Research Article
Austin J Pharmacol Ther. 2014; 2 (8).1044
Serotonin 5-HT7 Receptor Regulates Endothelial Cell Migration via Protein Kinase A
Elena Strekalova1,2 and Jasmina Profirovic3*
1Department of Medicine, University of Wisconsin Carbone Cancer Center, University of Wisconsin School of Medicine and Public Health, USA
2Institute of General Pathology and Pathophysiology, Russian Academy of Medical Sciences, Russia
3Pharmaceutical and Administrative Sciences, St. Louis College of Pharmacy, St. Louis, MO, USA
*Corresponding author: : Jasmina Profirovic, Pharmaceutical and Administrative Sciences, St. Louis College of Pharmacy, 4588 Parkview Place, St. Louis, MO, 63110, USA
Received: July 31, 2014; Accepted: September 24, 2014; Published: September 25, 2014
Abstract
The 5-hydroxytryptamine type 7 receptor (5HT7R) regulates many physiological processes, including learning and memory, circadian rhythm, and behavior. Its role is also implicated in psychiatric disorders. Little is known about the 5HT7R function outside the CNS. Here, we report that 5HT7R, endogenously expressed in endothelial cells (ECs), may promote cell migration and adhesion. Using Boyden chamber migration assay and wound healing “scratch” assay we demonstrated that stimulation of the receptor with 5HT7R agonists 5-CT and AS 19 significantly increased EC migration. In addition, 5-CT and AS 19 treatment increased EC adhesion to extracellular matrix. Downregulation of 5HT7R using specific siRNA significantly inhibited baseline and 5-HT-induced EC migration. Additionally, pretreatment of ECs with PKA inhibitor 14-22 amide significantly reduced 5-CT- or AS 19-induced EC migration, suggesting that PKA is involved in the regulation of EC migration mediated by 5HT7R. Our results suggest a prominent role of 5HT7R in promoting cell migration and adhesion and identify 5HT7R as a potential regulator of physiological and pathophysiological processes involving cell migration and adhesion.
Keywords: 5-hydroxytryptamine type 7 receptor; Endothelial cells; Cell migration; Cell adhesion
Abbreviations
5-HT: 5-hydroxytryptamine; 5HT7R: 5-hydroxytryptamine type 7 receptor; 5HT4R: 5- hydroxytryptamine type 4 receptor; 5-CT: 5-carboxamidotryptamine maleate salt; EC: Endothelial Cell; HUVECs: Human Umbilical Vein Endothelial Cells; PKA: Protein Kinase A; PKI: Protein Kinase A Inhibitor; siRNA: Small Interfering RNA; FBS: Fetal Bovine Serum; BSA: Bovine Serum Albumin; PBS: Phosphate-Buffered Saline; HBSS: Hanks’ Balanced Salt Solution; SDS-PAGE: Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis; RT-PCR: Reverse Transcriptase Polymerase Chain Reaction; qPCR: Quantitative Polymerase Chain Reaction; GAPDH: Glyceraldehydes-3-Phosphate Dehydrogenase; EGM-2: Endothelial Growth Medium 2
Introduction
Cell migration is a complex process that involves coordinate changes in cell adhesion, signal transduction and cytoskeletal organization. It is an essential process in the development and homeostatic processes such as angiogenesis, tissue repair and immune responses. Furthermore, it contributes to processes such as vascular disease, chronic inflammatory diseases, tumor formation and metastasis [1]. Therefore, it is of a considerable clinical interest to understand molecular mechanisms involved in regulation of cell migration.
Serotonin (5-hydroxytryptamine, 5-HT) functions as a neurotransmitter that regulates multiple physiological and pathophysiological functions in the human body (reviewed in [2- 4]). Although most of body serotonin is found in the intestinal enterochromaffin cells, serotonin is involved in regulation of all major organ system functions [5]. In endothelial cells (ECs), serotonin promotes cell migration [6], proliferation [7], induces endothelial nitric oxide synthase (eNOS) expression [8] and activation [9].
The actions of serotonin are mediated by over a dozen of cell membrane receptors divided into seven families of receptors that consist of multiple subtypes of receptors with distinct tissue and cell expression and signaling. All the receptors, except for 5-HT3 that is a ligand-gated ion channel, are G-protein coupled receptors [10].
