
Research Article
Austin J Mol & Cell Biol. 2014;1(1): 1003.
RAPD-PCR Based Biomarker Study in Fish Species (Family: Cyprinidae) of Madhya Pradesh, India
Neekhra B¹, Mansoori AA¹, Verma S¹, Koiri RK² and Jain SK¹*
1Department of Biotechnology, Dr. Hari Singh Gour Central University, India
2Department of Zoology, Dr. Hari Singh Gour Central University, India
*Corresponding author: Jain SK, Department of Biotechnology, School of Biological Sciences, Dr. H. S. Gour Central University, Sagar – 470003, India,
Received: October 14, 2014; Accepted: December 01, 2014; Published: December 31, 2014
Abstract
Random Amplified Polymorphic DNA (RAPD) analysis was conducted for molecular identification of most commonly occurring fish species in Central India. In the study, male and female species of Labeo rohita, Catla catla and Cirrhina mrigala (family: Cyprinidae) were randomly collected from various locations of central Indian region. For polymerase chain reaction, ten decanucleotide primers (RAn primer series) were used. On the basis of interpretability, simplicity and reproducibility, six random primers RAn 1 (GATGACCGCC), RAn 3 (GGCACGTAAC), RAn 4 (GGCATGACCT), RAn 5 (GGGTAACGCC), RAn 9 (GTGCCAAATG) and RAn11 (GTGCCCGTTA) were considered for genetic differentiation and band analysis. We observed varied size of amplified products depending upon the sequence of random primers and genotypes used. A total of 55 discrete amplified products were obtained (size 200 to 2000 bp approximately). Out of 55 products, 29 were species specific markers indicating high level polymorphism among species. The highest and lowest number of RAPD bands detected for primers RAn 5 and RAn 9 was 12 and 6 respectively. 16 bands were most relevant as found common in all three species of family: Cyprinidae. Highest monomorphism observed for C. catla and C. mrigala (5 bands) and lowest for L. rohita & C. mrigala (1 band). Similar band patterns were obtained for both male and female of all three fish species showing 100 percent sexual genetic similarity. The diverse nature of DNA bands indicated the genetic distance between fish species and presence of common bands attributed to an evolutionary relationship. Pattern of species specific unique bands observed might be useful tools for molecular identification. Our study re-establishes the importance of RAPD-PCR technique for molecular taxonomy studies in diverse fish species.
Keywords: Genetic differentiation; Polymorphism; RAPD- PCR; Random primers
Introduction
India has about 4.4 percent of the global fish production; this sector contributes to 1.1 percent of the GDP and 4.7 percent of the agricultural GDP. The total fish production of India (2010-11) is around 7.50 million tones including 4.50 million tones of inland fish and 3.00 million tones of marine fish [1]. The application of genetics in aquaculture and fisheries resource management has been very limited as compared to agriculture, forestry and animal husbandry.
Information on the molecular structure of fish species is useful for optimizing identification of stocks, stock enhancement, breeding programs, management for sustainable yield and preservation of genetic diversity [2-5].
Molecular techniques based on DNA sequence polymorphism are now used in population genetics studies, systematic and molecular taxonomy to get an answer to systematic related problems. Molecular techniques played an important role to understand the basis of polymorphism of a species, species diagnostics and population differentiation [6].
Within the last decade, scientific progression has increasingly supported the use of molecular techniques in determining population diversity. Many molecular techniques are now available, which allow ecologists and evolutionary biologists to determine the genetic architecture of a wide variety of closely related individuals. The discovery of the PCR had a major impact on the research of eukaryotic genomes and contributed to the development and application of various DNA markers [7-9].
RAPDs are particularly useful to study the genetic structure of populations because they reveal polymorphisms in non-coding regions of the genome. Random primers produce RAPDs that have been used extensively as molecular markers [10-11]. RAPDs also have the advantage that no prior knowledge of the genome is necessary for successful application [12-13].
