes following activation of NLRP3 inflammasome.
61
0.986553364
2.153150265
3047
HBG1
hemoglobin, gamma A
62
0.87493697
2.150450624
100507607
NPIPB9
nuclear pore complex interacting protein family, member B9
63
1.201327908
2.147514699
259292
TAS2R46
taste receptor, type 2, member 46
64
0.885483295
2.144478729
51574
LARP7
La ribonucleoprotein domain family, member 7
65
0.970156229
2.132807866
9839
ZEB2
zinc finger E-box binding homeobox 2
66
0.700126731
2.102345827
100133941
CD24
CD24 molecule
67
1.471640204
2.097753274
6303
SAT1
spermidine/spermine N1-acetyltransferase 1
68
0.796744464
2.080051151
9572
NR1D1
nuclear receptor subfamily 1, group D, member 1
69
1.754590053
2.069409283
10129
FRY
furry homolog (Drosophila)
70
1.117049405
2.06451372
5586
PKN2
protein kinase N2
71
1.084905208
2.058951728
339883
C3orf35
chromosome 3 open reading frame 35
72
1.007649566
2.047104863
1195
CLK1
CDC-like kinase 1
73
1.001286612
2.046307571
1185
CLCN6
chloride channel 6
74
1.005938423
2.043756057
338442
HCAR2
hydroxycarboxylic acid receptor 2
75
0.88066058
2.04297423
6144
RPL21
ribosomal protein L21
76
1.048011825
2.039547357
1844
DUSP2
dual specificity phosphatase 2
77
1.361895488
2.039480914
3092
HIP1
huntingtin interacting protein 1
78
0.951119813
2.038925421
388022
LOC388022
hypothetical gene supported by AK131040
79
0.888482949
2.018363478
144132
DNHD1
dynein heavy chain domain 1
80
0.972189862
2.012125102
23049
SMG1
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
81
0.89112764
2.007348359
6181
RPLP2
ribosomal protein, large, P2
82
0.798221473
2.005195646
23329
TBC1D30
TBC1 domain family, member 30
83
1.206469961
2.003702064
3726
JUNB
jun B proto-oncogene
Table 1 part 3 of 3: The set of 83 up-regulated genes in THP-1 monocytes following activation of NLRP3 inflammasome.
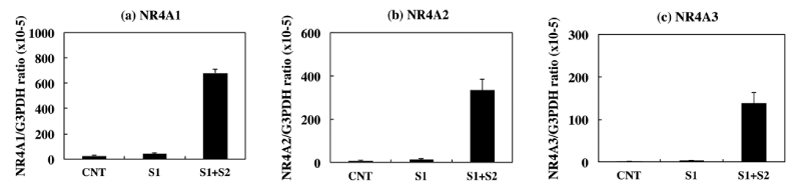
Most notably, three members of NR4A nuclear receptor family, such as NR4A1 (NUR77), NR4A2 (NURR1), and NR4A3 (NOR1), were identified as those ranked within top 10 genes. Coordinated up regulation of NR4A1, NR4A2, and NR4A3 in NLRP3 inflammasome-activated THP-1 cells was validated by qPCR (Figure 3, panels a-c). Signal 1 alone mildly elevated expression of these mRNA levels, whereas introduction of Signal 2 after Signal 1 markedly elevated the levels of NR4A1, NR4A2, and NR4A3 transcripts with a 16-fold, 25-fold, or 51-fold increase, respectively. We also identified Early Growth Response (EGR) family members, such as EGR1, EGR2, and EGR3, which belong to a family of zinc finger transcription factors involved in the regulation of cell growth, differentiation, and survival, NF-κB inhibitor (IκB) family members, such as NFKBIZ, NFKBID, and NFKBIA, along with a panel of pro inflammatory cytokines and chemokines, including CCL3, CCL3L3, IL8, CXCL2, CCL20, IL23A, and TNFSF9, as a subgroup of NLRP3 inflammasome activation-responsive genes.
Figure 3: Upregulated expression of NR4A family members in THP-1 cells during NLRP3 inflammasome activation. The levels of expression of NR4A1, NR4A2, and NR4A3 transcripts in THP-1 cells following exposure to 0.2 μg/ml LPS for 3 hours (Signal 1), followed by exposure to 10 μM nigericin for 2 hours (Signal 2) were determined by qPCR. They were standardized against the levels of G3PDH detected in the corresponding cDNA samples. The panels (a-c) indicate qPCR of (a) NR4A1, (b) NR4A2, and (c) NR4A3. The bars represent CNT (LPS -, nigericin -), S1 (LPS +, nigericin -), and S1+S2 (LPS+, nigericin +).