5-HT7R was the newest addition to the serotonin receptor family, cloned in 1993 [11-15]. It is expressed in several regions of CNS as well as in peripheral tissues, particularly in gastrointestinal tract and vascular smooth muscle cells [13]. The 5-HT7R is coupled to Gs protein, leading to activation of adenylyl cyclase [13,16] and to G12 protein, leading to Cdc42-mediated filopodia formation [17]. A great number of studies have suggested that 5-HT7R regulates many physiological and pathophysiological processes, including learning and memory [18,19], sleep and circadian rhythm [12], body temperature [20] and development of migraine [21]. Although the expression of 5-HT7Rs was demonstrated in ECs [22], their role in these cells has not been understood.
Our previous data indicate that another serotonin receptor, 5-HT4 receptor (5-HT4R), promotes migration and adhesion of ECs. It also stimulates angiogenesis both in ECs and in mouse model of ischemia [23]. The present study was aimed to characterize the role of 5-HT7R in EC migration and adhesion. Using specific agonists and siRNA-mediated downregulation of 5-HT7R in two different migration assays, we demonstrated a critical role of endogenous 5-HT7R in regulation of migration. Additionally, our results suggested that 5-HT7R is essential for adhesion as well. Finally, we proposed the mechanism of 5-HT7R- mediated regulation of migration involving protein kinase A (PKA). These data suggest a prominent role of 5-HT7R in promoting cell migration and adhesion of ECs.
Materials and Methods
Reagents
Rabbit polyclonal anti-5-HT7R-receptor antibody was obtained from Imgenex (Imgenex Corporation, San Diego, CA). 5-HT7R receptor agonists, 5-CT salt (5-carboxamidotryptamine maleate) and AS 19 ((2S)-(+)-5-(1,3,5-Trimethylpyrazol-4-yl)-2-(dimethylamino) tetralin) were from Tocris (R&D Systems, Inc., Minneapolis, MN). Serotonin (5-hydroxytryptamine, 5-HT) and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (St. Louis, MO). Protein kinase A inhibitor 14-22 amide, cell-permeable, myristoylated - Calbiochem was from EMD Millipore (Billerica, MA). Fetal bovine serums (FBS), Phosphate-Buffered Saline (PBS) and Hanks’ balanced salt solution (HBSS) were from Invitrogen (Life Technologies, Grand Island, NY). The human umbilical vein ECs (HUVECs) and endothelial growth medium (EGM-2) BulletKit were purchased from Lonza (Walkersville, MD).
Cell culture and immunoblotting
HUVECs were cultured in the EGM-2 BulletKit supplemented with 10% FBS up to 8 passages. Confluent HUVECs were lysed in the lysis buffer containing 25 mM HEPES (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Triton X-100, 0.5% Na-deoxycholate, 0.1% SDS, 5 mM EDTA, 1 mM dithiothreitol (DTT), and 5 μL/mL mammalian protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) and homogenized by sonication. The insoluble material was removed from the lysates by centrifugation at 15,000×g for 10 min. Cleared lysates were subjected to SDS-polyacrylamide gel electrophoresis (SDS- PAGE), transferred to polyvinylidene difluoride membrane (PVDF) membrane, and analyzed by immunoblotting with anti-5-HT7R antibodies.
Reverse transcription-polymerase chain reaction (RTPCR)
Detection of 5-HT7R receptor mRNA in HUVECs was performed using RT-PCR. Total RNA was isolated from HUVECs using RN easy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Primer sequences for human 5-HT7R were as follows: sense primer, 5’- GAAGGAGGTGGAAGAGTGTGCAA-3’; antisense primer, 5’-ACAGAAGCTGCATTCCATTCTGC-3’. This primer set produces a PCR product of 516 bp. RT-PCR was performed using Easy-A One-Tube RT-PCR Kit (Agilent Technologies, Santa Clara, CA). RT-PCR reaction was performed for 1 cycle of 15 min at 42 °C and 1 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 30 s at 58 °C and 2 min at 68 °C, and 1 cycle of 5 min at 68 °C. PCR products were analyzed by 1.5% agarose gel electrophoresis.