The RAPD markers method has been reported to be an efficient tool to differentiate geographically and genetically isolated population, and has been used to verify the existence of population of species that have arisen either through genetic selection under different environmental conditions or as a result of genetic drift [14]. During the last few years, RAPD-PCR has been shown wide range of applications in fisheries and poultry [15-21].
In view of the above facts, the present study was aimed to develop RAPD fingerprinting in Labeo rohita, Catla catla and Cirrhina mrigala collected from Sagar region, Madhya Pradesh and also aimed to generate species-specific RAPD marker as a reference database for species recognition and characterization. Thus, the investigation will provide valuable tool in the form of diagnostic markers for fisheries conservation and management of these species.
Material and Methods
Sample collection
Three species Labeo rohita, Catla catla and Cirrhina mrigala (male and female) were collected from local rivers and lakes of Sagar region and preserved at -20°C. The most commonly used sources of fish DNA are scales, fin and muscles tissue. A rapid DNA extraction method developed by using modified lysis buffer that require about 2 hours duration. This methodology is non-invasive, less expensive and reproducible with high efficiency of DNA recovery [22].
DNA extraction
Approximately, 50 mg of scales were taken from each species and dried on a filter paper. The scales were then cut into small pieces and placed in a 2 ml-Eppendorf tube containing 940 μl lysis buffer (200mM Tris-HCl, pH 8.0; 100mM EDTA, pH 8.0; 250 mM NaCl), 30 μl proteinase K (10 mg/ml) and 30 μl 20 percent SDS. The content in the tubes were incubated at 48°C for 45-50 min in a water bath. After incubation, 500 μl volume of phenol: chloroform: isoamyl alcohol (25:24:1) was added to the tube containing lysed scales cells. The contents were then mixed properly by gently inverting the tube for 10 min to precipitate the proteins and other part of the nucleic acids. The tube was then rotated for 10 min at 9,200 g. The top aqueous layer was transferred to a new 1.5 ml-Eppendorf tube, leaving interface and lower phase. The DNA was precipitated by adding equal volume of isopropanol and 0.2 volumes of 10 M ammonium acetate and inverting the tubes gently several times. The precipitated DNA was then pelleted by centrifugation at 13,200 g for 10 min. The supernatant was removed by pouring out gently, taking care to avoid loss of DNA pellet. The pellet was then washed briefly in 500 μl chilled 70 percent ethanol, air-dried and resuspended in 200 μl sterile water/ TE buffer.
After ensuring complete solubility of DNA, the purity factor (A260/A280 nm) was measured by UV spectrophotometer (Cole Parmer Ins. Company, US) and its integrity was checked by loading 10 μl DNA preparation (2 μl extracted DNA, 2 μl dye and 6 μl sterile water) on 0.8 percent agarose gel and stained with ethidium bromide. The extracted DNA samples were then stored at -20°C till their further use. These DNAs were used as templates in a PCR based search for producing RAPD markers.
DNA amplification by PCR
Prior to the experimentation pilot study was performed in 10 fishes of each species (both female and male). The data thus observed showed similarity in banding pattern (considering all bands) and therefore only one male and female from each species considered during experimentation.
The DNA was amplified by using 50 μl of reaction mixture containing sterile water 39.0 μl, 10X Taq Buffer A 5.0 μl, 10mM dNTP mix 2.0 μl, RAPD primer 2.0 μl (RAn series supplied by Bangalore genei company), DNA template (10ng/μl) 1.0 μl, Taq DNA polymerase (3 U/μl) 1.0 μl. The amplification was carried out in thermal-cycler (Eppendorf pro) under the PCR conditions were predenaturation at 94°C for 5 min followed by two loops. First loop of 10 cycles of denaturation at 94°C for 45 sec, annealing at 35 °C for 1 min and extension at 72 °C for 1.5 min and, second loop of 40 cycles of denaturation at 94 °C for 45 sec, annealing at 37 °C for 45 sec and extension at 72 °C for 1 min. The final extension was performed at 72 °C for 10 min. On the basis of results of initial RAPD analysis on pooled DNA, 6 out of 10 primers were chosen for further analysis, based on band pattern quality, reproducibility and the presence of diagnostic bands. Finally the RAPD-PCR was done using six primers (Table 1).