Molecular network of NLRP3 inflammasome activation responsive genes
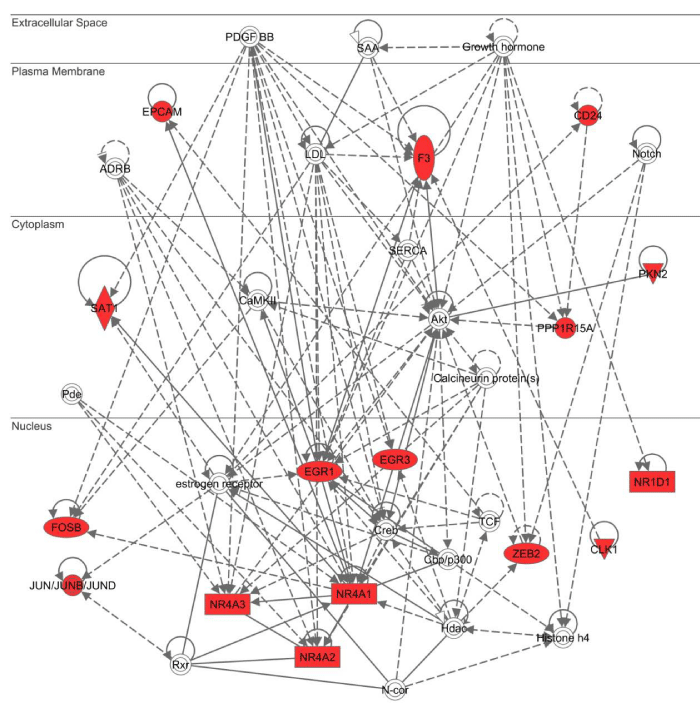
Next, by using three different bioinformatics tools for molecular network analysis based on knowledgebase, we studied biologically relevant molecular networks for the set of 83 NLRP3 inflammasome activation-responsive genes in THP-1 cells. The core analysis of IPA identified the networks defined as “Auditory and Vestibular System Development and Function, Embryonic Development, Organ Development” (p = 1.00E-32), “Cell Cycle, Cellular Development, Cell Death and Survival” (p = 1.00E-30) (Figure 4), and “Connective Tissue Disorders, Immunological Disease, Inflammatory Disease” (p = 1.00E-26) as top three most relevant functional networks. These results suggest that NLRP3 inflammasome activation-responsive genes play a pivotal role in cell development, death, and immune and inflammatory responses. KeyMolnet by the neighboring network-search algorithm operating on the core contents extracted the highly complex molecular network composed of 455 molecules and 529 molecular relations. The network showed the most statistically significant relationship with canonical pathways termed as “transcriptional regulation by AP-1” (p = 3.82E-184), “transcriptional regulation by NR4A” (p = 2.28E-105), and “transcriptional regulation by EGR” (p = 2.78E-99) (Figure 5). These results suggest a central role of transcription factors AP-1, NR4A, and EGR in regulation of expression of NLRP3 inflammasome activation-responsive genes, by acting as a hub of the molecular network.
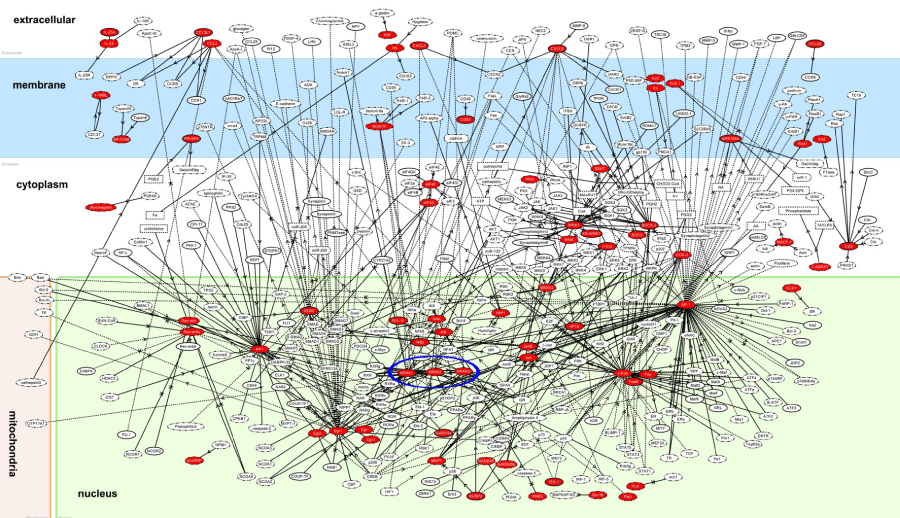
Figure 4: IPA molecular network of NLRP3 inflammasome activation-responsive genes. Entrez Gene IDs corresponding to the set of 83 NLRP3 inflammasome activation-responsive genes in THP-1 cells (Table 1) were imported into the core analysis tool of IPA. The functional network defined as “Cell Cycle, Cellular Development, Cell Death and Survival” is shown. Red nodes indicate NLRP3 inflammasome activation-responsive genes.