Boyden chamber migration assay
Migration assays were performed using a 48-well Boyden chamber (Neuroprobe Inc., Gaithersburg, MD) with 8-μm-pore size polycarbonate Nuclepore TM membrane (Whatman, Clifton, NJ) as described previously [23]. Briefly, HUVECs, grown until confluent on gelatin-coated plate, were incubated in serum free medium containing 0.1% BSA overnight. Next day, the cells were trypsinized, resuspended and placed in the lower wells of the chamber. After 90 min of upside down incubation at 37°C, 1μmol/L 5-CT or 1 μmol/L AS 19 in 0.1% BSA was applied to the upper wells. After 4 h of incubation at 37°C, the filter was fixed and stained with Diff Quick kit (Dade Behring, Newark, DE). Thereafter, the cells that migrated through the pores to the upper side of the filter were quantified using light microscopy at magnification of 100 ×. The average number of migrating cells in 10 fields was taken as a number of migrated cells of the group. Each experiment was performed at least three times, and all samples were tested in quadruplicates.
In vitro wound healing assay
We performed in vitro wound healing assay as we described previously [23]. HUVECs seeded onto a 6- well plate were incubated for 24 h and maintained in serum free medium for 8 h. After wounding in a straight line using a sterile 200 μL tip, the cells were washed with PBS and incubated with 5-HT7R receptor agonist 1 μmol/L 5-CT or 1 μmol/L AS 19 in serum free medium for 17 h. Images were taken at the time of the wounding and at 17 h after wounding using Nikon eclipse TE 300 microscope equipped with 4 × objective and Nikon cool pix 990 digital camera. The number of cells within the wounded region represented the number of migrated cells. In each well, four measurements were taken from four fields. At least three independent experiments were performed.
Cell viability assay
An MTS assay was used to analyze the effect of 5-CT (1 μmol/L) or AS19 (1 μmol/L) on cell viability and assess if these agents affect cell proliferation. Cells seeded at 2.5 x 103 cells/well in 96-well plates were grown for 4, 24 or 48 hours. The number of viable cells was determined by measuring the absorbance at 490 nm 1 h after addition of the MTS reagent (Promega) as described by the manufacturer. The experiment was performed in triplicate. Cell viability was expressed as the percentage of viable cells: Aexp group/Acontrol × 100.
Adhesion assay
We used the method we described previously [23]. Briefly, gelatin-coated 24-well plates were washed and blocked with 1% BSA for 1 h at 37°C. HUVECs, serum-starved overnight in a medium containing 1% BSA, were seeded onto the 24-well plate (1 × 105 cells per well) and incubated in the presence of 5- HT7 R agonists 1 μmol/L 5-CT, 1 μmol/L AS 19 or medium alone for 2 h at 37°C. Thereafter, the cells were washed three times with PBS, fixed and stained with 0.5% solution of crystal violet. Light microscopy at magnification of 10 × was used to count the number of cells adhered to the extracellular matrix in four fields of each well. Adhesion experiments were done in triplicates and repeated at least three times.
Transfection of siRNAs
HUVECs were transfected using siRNA transfection reagent and medium (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) according to the manufacturer’s protocol. siRNA oligonucleotides (SMART pool) were purchased from Dharmacon (Thermo Fisher Scientific, Waltham, MA ) to silence human 5-HT7R. The non-silencing control siRNA was from Qiagen (Valencia, CA). The effects of siRNAs were confirmed by real-time qPCR 24 h after transfection.
Quantitative PCR (qPCR)
Total RNA was isolated from HUVECs using RNeasy Mini kit with DNaseI treatment following the manufacturer’s instructions (Qiagen, Valencia, CA). Using random primers and Superscript III transcriptase (Invitrogen, Life Technologies, Grand Island, NY), 1 μg total RNA was converted into cDNA. The housekeeping gene glyceraldehydes-3-phosphate dehydrogenase (GAPDH) was used as a reference gene for quantification. Specific primers for 5-HT7R receptor and GAPDH primers were synthesized by Integrated DNA Technologies (Coralville, IA). Quantitative PCR was performed with 50 ng cDNA in a 25 μl reaction volume containing a SYBR Green Master Mix (Applied Biosystems, Life Technologies, Grand Island, NY). Amplification was carried in an ABI PRISM 7000 sequence detection system (Applied Biosystems, Life Technologies, Grand Island, NY). Cycling conditions were 50°C for 2 min, 95°C for 10 min followed by a 40-cycle amplification at 95°C for 15 s, and 57°C for 1 min.