Primer
Nucleotide sequence 5’ to 3’
Size range of amplified bands (base pairs)
Total number of bands
No. of selected species-specific RAPD fragments characteristic for:
L.rohita
C.catla
C.mrigala
L. rohita
C. catla
C. mrigala
RAn1
GATGACCGCC
200-1185
1
5
5
0
0
2
RAn3
GGCACGTAAC
350-1500
4
3
3
2
0
2
RAn4
GGCATGACCT
200-1500
2
3
3
2
3
2
RAn5
GGGTAACGCC
300-2000
4
3
5
3
2
2
RAn9
GTGCCAAATG
200-900
1
3
2
1
2
1
RAn11
GTGCCCGTTA
350-2000
4
3
1
3
2
0
Total
16
20
19
11
9
9
55
29
Table 1: Total number of amplified band fragments and number of selected species specific fragments with six primers in three fish species. (Only male specimens were considered).
The amplified products were run on 2 percent agarose gel (stained with Ethidium bromide) with 100bp DNA ladder and Low range DNA ruler ranges from 100 bp to 3000 bp (LRDR). Gels were photographed under gel documentation (MultiDoc-It, Labmate, UK). RAPD patterns were visually analyzed and scored from photographs.
Results
A series of discrete bands were obtained after amplification of DNA samples of all three fish species with six primers (Table 1). The different primers produced different banding patterns (Figure 1-6). For the comparison of these patterns, a set of distinct, well separated bands were selected, avoiding the weak and unresolved bands [23] to find out consistent banding pattern among the individuals within specific populations of the fish. Majority of primers produced potential markers.
The data of inter species specific analytical study indicates that a total of 55 DNA markers (considering all bands) were detected in all three fish species, out of which, 40 bands were polymorphic, can be considered as useful RAPD markers (Table 1). The remaining 15 bands were monomorphic suggesting that all three fish species have a common ancestor and are genetically closer to each other. During the analysis, only males were considered as sexual differentiation was not observed during the pilot study.
The largest number of RAPD bands were detected for primer RAn 5 (12 bands, Figure 4), while the lowest number was scored for primer RAn 9 (6 bands, Figure 5). Such a wide variation in the number of markers produced by these arbitrary primers attributed to the difference in the binding sites throughout the genome of the fish species under study. The molecular weight of scorable bands generated by these primers ranged from 200 to 2000 bp. Highest molecular weight range was exhibited for RAn 5 (300 bp - 2000 bp) while it was lowest for RAn 9 (200 bp - 900 bp). Number of polymorphic bands produced per primer ranged from one to five with an average of 3.06 bands per primer.
Seven scorable bands were obtained by these primers having molecular weight more than 1000 bp and 29 scorable bands were having molecular weight of more than 500 bp (52.73 %).
Large numbers of family and species specific bands were scored within RAPD profiles produced by these primers. Out of 40 polymorphic bands, 29 bands were counted as species specific (52.73 percent). 11 species specific markers were scored for Labio rohita while 9 markers each for Catla catla and C. mrigala species (Table 1). The largest number of RAPD species specific markers were scored by Primer RAn 4 (7 markers) and RAn 5 (7 markers) while the lowest number was scored by the primers RAn 1 (2 markers). On the basis of amplification pattern obtained, the six primers can be classified as follows:
100 percent polymorphic primers: RAn 4 (8 bands) and Ran 9 (6 bands) primers successfully amplified 100 percent polymorphic band pattern for differentiation of all three species of fish.