Figure 5: KeyMolnet molecular network of NLRP3 inflammasome activation-responsive genes. Entrez Gene IDs corresponding to the set of 83 NLRP3 inflammasome activation-responsive genes in THP-1 cells (Table 1) were imported into KeyMolnet. The neighboring network-search algorithm operating on the core contents extracted the highly complex molecular network. Red nodes represent NLRP3 inflammasome activation-responsive genes, while white nodes exhibit additional nodes extracted automatically from the core contents of KeyMolnet to establish molecular connections. The molecular relation is indicated by solid line with arrow (direct binding or activation), solid line with arrow and stop (direct inactivation), solid line without arrow (complex formation), dash line with arrow (transcriptional activation), and dash line with arrow and stop (transcriptional repression). The cluster of NR4A1, NR4A2, and NR4A3 is highlighted by blue circle.
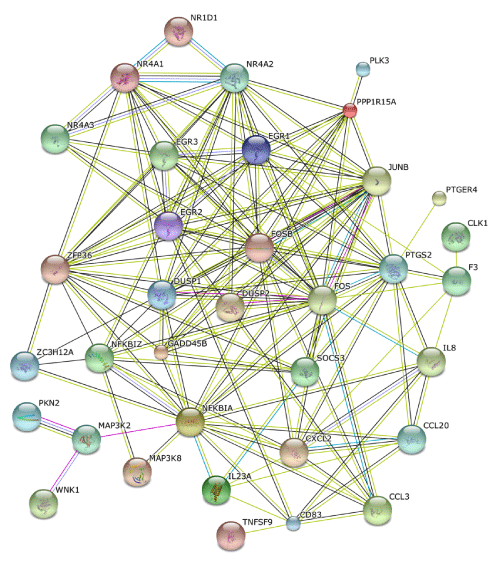
Finally, STRING extracted a protein-protein interaction network, composed of 35 core molecules derived from the set of 83 NLRP3 inflammasome activation-responsive genes in THP-1 cells. In this network, both the set of NR4A family members NR4A1, NR4A2, and NR4A3 and EGR transcription factors EGR1, EGR2, and EGR3 constituted a close and intense protein interaction sub network (Figure 6).
Figure 6: STRING molecular network of NLRP3 inflammasome activation-responsive genes. Gene Symbols corresponding to the set of 83 NLRP3 inflammasome activation-responsive genes in THP-1 cells (Table 1) were imported into STRING. The set of 35 molecules constructing the protein-protein interaction network are shown on the evidence view of STRING.
Discussion
By genome-wide gene expression profiling, we identified the set of 83 NLRP3 inflammasome activation-responsive genes in THP-1 cells sequentially given two-step signals. Among them, we found three members of NR4A nuclear receptor family, such as NR4A1, NR4A2, and NR4A3, three members of EGR family, such as EGR1, EGR2, and EGR3, three members of IκB family, such as NFKBIZ, NFKBID, and NFKBIA as a noticeable subset of NLRP3 inflammasome activation-responsive genes. By molecular network analysis, we found that they play a central role in cellular development and death, and immune and inflammatory responses, where transcription factors AP-1, NR4A, and EGR serve as a hub in the molecular network. Because THP-1 is a spontaneously immortalized human monocytic cell line derived from an acute monocytic leukemia patient, the possibility could not be excluded that the molecular network we identified does not represent the physiological network of non-malignant human monocytes.