Experiments were repeated two times and samples were analyzed in triplicate. qPCR results were presented as Ct values, where Ct is defined as the threshold cycle of PCR at which amplified product was first detected. To compare the different RNA samples, we used the comparative Ct method and compared the RNA expression in samples to that of the control in each experiment.
Statistical analysis
All experiments were carried out at least in triplicates. The results are expressed as the mean value ± SE. The statistical differences between groups were evaluated using Student’s t-test. A value of P < 0.05 was considered statistically significant.
Results and Discussion
Stimulation of endogenous 5-HT7R promotes EC migration and adhesion
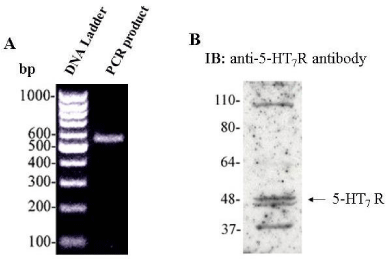
We confirmed the expression of 5-HT7R in HUVECs using two approaches: (I) detection of 5-HT7R mRNA by RT-PCR, and (II) detection of 5-HT7R protein expression by immunoblotting (Figure 1A and B). These findings were in line with the previous report that 5-HT7R mRNA is expressed in HUVECs [22].
Figure 1: 5-HT7R is expressed in HUVECs. A: RT-PCR analysis of the 5-HT7R expression in HUVECs. Gene-specific cDNA primers for 5-HT7R were used as described in “Materials and Methods”. B: Detection of 5-HT7R by Western blot analysis of the HUVEC lysates. Confluent HUVECs were lysed and cleared cell lysates were subjected to 9% SDS-PAGE and immunoblotting with anti-5-HT7R antibody.
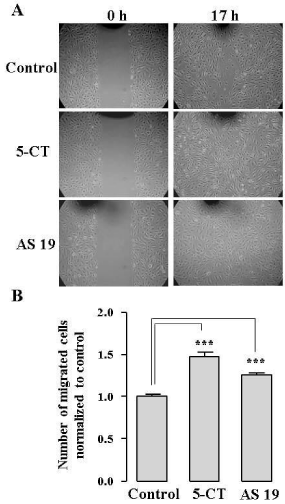
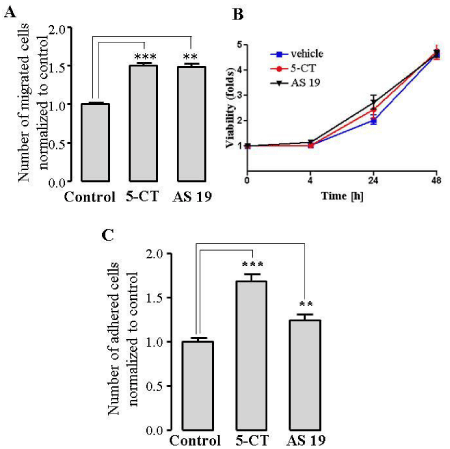
We employed two methods to assess the effects of 5-HT7R stimulation on HUVEC migration: (I) wound healing “scratch” assay, and (II) Boyden chamber migration assay. We used in vitro wound healing assay in which cells migrate into a wound produced on a cell monolayer [24]. We stimulated the endogenous 5- HT7R using two different 5-HT7R agonists, 5-CT and AS 19. Significantly enhanced healing of the wounded area was observed in the cells treated with either 5-CT or AS 19 in comparison to untreated cells (Figure 2A and 2B). To further confirm the effect of 5-HT7R stimulation on EC migration, we performed Boyden chamber migration assay to quantify HUVEC migration through a porous membrane [25]. Stimulation of 5-HT7R with 5-CT or AS 19 significantly increased the number of migrated cells (Figure 3A). Importantly, we did not observe any significant difference in cell viability between the control and 5-CT- or AS 19-stimulated cells 4, 24 and 48 h after cell stimulation in cell viability (MTS) assay (Figure 3B).