Monomorphic primers: Four primers RAn 1, RAn 3, RAn 5 and RAn 11 exhibited homologous banding patterns along with polymorphism. Total 7 homologous bands obtained (Table 2). The monomorphic bands obtained between fish species were as follows:
S. No.
Fish Species
Band size (bp)
Primer used
1.
C.catla & C.mrigala (4 bands)
800
RAn1
700
RAn3
650
RAn5
250
RAn1
2.
C.catla & L.rohita (2 bands)
550
RAn3
350
RAn3
3.
L. rohita, C. Catla & C.mrigala (1 band)
450
RAn11
Table 2: Number of selected homologous bands between fish species.
• Between C. catla and C. mrigala: Three primers showed homology between C. catla and C. Mrigala (Figure 1,2 and 4). Primer RAn 1 amplified 2 (250 bp and 800 bp), primer RAn 3 amplified 1 (700 bp) and primer RAn 5 amplified 1 monomorphic band (650 bp).
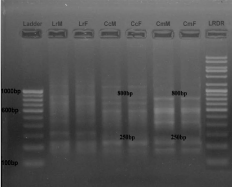
Figure 1: RAPD banding pattern amplified by RAn 1 primer (GATGACCGCC)
Lane 1 100bp DNA ladder, Lane 2 Labeo rohita (Male), Lane 3 Labeo rohita
(Female), Lane 4 Catla catla (Male), Lane 5 Catla catla (Female), Lane 6
Cirrhina mrigala (Male), Lane 7 Cirrhina mrigala (Female), Lane 8 Low range
DNA ruler.
• Between C. catla and L. rohita: Only one primer RAn 3 (Figure 2) amplified 2 monomorphic band pairs (800 bp and 250 bp).
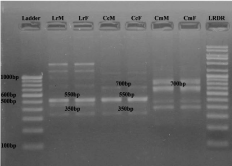
Figure 2: RAPD banding pattern amplified by RAn 3 primer (GGCACGTAAC)
Lane 1 100bp DNA ladder, Lane 2 Labeo rohita (Male), Lane 3 Labeo rohita
(Female), Lane 4 Catla catla (Male), Lane 5 Catla catla (Female), Lane 6
Cirrhina mrigala (Male), Lane 7 Cirrhina mrigala (Female), Lane 8 Low range
DNA ruler.

Figure 3: RAPD banding pattern amplified by RAn 4 primer (GGCATGACCT)
Lane 1 100bp DNA ladder, Lane 2 Labeo rohita (Male), Lane 3 Labeo rohita
(Female), Lane 4 Catla catla (Male), Lane 5 Catla catla (Female), Lane 6
Cirrhina mrigala (Male), Lane 7 Cirrhina mrigala (Female), Lane 8 Low range
DNA ruler.
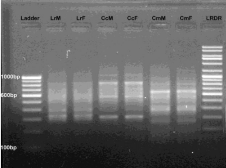
Figure 4: RAPD banding pattern amplified by RAn 5 primer (GGGTAACGCC)
Lane 1 100bp DNA ladder, Lane 2 Labeo rohita (Male), Lane 3 Labeo rohita
(Female), Lane 4 Catla catla (Male), Lane 5 Catla catla (Female), Lane 6
Cirrhina mrigala (Male), Lane 7 Cirrhina mrigala (Female), Lane 8 Low range
DNA ruler.
• Between L. rohita, C. Catla and C. mrigala: Primer RAn 11 (Figure 5) specifically amplified 1 band (450 bp) homologous to all three fish species. Therefore considered as family specific diagnostic marker.
100 percent sexual and intra specific genetic similarity: Similar band patterns were obtained for both male and female individuals of all three species of fishes (Figure 1-6).