NR4A1, NR4A2, and NR4A3 are three closely related, highly homologous nuclear transcription factors of the steroid/thyroid hormone receptor superfamily, categorized as orphan nuclear receptors because of lack of their cognate ligands [15]. They are encoded by immediate early genes, rapidly induced by exposure of the cells to the serum, growth factors, cytokines, and peptide hormones. NR4A receptors act as a transcription factor for a battery of downstream genes involved in cell proliferation, apoptosis, DNA repair, inflammation, and angiogenesis [16]. Accumulating evidence indicates that NR4A family exerts not only proinflammatory but also anti-inflammatory effects on various cell types. NR4A receptors play a pivotal role in development of regulatory T (Treg) cells in the thymus [17]. Knockdown of either NR4A1 or NR4A3 elevates the levels of production of IL-1β, IL-8, and MCP-1 in THP-1 cells [18]. By binding directly to NF-κB p65, a central regulator of innate and adaptive immune response, NR4A1 recruits the Co REST corepressor complex on gene promoter and inhibits transcription of pro inflammatory genes in mouse microglia and astrocytes [19]. Adenosine monophosphate released from apoptotic cells, when metabolized to adenosine, activates macrophages to express NR4A1, NR4A2, and NR4A3 that play a role in suppression of inflammation during engulfment of apoptotic cells [20]. Recently, we found that NR4A2 is one of vitamin D receptor-target genes with protective function against development of MS by analyzing a chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq) dataset derived from immortalized B cells and THP-1 cells [21]. All of these observations suggest that NR4A proteins, whose expression is induced by pro inflammatory mediators, serve as a safety valve for shutting down sustained inflammation that is amplified by NLRP3 inflammasome activation. Consistent with this view, IκB family members acting as a negative regulator of NF-κB activation, such as NFKBIZ, NFKBID, and NFKBIA [22-24], are coordinately induced along with enhanced expression of NR4A family, suggesting that these molecules constitute a negative feedback loop for NLRP3 inflammasome activation.
EGR family constitutes a family of zinc finger transcription factors very rapidly and transiently induced in various cell types without de novo protein synthesis following exposure to mitogenic signals [25,26]. EGR1 functions as a positive regulator for T and B cell functions, by regulating transcription of the genes encoding key cytokines and costimulatory molecules, while EGR2 and EGR3 act as a negative regulator essential for induction of anergy [27]. EGR1 down regulates the expression of itself by binding to an EGR1- binding site located on its own promoter [28]. Furthermore, EGR1 directly activates transcription of NR4A1 (nur77) in mouse IgM+ B cells [29]. Deletion of EGR2 and EGR3 in mouse T and B cells causes a lethal autoimmune syndrome characterized by excessive production of pro inflammatory cytokines accompanied by over activation of STAT1 and STAT3 [30]. Importantly, we identified SOCS3, a potent inhibitor of STAT3 activation [31], as one of NLRP3 inflammasome activation-responsive genes (Rank 26 in Table 1). These observations suggest the working hypothesis that the EGR family members are actively involved in resolution of sustained inflammation amplified by NLRP3 inflammasome activation.
Conclusion
By genome-wide gene expression profiling, we identified the set of 83 NLRP3 inflammasome activation-responsive genes in THP-1 cells. Among them, we found NR4A nuclear receptor family, EGR family, and IκB family as a group of the genes that possibly constitute a negative feedback loop for shutting down sustained inflammation following NLRP3 inflammasome activation. By molecular network analysis, we found that NLRP3 inflammasome activation-responsive genes play a pivotal role in cellular development and death, and immune and inflammatory responses, where transcription factors AP-1, NR4A, and EGR act as a hub in the molecular network.
Acknowledgement
This work was supported by the JSPS KAKENHI (C25430054), and the Intractable Disease Research Center (IDRC) project, the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, and the grant from the National Center for Geriatrics and Gerontology (NCGC 26-20). The authors would thank Ms. Aki Takaoka for her invaluable help in microarray analysis. The microarray data are available from the Gene Expression Omnibus (GEO) under the accession number GSE58959.
References
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010; 140: 821-832.
- Menu P, Vince JE. The NLRP3 inflammasome in health and disease: the good, the bad and the ugly. Clin Exp Immunol. 2011; 166: 1-15.
- Wang H, Mao L, Meng G. The NLRP3 inflammasome activation in human or mouse cells, sensitivity causes puzzle. Protein Cell. 2013; 4: 565-568.
- Walsh JG, Muruve DA2, Power C1. Inflammasomes in the CNS. Nat Rev Neurosci. 2014; 15: 84-97.
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011; 469: 221-225.
- Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012; 36: 401-414.
- Di Virgilio F. Liaisons dangereuses: P2X(7) and the inflammasome. Trends Pharmacol Sci. 2007; 28: 465-472.
- Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008; 9: 857-865.