Figure 2: 5-HT7R agonists, 5-CT and AS 19, promote wound healing in ECs. A: Representative phase-contrast images of the cell monolayers taken at the time of wounding and 17 h after wounding. B: Cell migration was quantified by counting the number of cells within the wounded region. The data represent mean number of migrated cells normalized to control ± S.E. (n = 3, ***P < 0.001).
Figure 3: 5-HT7R agonists, 5-CT and AS 19, promote EC migration in a Boyden chamber migration assay and enhance EC adhesion. A: Cell migration was quantified by counting the migrated cells in 10 fields of each well. The data represent mean number of migrated cells normalized to control of three experiments done in quadruplicates ± S.E. (***P < 0.001). B: Cell viability (MTS) assay of HUVECs treated with 5-CT or AS 19. MTS assay was performed according to manufacturer’s instructions. The data represent mean ± S.E. C, 5-CT and AS 19 enhance EC adhesion. HUVECs were incubated with 1 μmol/L 5-CT or AS 19 for 2 h at 37°C. The data represent mean number of adhered cells normalized to control of three experiments performed in duplicates ± S.E. (** P< 0.01,***P < 0.001).
Because cell attachment to extracellular matrix is a critical step for the process of cell migration (1), we next investigated if 5-HT7R activation would affect cell adhesion. Treatment of HUVECs with 1 μmol/L 5-CT or AS 19 for 2 h caused a significant increase in the number of adhered cells compared to untreated cells (Figure 3C). Together, our results suggest that 5-HT7R activation promotes migration and adhesion of ECs.
Although serotonin has been first isolated and studied in the vasculature [26], many effects of serotonin in the cardiovascular system have not been well understood [27]. It has been shown that serotonin may promote angiogenesis [7, 28]. Several studies have suggested that the angiogenic effects of serotonin may be mediated by 5-HT1 and 5-HT2 receptors [7,29,30]. Our previous study has demonstrated that 5- HT4R promotes angiogenesis both in vitro and in vivo as well as migration and adhesion of ECs [23].
Our present study identified yet another serotonin receptor, 5-HT7R, which promotes processes of EC migration and adhesion.
Serotonin 5-HT7R is expressed in the brain [11,12], gastrointestinal tract and blood vessels [13]. The role of 5-HT7R in CNS has been extensively studied and some of the CNS functions are better understood. Despite the fact that 5-HT7R expression in ECs was demonstrated almost two decades ago [22], the role of 5-HT7R in ECs remained unknown. For the first time, the present study demonstrates that 5-HT7R may serve as a positive regulator of EC migration and adhesion, and presumably the other processes in which migration and adhesion are part of.
Downregulation of endogenous 5-HT7R receptor in ECs
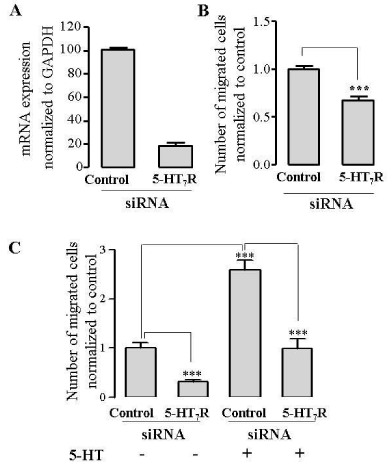
In our previous experiments, we used pharmacological stimulation of 5-HT7R to examine its effects on EC migration. We next used small interfering RNA (siRNA)-dependent gene downregulation to knock down the expression of 5-HT7R. HUVECs were transfected with control or 5-HT7R-specific siRNA. Expression of mRNA was examined using qPCR, whereby GAPDH, the housekeeping gene, was used as a reference gene for quantification. Twenty-four hours after transfection, 5-HT7R mRNA level was decreased by 85% in 5-HT7R siRNA-transfected cells compared to control siRNA-transfected cells (Figure 4A).