Figure 5: RAPD banding pattern amplified by RAn 9 primer (GTGCCAAATG)
Lane 1 100bp DNA ladder, Lane 2 Labeo rohita (Male), Lane 3 Labeo rohita
(Female), Lane 4 Catla catla (Male), Lane 5 Catla catla (Female), Lane 6
Cirrhina mrigala (Male), Lane 7 Cirrhina mrigala (Female), Lane 8 Low range
DNA ruler.

Figure 6: RAPD banding pattern amplified by RAn 11 primer (GTGCCCGTTA)
Lane 1 100bp DNA ladder, Lane 2 Labeo rohita (Male), Lane 3 Labeo rohita
(Female), Lane 4 Catla catla (Male), Lane 5 Catla catla (Female), Lane 6
Cirrhina mrigala (Male), Lane 7 Cirrhina mrigala (Female), Lane 8 Low range
DNA ruler.
The diverse nature of DNA bands indicates the genetic distance between fish species but the presence of common bands indicates evolutionary relationship. RAPD fingerprinting offers a rapid and efficient method for generating a new series of DNA markers in fish [24-26]. A variety of published studies have been utilized the RAPD assay for solving numerous problems of systematic as well as for reconstruction of phylogenetic relationships in fish [27,28]. RAPD has also been used to estimate genetic diversity and variations required studying fish management and conservation practices, even with endangered species [29,30].
RAPD markers have been proved for pedigree identification, pathogenic diagnostics, and trait improvement in genetics and breeding programmes [31]. RAPD markers have been used for gynogenetic fish identification [32,33] and for gene mapping studies in fish [34-36].
Many RAPD-PCR studies have been analysed at molecular level in fisheries and can be used efficiently for variation analysis of populations with differential degrees of geographic isolation. Garg et al. [37] analyzed the genetic variation of the 2 populations of Mystus vittatus (Bloch) of Madhya Pradesh, India using RAPD-PCR. Fundamental DNA marker studies on genetic variation in riverine and hatchery populations of C. catla in Bangladesh have been carried out using microsatellites [38] and RAPD markers [26]. Brahmane et al. [39] studied about the RAPD genetic variation between 4 different populations of Hilsa Shad fish from Ganga, Yamuna, Hoogly and Narmada rivers of India. Rahman et al. [40] used RAPD markers to assess genetic variation in three wild populations of the Catla carp in the rivers and one hatchery population in Bangladesh. Da Silva Cortinhas et al. [41] examined RAPD markers to authenticate the genetic diversity in A. brasiliensis from two different places. Genetic structure of four wild populations of two hill stream fish Barilius bendelisis and B. barna was studied using RAPD markers in Uttarakhand, India [42]. RAPD and Inter-Simple Sequence Repeat (ISSR) markers were used to investigate the genetic structure of four subpopulations of Mystus nemurus in Thailand [43]. RAPD assay polymorphisms have also been applied to characterize markers within and among Tilapia from Epe, Badore water bodies and Carboom fish farm hatchery through selective breeding programs [44]. Thus, the sequence analysis is helpful in calculating the exact genetic distance between fish species. Usefulness of RAPD-PCR technique in differentiating species, subspecies and strains in fish has been demonstrated by many researchers worldwide [45-50].
Conclusion
The study indicates effectiveness of RAPD markers in detecting the ratio of polymorphism, monomorphism and estimating genetic distance among three fish species under study. Both monomorphic and polymorphic DNA bands were identified based on their presence or absence that could be used for species differentiation. Considering the above results, RAPD markers may be recommended as quick and reliable discrimination technique, compared with other marker approaches, for detecting polymorphism and establishment of genetic relationship among L. rohita, C. catla and C. mrigala species.
Once the population structure is known, scientific management for optimal harvest and conservation of the fishery resource can be undertaken. This method of DNA fingerprinting is important since it is relatively easy to obtain valuable data. It allows for a more introspective interpretation of diversity within a population. Therefore, the present study may serve as a reference point for future examinations of genetic variations between the species of fish which are commercially important and play a significant role in food chain in lentic as well as lotic habitats.