- Jourdan T, Godlewski G, Cinar R, Bertola A, Szanda G, Liu J, et al. Activation of the Nlrp3 inflammasome in infiltrating macrophages by endocannabinoids mediates beta cell loss in type 2 diabetes. Nat Med. 2013; 19: 1132-1140.
- Heneka MT, Kummer MP, Stutz A, Delekate A, Schwartz S, Vieira-Saecker A, Griep A. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature. 2013; 493: 674-678.
- Kawana N, Yamamoto Y, Ishida T, Saito Y, Konno H, Arima K, et al. Reactive astrocytes and perivascular macrophages express NLRP3 inflammasome in active demyelinating lesions of multiple sclerosis and necrotic lesions of neuromyelitis optica and cerebral infarction. Clin Exp Neuroimmunol. 2013; 4: 296-304.
- Gris D, Ye Z, Iocca HA, Wen H, Craven RR, Gris P, Huang M. NLRP3 plays a critical role in the development of experimental autoimmune encephalomyelitis by mediating Th1 and Th17 responses. J Immunol. 2010; 185: 974-981.
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013; 41: D808-815.
- Satoh J. Bioinformatics approach to identifying molecular biomarkers and networks in multiple sclerosis. Clin Exp Neuroimmunol. 2010; 1: 127–140.
- Zhao Y, Bruemmer D. NR4A orphan nuclear receptors: transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler Thromb Vasc Biol. 2010; 30: 1535-1541.
- Mohan HM, Aherne CM, Rogers AC, Baird AW, Winter DC, Murphy EP. Molecular pathways: the role of NR4A orphan nuclear receptors in cancer. Clin Cancer Res. 2012; 18: 3223-3228.
- Sekiya T, Kashiwagi I, Yoshida R, Fukaya T, Morita R, Kimura A, Ichinose H. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat Immunol. 2013; 14: 230-237.
- Bonta PI, van Tiel CM, Vos M, Pols TW, van Thienen JV, Ferreira V, et al. Nuclear receptors Nur77, Nurr, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler Thromb Vasc Biol. 2006; 26: 2288-2294.
- Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009; 137: 47-59.
- Yamaguchi H, Maruyama T, Urade Y, Nagata S. Immunosuppression via adenosine receptor activation by adenosine monophosphate released from apoptotic cells. Elife. 2014; 3: e02172.
- Satoh J, Tabunoki H. Molecular network of chromatin immunoprecipitation followed by deep sequencing-based vitamin D receptor target genes. Mult Scler. 2013; 19: 1035-1045.
- Muta T, Yamazaki S, Eto A, Motoyama M, Takeshige K. IkappaB-zeta, a new anti-inflammatory nuclear protein induced by lipopolysaccharide, is a negative regulator for nuclear factor-kappaB. J Endotoxin Res. 2003; 9: 187-191.
- Tergaonkar V, Correa RG, Ikawa M, Verma IM. Distinct roles of IkappaB proteins in regulating constitutive NF-kappaB activity. Nat Cell Biol. 2005; 7: 921-923.
- Kuwata H, Matsumoto M, Atarashi K, Morishita H, Hirotani T, Koga R, et al. IkappaBNS inhibits induction of a subset of Toll-like receptor-dependent genes and limits inflammation. Immunity. 2006; 24: 41-51.
- Christy B, Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci U S A. 1989; 86: 8737-8741.
- Beckmann AM, Wilce PA. Egr transcription factors in the nervous system. Neurochem Int. 1997; 31: 477-510.
- Gómez-Martín D, Díaz-Zamudio M, Galindo-Campos M, Alcocer-Varela J. Early growth response transcription factors and the modulation of immune response: implications towards autoimmunity. Autoimmun Rev. 2010; 9: 454-458.
- Cao X, Mahendran R, Guy GR, Tan YH. Detection and characterization of cellular EGR-1 binding to its recognition site. J Biol Chem. 1993; 268: 16949-16957.
- Dinkel A, Warnatz K, Ledermann B, Rolink A, Zipfel PF, Bürki K, et al. The transcription factor early growth response 1 (Egr-1) advances differentiation of pre-B and immature B cells. J Exp Med. 1998; 188: 2215-2224.
- Li S, Miao T, Sebastian M, Bhullar P, Ghaffari E, Liu M, et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity. 2012; 37: 685-696.
- Carow B, Rottenberg ME. SOCS3, a Major Regulator of Infection and Inflammation. Front Immunol. 2014; 5: 58.