Figure 4: Downregulation of endogenous 5-HT7R in HUVECs by siRNA inhibits EC migration. A: HUVEC were grown to confluence and transfected with 60 nmol/L either 5-HT7R siRNA or control siRNA. 24 h after transfection with siRNA, mRNAs were quantified using qPCR. Expression was normalized to GAPDH expression (100%). B,C: Downreguation of 5-HT7R inhibits EC migration in a wound healing assay (B), or a Boyden chamber migration assay (C). HUVECs were transfected with either 5-HT7R siRNA or control siRNA and used in wound healing assay, (B) or in Boyden chamber migration assay with or without stimulation with 100 µmol/L serotonin (5-HT) (C). The data represent mean number of migrated cells normalized to control ± S.E. (n = 3, ***P<0.001). 5-HT, serotonin.
Because pharmacological tools used to modulate the receptor function may have known and/or unknown off-target actions, to corroborate our findings obtained by using 5-HT7R agonists 5-CT and AS 19, we used siRNA-dependent gene expression silencing. We tested if downregulation of 5-HT7R affects EC migration. HUVECs were transfected with control or 5-HT7R siRNA and used in wound healing assay. Downregulation of 5-HT7R significantly inhibited HUVEC migration (Figure 4B). Similar results were obtained from Boyden chamber migration assay in which depletion of 5-HT7R resulted in significantly reduced migration of ECs compared to control siRNA-transfected cells (Figure 4C). Furthermore, serotonin-induced migration was abrogated in 5-HT7R siRNA-transfected cells (Figure 4C), supporting the important role of this receptor in cell migration.
These results provide evidence that 5-HT7R deficiency decreases both baseline and agonist-induced EC migration. The finding that 5-HT7R deficiency alone may significantly reduce EC migration may suggest that 5-HT7R exerts constitutive receptor activity in the absence of an agonist, which is in line with the previous studies [17,31].
PKA is involved in regulation of EC migration mediated by 5-HT7R
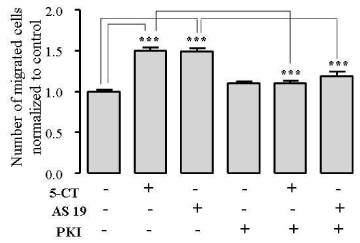
Because 5-HT7R is coupled to Gs protein leading to activation of adenylyl cyclase and accumulation of cAMP in COS-7 cells [13,16], ECs [32], which may promote cell migration [33], we hypothesized that 5-HT7R-dependent EC migration may be mediated by PKA. We addressed the question whether PKA is essential for 5-HT7R-induced migration by using specific cell-permeable PKA inhibitor 14-22 amide (PKI), which acts as PKA pseudosubstrate [34]. Under our experimental conditions, inhibition of PKA with PKI, did not significantly affect cell migration compared to control. Pretreatment of HUVECs with PKI followed by treatment with 5-HT7R agonists 5-CT or AS 19 significantly reduced EC migration when compared to the cells treated with 5-CT or AS 19 alone (Figure 5) indicating that PKA inhibition abolished EC migration mediated by 5-HT7R activation. These data suggest that 5-HT7R promotes EC migration via PKA pathway.
Figure 5: PKI mediates 5-HT7R–dependent EC migration in a Boyden chamber assay. HUVECs were stimulated with 1 μmol/L 5-CT or 1 µmol/L AS 19 alone or together with 10 μmol/L PKI and used in Boyden chamber migration assay. The data represent mean number of migrated cells normalized to control ± S.E. (n = 3, ***P < 0.001).
Interestingly, using the same assay in our previous study, we demonstrated that PKA is not involved in 5- HT4R-dependent EC cell migration despite 5-HT4R’s coupling to Gs protein [23]. Unlike 5-HT7R, which has been demonstrated to couple to Gs in bovine ECs [32], 5-HT4R has not been shown to couple to Gs in ECs of any kind yet although this was shown in neuronal cells [35] and insect Sf.9 cells [36].
In addition to Gs, 5-HT7R has been shown to couple to G12 protein, leading to Cdc42-mediated filopodia formation in NIH3T3 cells and possibly neurite elongation in mouse hippocampal neurons [17]. The question remains if this pathway may function in ECs as well and if it may contribute to 5-HT7R- dependent EC migration and adhesion.