It is important to mention that data results from RAPD assays can be extended to further dissect traits in a more refined way. There is also the opportunity and need to study sequences of specific polymorphic bands, to determine the genes detected by RAPD experiments. Further studies with other molecular methodologies are essential to clarify and confirm genetic relationships among fish species depicted using RAPDs.
Acknowledgement
Authors are thankful to Madhya Pradesh Council of Science and Technology, Bhopal, India for financial assistance.
References
- Tripathi SD. Aquaculture: A panacea for the future. Fishing Chimes. 2011; 31: 12-15.
- Dinesh KR, Lim TM, Chua KL, Chan WK, Phang VP. RAPD analysis: an efficient method of DNA fingerprinting in fishes. Zoolog Sci. 1993; 10: 849-854.
- Garcia DK, Benzie JAH. RAPD markers of potential use in penaeid prawn (Penaeus monodon) breeding programs. Aquaculture. 1995; 130: 137–144.
- Tassanakajon, Pongsomboon S, Rimphanitchayakit V, Jarayabhand P, Boonsaeng V. Random Amplified Polymorphic DNA (RAPD) markers for determination of genetic variation in wild populations of the black tiger prawn (Penaeus monodon) in Thailand. Mol Mar Biol Biotechnol. 1997; 6: 110–115.
- Tassanakajon, Pongsomboon S, Jarayabhand P, Klinbunga S, Boonsaeng VV. Genetic structure in wild populations of black tiger shrimp (Penaeus monodon) using randomly amplified polymorphic DNA analysis. J Mar Biotechnol. 1998; 6: 249–254.
- Avise JC. Introduction: the scope of conservation genetic. Avise JC, Hamrick JL, editors. In: Conservation genetics: Case histories from nature. Chapman & Hall. 1996; 1-9.
- Mulcahy DL, Cresti M, Sansavini S, Douglas GC, Linskens HF, Bergamini G, et al. The use of random amplified polymorphic DNAs to fingerprint apple genotypes. Science Horticulture. 1993; 54: 89–96.
- Marle-Koster EV, Nel LH. Genetic markers and their application in livestock breeding in South Africa: A review. South African Journal of Animal Science. 2003; 33: 1–10.
- Schlötterer C1. The evolution of molecular markers--just a matter of fashion? Nat Rev Genet. 2004; 5: 63-69.
- Koh TL, Khoo G, Fan LQ, Phang VPE. Genetic diversity among wild forms and cultivated varieties of Discuc Symphysodon spp. as revealed by random amplified polymorphic DNA RAPD fingerprinting. Aquaculture. 1999; 173: 485–497.
- Shikano T, Taniguchi N. Using microsatellite and RAPD markers to estimate the amount of heterosis in various strain combinations in the guppy Poecilia reticulate as a fish model. Aquaculture. 2002; 204: 271–281.
- Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990; 18: 7213-7218.
- Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990; 18: 6531-6535.
- Fuchs H, Gross R, Stein H, Rottamann O. Application of molecular markers for the differentiation of bream (Abramis brama L.) populations from the rivers Main and Danube. Journal of Applied Ichthyology. 1998; 14: 49-55.
- Liu ZJ, Li P, Argue BJ, Dunham RA. Random amplified polymorphic DNA markers: usefulness for gene mapping and analysis of genetic variation of catfish. Aquaculture. 1999; 174: 59–68.
- Ali BA, Ahmed MMM, Aly OM. Relationship between genetic similarity and some productive traits in local chicken strains. African Journal of Biotechnology. 2003; 2: 46–47.
- Ali BA. Detection of DNA alteration in abnormal phenotype of broiler chicken male by random amplified polymorphic DNA (RAPD). African Journal of Biotechnology. 2003; 2:153-156.
- Ali BA, Ahmed MMM, El-Zaeem SY. Application of RAPD markers in fish: Part II – Among and within families; Cichlidae (freshwater), Mugilidae (Catadromus), Sparidae and Serranidae (marine). Int J of Biotechnology. 2004; 6: 393–401.