Conclusion
While the expression of 5-HT7R in ECs was demonstrated almost twenty years ago [22], its role in ECs was not studied. Using pharmacological stimulation and siRNA-mediated downregulation of 5-HT7R, in our present study we have demonstrated that endogenously expressed 5-HT7R promotes EC migration in two different migration assays. We have also shown that 5-HT7R is essential for EC adhesion to extracellular matrix. Finally, we have proposed that 5-HT7R-mediated regulation of migration depends on activation of PKA. For the first time, these data represent the evidence that 5-HT7R may play a critical role in EC function. Based on our data, 5-HT7R has a prominent role in promoting cell migration and adhesion of ECs and may serve as a potential molecular target for pharmacological interventions intended to modulate the processes of migration and adhesion, and possibly other complex processes such as angiogenesis, in which migration and adhesion are critical steps.
Acknowledgement
We thank our former mentor Dr. Tatyana Voyno-Yasenetskaya for her contribution to this study.
References
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back.Science. 2003; 302: 1704-1709.
- Nichols DE, Nichols CD. Serotonin receptors.Chem Rev. 2008; 108: 1614-1641.
- Berger M, Gray JA, Roth BL. The expanded biology of serotonin.Annu Rev Med. 2009; 60: 355-366.
- Machida T, Iizuka K, Hirafuji M. 5-hydroxytryptamine and its receptors in systemic vascular walls.Biol Pharm Bull. 2013; 36: 1416-1419.
- Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders.Gastroenterology. 2007; 132: 397-414.
- Matsusaka S, Wakabayashi I. 5-Hydroxytryptamine as a potent migration enhancer of human aortic endothelial cells.FEBS Lett. 2005; 579: 6721-6725.
- Pakala R, Willerson JT, Benedict CR. Mitogenic effect of serotonin on vascular endothelial cells.Circulation. 1994; 90: 1919-1926.
- Asada M, Ebihara S, Yamanda S, Niu K, Okazaki T, Sora I, et al. Depletion of serotonin and selective inhibition of 2B receptor suppressed tumor angiogenesis by inhibiting endothelial nitric oxide synthase and extracellular signal-regulated kinase 1/2 phosphorylation. Neoplasia. 2009; 11: 408-417.
- Iwabayashi M, Taniyama Y, Sanada F, Azuma J, Iekushi K, Kusunoki H, et al. Role of serotonin in angiogenesis: induction of angiogenesis by sarpogrelate via endothelial 5-HT1B/Akt/eNOS pathway in diabetic mice.Atherosclerosis. 2012; 220: 337-342.
- Hannon J, Hoyer D. Molecular biology of 5-HT receptors.Behav Brain Res. 2008; 195: 198-213.
- Ruat M, Traiffort E, Leurs R, Tardivel-Lacombe J, Diaz J, Arrang JM, et al. Molecular cloning, characterization, and localization of a high-affinity serotonin receptor (5-HT7R) activating cAMP formation. Proceedings of the National Academy of Sciences of the United States of America. 1993; 90: 8547-8551.
- Lovenberg TW, Baron BM, de Lecea L, Miller JD, Prosser RA, Rea MA, et al. A novel adenylyl cyclase-activating serotonin receptor (5-HT7R) implicated in the regulation of mammalian circadian rhythms.Neuron. 1993; 11: 449-458.
- Bard JA, Zgombick J, Adham N, Vaysse P, Branchek TA, Weinshank RL. Cloning of a novel human serotonin receptor (5-HT7R) positively linked to adenylate cyclase.J Biol Chem. 1993; 268: 23422-23426.
- Plassat JL, Amlaiky N, Hen R. Molecular cloning of a mammalian serotonin receptor that activates adenylate cyclase.Mol Pharmacol. 1993; 44: 229-236.
- Meyerhof W, Obermuller F, Fehr S, Richter D. A novel rat serotonin receptor: primary structure, pharmacology, and expression pattern in distinct brain regions. DNA and cell biology. 1993; 12: 401-409.
- Adham N, Zgombick JM, Bard J, Branchek TA. Functional characterization of the recombinant human 5-hydroxytryptamine7(a) receptor isoform coupled to adenylate cyclase stimulation. The Journal of pharmacology and experimental therapeutics. 1998; 287: 508-514.
- Kvachnina E, Liu G, Dityatev A, Renner U, Dumuis A, Richter DW, et al. 5-HT7R receptor is coupled to G alpha subunits of heterotrimeric G12-protein to regulate gene transcription and neuronal morphology.J Neurosci. 2005; 25: 7821-7830.