- Ali BA, Huang TH, Qin DN, Wang XM. A review of Random Amplified Polymorphic DNA (RAPD) markers in fish. Reviews in Fish Biology and Fisheries. 2004; 14: 443–453.
- Ahmed MMM, Ali BA, El-Zaeem SY. Application of RAPD markers in fish: Part I – some genera (Tilapia, Sarotherodon and Oreochromis) and species (Oreochromis aureus and Oreochromis niloticus) of Tilapia. International Journal of Biotechnology. 2004; 6: 86–93.
- Salem HH, Ali BA, Huang TH, Qin DN. Use of randomly amplified polymorphic DNA (RAPD) markers in poultry research. International Journal of Poultry Science. 2005; 4: 804–811.
- Kumar R, Singh PJ, Nagpure NS, Kushwaha B, Srivastava SK, Lakra WS. A non-invasive technique for rapid extraction of DNA from fish scales. Indian J Exp Biol. 2007; 45: 992-997.
- Kambhampati S, Black WC 4th, Rai KS. Random amplified polymorphic DNA of mosquito species and populations (Diptera: Culicidae): techniques, statistical analysis, and applications. J Med Entomol. 1992; 29: 939-945.
- Foo CL, Dinesh KR, Lim TM, Chan WK, Phang VP. Inheritance of RAPD markers in the guppy fish, Poecilia reticulata. Zoolog Sci. 1995; 12: 535-541.
- Islam MS, Alam MA. Randomly amplified polymorphic DNA analysis of four different populations of the Indian major carp, Labeo rohita (Hamilton). Journal of Applied Ichthyology. 2004; 20: 407-412.
- Islam MS, Ahmed ASI, Azam MS, Alam MS. Genetic analysis of three river populations of Catla catla (Hamilton) using randomly amplified polymorphic DNA markers. Asian Aust J Anim Sci. 2005; 18: 453-457.
- Barman HK, Barat A, Yadav BM, Banerjee S, Meher PK, Reddy PVGK, et al. Genetic variation between four species of Indian major carps as revealed by random amplified polymorphic DNA assay. Aquaculture. 2003; 62093: 1-9.
- Jayasankar P. Random amplified polymorphic DNA (RAPD) fingerprinting resolves species ambiguity of domesticated clown fish (genus: Amphiprion, family: Pomacentridae) from India. Aquaculture Research. 2004; 35: 1006-1009.
- Lopera-Barrero NM, Ribeiro RP, Sirol RN, Povh JA, Gomes PC, Vargas L, et al. Genetic diversity in piracanjuba populations (Bryconorbignyanus) with the RAPD (Random Amplified Polymorphic DNA) markers. Journal Animal Science. 2006; 84: 170.
- Shair OHM, Al- Ssum RM, Bahkali AH. Genetic variation investigation of tilapia grown under Saudi Arabian controlled environment. American Journal of Biochemistry and Molecular Biology. 2011; 1: 89-94.
- Yoon JM, Kim GW. Randomly amplified polymorphic DNA-polymerase chain reaction analysis of two different populations of cultured Korean catfish Silurus asotus. J Biosci. 2001; 26: 641–647.
- Chen H, Leibenguth F. Studies on multilocus fingerprints, RAPD markers, and mitochondrial DNA of a gynogenetic fish (Carassius auratus gibelio). Biochem Genet. 1995; 33: 297-306.
- Corley-Smith GE, Lim CJ, Brandhorst BP. Production of androgenetic zebrafish (Danio rerio). Genetics. 1996; 142: 1265-1276.
- Postlethwait JH, Johnson SL, Midson CN, Talbot WS, Gates M, Ballinger EW, et al. A genetic linkage map for the zebrafish. Science. 1994; 264: 699-703.
- Postlethwait JH, Yan YL, Gates MA. Using random amplified polymorphic DNAs in zebrafish genomic analysis. Methods Cell Biol. 1999; 60: 165-179.
- Kazianis S, Morizot DC, McEntire BB, Nairn RS, Borowsky RL. Genetic mapping in Xiphophorus hybrid fish: assignment of 43 AP-PCR/RAPD and isozyme markers to multipoint linkage groups. Genome Res. 1996; 6: 280-289.
- Garg RK, Silawat N, Sairkar P, Neetu Vijay, Mehrotra NN. RAPD analysis for genetic diversity of two populations of Mystus vittatus (Bloch) of Madhya Pradesh, India. African Journal of Biotechnology. 2009; 8: 4032-4038.
- Alam MS, Islam MS. Population genetic structure of Catla catla (Hamilton) revealed by microsatellite DNA markers. Aquaculture. 2005; 246: 151-160.
- Brahmane MP, Das MK, Sinha MR, Sugunan VV, Mukherjee A, Singh SN, et al. Use of RAPD fingerprinting for delineating populations of hilsa shad Tenualosa ilisha (Hamilton, 1822). Genet Mol Res. 2006; 5: 643-652.
- Rahman SMZ, Khan MR, Islam S, Alam S. Genetic variation of wild and hatchery populations of the catla Indian major carp (Catla catla Hamilton 1822: Cypriniformes, Cyprinidae) revealed by RAPD markers. Genetics and Molecular Biology. 2009.
- Da Silva Cortinhas MC, Glienke C, Prioli AJ, Noleto RB, Matoso DA, Cestari MM. A prime inference on genetic diversity (RAPDs) in the marine fish Atherinella brasiliensis (Teleostei, Atherinopsidae) from Southern Brazil. Acta Zoologica. 2010; 91: 242–248.
- Mishra AK, Lakra WS, Bhatt JP, Goswami M, Nagpure NS. Genetic characterization of two hill stream fish species Barilius bendelisis (Ham.1807) and Barilius barna (Ham.1822) using RAPD markers. Mol Biol Rep. 2012; 39: 10167-10172.
- Kumla S, Doolgindachbaporn S, Sudmoon R, Sattayasai N. Genetic variation, population structure and identification of yellow catfish, Mystus nemurus (C&V) in Thailand using RAPD, ISSR and SCAR marker. Mol Biol Rep. 2012; 39: 5201-5210.
- Mojekwu TO, Oguntade OR, Oketoki TO, Usman AB, Omidiji O. Molecular characterization of tilapia in different water bodies using RAPD markers. African Journal of Applied Biotechnology Research. 2013; 1: 1-12.
- Bardakci F, Skibinski DO. Application of the RAPD technique in tilapia fish: species and subspecies identification. Heredity (Edinb). 1994; 73 : 117-123.
- Bardakci F. The Use of Random Amplified Polymorphic DNA (RAPD) Markers in Sex Discrimination in Nile Tilapia, Oreochromis niloticus (Pisces: Cichlidae). Turkish Journal of Biology. 2000; 24: 169–175.
- Borowsky RL, McClelland M, Cheng R, Welsh J. Arbitrarily primed DNA fingerprinting for phylogenetic reconstruction in vertebrates: the Xiphophorus model. Mol Biol Evol. 1995; 12: 1022-1032.
- Sültmann H, Mayer WE, Figueroa F, Tichy H, Klein J. Phylogenetic analysis of cichlid fishes using nuclear DNA markers. Mol Biol Evol. 1995; 12: 1033-1047.
- Partis L, Wells RJ. Identification of fish species using random amplified polymorphic DNA (RAPD). Mol Cell Probes. 1996; 10: 435-441.
- Jong-Man Y. Genetic similarity and difference between common carp and Israeli carp (Cyprinus carpio) based on Random Amplified Polymorphic DNA analyses. Korean Journal of Biological Sciences. 2001; 5: 333-339.