- Meneses A. Effects of the 5-HT7R receptor antagonists SB-269970 and DR 4004 in autoshaping Pavlovian/instrumental learning task.Behav Brain Res. 2004; 155: 275-282.
- Roberts AJ, Hedlund PB. The 5-HT(7) receptor in learning and memory.Hippocampus. 2012; 22: 762-771.
- Hedlund PB, Danielson PE, Thomas EA, Slanina K, Carson MJ, Sutcliffe JG. No hypothermic response to serotonin in 5-HT7R receptor knockout mice.Proc Natl Acad Sci U S A. 2003; 100: 1375-1380.
- Terron JA, Falcon-Neri A. Pharmacological evidence for the 5-HT7R receptor mediating smooth muscle relaxation in canine cerebral arteries. British journal of pharmacology. 1999; 127: 609-616.
- Ullmer C, Schmuck K, Kalkman HO, Lübbert H. Expression of serotonin receptor mRNAs in blood vessels.FEBS Lett. 1995; 370: 215-221.
- Profirovic J, Strekalova E, Urao N, Krbanjevic A, Andreeva AV, Varadarajan S, et al. A novel regulator of angiogenesis in endothelial cells: 5-hydroxytriptamine 4 receptor.Angiogenesis. 2013; 16: 15-28.
- Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro.Nat Protoc. 2007; 2: 329-333.
- Chen HC. Boyden chamber assay. Methods Mol Biol. 2005; 294: 15-22.
- Rapport MM, Green AA, Page IH. Serum vasoconstrictor, serotonin; isolation and characterization.J Biol Chem. 1948; 176: 1243-1251.
- Watts SW, Morrison SF, Davis RP, Barman SM. Serotonin and blood pressure regulation.Pharmacol Rev. 2012; 64: 359-388.
- Nocito A, Dahm F, Jochum W, Jang JH, Georgiev P, Bader M, et al. Serotonin regulates macrophage-mediated angiogenesis in a mouse model of colon cancer allografts.Cancer Res. 2008; 68: 5152-5158.
- Siddiqui EJ, Shabbir MA, Mikhailidis DP, Mumtaz FH, Thompson CS. The effect of serotonin and serotonin antagonists on bladder cancer cell proliferation.BJU Int. 2006; 97: 634-639.
- Nemecek GM, Coughlin SR, Handley DA, Moskowitz MA. Stimulation of aortic smooth muscle cell mitogenesis by serotonin.Proc Natl Acad Sci U S A. 1986; 83: 674-678.
- Krobert KA, Levy FO. The human 5-HT7R serotonin receptor splice variants: constitutive activity and inverse agonist effects.Br J Pharmacol. 2002; 135: 1563-1571.
- Grueb M, Rohrbach JM, Schlote T, Mielke J. Serotonin (5-HT7R) receptor-stimulated activation of cAMP-PKA pathway in bovine corneal epithelial and endothelial cells.Ophthalmic Res. 2012; 48: 22-27.
- Plopper GE, Huff JL, Rust WL, Schwartz MA, Quaranta V. Antibody-induced activation of beta1 integrin receptors stimulates cAMP-dependent migration of breast cells on laminin-5. Molecular cell biology research communications: MCBRC. 2000; 4: 129-135.
- Harris TE, Persaud SJ, Jones PM. Pseudosubstrate inhibition of cyclic AMP-dependent protein kinase in intact pancreatic islets: effects on cyclic AMP-dependent and glucose-dependent insulin secretion.Biochem Biophys Res Commun. 1997; 232: 648-651.
- Bockaert J, Sebben M, Dumuis A. Pharmacological characterization of 5-hydroxytryptamine4(5-HT4) receptors positively coupled to adenylate cyclase in adult guinea pig hippocampal membranes: effect of substituted benzamide derivatives.Mol Pharmacol. 1990; 37: 408-411.
- Ponimaskin EG, Profirovic J, Vaiskunaite R, Richter DW, Voyno-Yasenetskaya TA. 5-Hydroxytryptamine 4(a) receptor is coupled to the Galpha subunit of heterotrimeric G13 protein.J Biol Chem. 2002; 277: 20812-20819.