
Review Article
Austin J Pharmacol Ther. 2015; 3(2).1069.
The Zebrafish as a Tool to Cancer Drug Discovery
Huiting LN¹, Laroche FJF¹ and Feng H¹*
¹Departments of Pharmacology and Medicine, Cancer Research Center, Section of Hematology and Medical Oncology, Boston University School of Medicine, Boston, MA, USA
*Corresponding author: : Feng H, Departments of Pharmacology and Medicine, Cancer Research Center, Section of Hematology and Medical Oncology, Boston University School of Medicine, Boston, MA, USA
Received: March 23, 2015; Accepted: April 23, 2015; Published: May 04, 2015
Abstract
The ability of zebrafish to faithfully recapitulate a variety of human cancers provides a unique in vivo system for drug identification and validation. Zebrafish models of human cancer generated through methodologies such as transgenesis, gene inactivation, transplantation, and carcinogenic induction have proven similar to their human counterparts both molecularly and pathologically. Suppression of cancer-relevant phenotypes provides opportunities to both identify and evaluate efficacious compounds using embryonic and adult zebrafish. After relevant compounds are selected, preclinical evaluation in mammalian models can occur, delivering lead compounds to human trials swiftly and rapidly. The advantages of in vivo imaging, large progeny, and rapid development that the zebrafish provides make it an attractive model to promote novel cancer drug discovery and reduce the hurdles and cost of clinical trials. This review explores the current methodologies to model human cancers in zebrafish, and how these cancer models have aided in formation of novel therapeutic hypotheses.
Keywords: Zebrafish; Cancer; Drug discovery; Small molecule screens; Efficacy; Toxicity
Abbreviations
MPNST: Malignant Plural Nerve Sheath Tumor; T-ALL: T-cell acute lymphoblastic leukemia; hpf: Hours Post-Fertilization; DHODH: Dihydroorotate Dehydrogenase; B-ALL: B-cell Acute Lymphoblastic Leukemia; AML: Acute Myeloid Leukemia; MPN: Myeloproliferative Neoplasm
Current Challenges in Drug Discovery
The pharmaceutical industry is experiencing lapses in drug development productivity. The predominant drug discovery methodologies for the past 50 years have been target centric. The drug development process in its entirety, from compound identification through preclinical animal models, takes approximately 10-15 years [1]. A high number of compounds are often filtered out during the preclinical animal testing stage, due to failure to meet standards of Absorption, Distribution, Metabolism, and Excretion (ADME). This is because multiple iterations of in vivo studies are performed on preclinical animal models in later stages of drug development (i.e., prior to compounds being relinquished for human trials). Specifically, more than 70% of compounds in oncology fail in phase II clinical trials, while 59% of the remaining compounds are discarded in phase III due to intolerable toxicities [2]. To increase success rates, it is extremely important to test compounds using inexpensive, whole organism vertebrate models during early stages of drug development. Whole organism testing not only provides information on tissue specificity and toxicity, but also determines compound bioavailability that may not be accurately accounted for in a small number of murine models. The zebrafish has emerged as an ideal complementary model system for drug discovery, as it is capable of high throughput screening for discoveryof novel therapeutic compounds or testing of candidate cancer modulators. Research in the past few years has proven the potential of zebrafish to significantly improve the capacity of predicting clinical efficacy and reduce the time and money lost in pushing ineffective drugs to market [3].
The Advantage of the Zebrafish System in Drug Discovery
Zebrafish have emerged as powerful models for drug discovery and biosafety studies because they develop most of the organs found in mammals including those of the nervous, digestive, reproductive, immune, excretory, and cardiovascular systems [4,5]. Zebrafish have a number of unique advantages positioning them for rapid drug discovery and toxicity testing: (i) zebrafish generate large numbers of progeny, offering high confidence in statistical analysis; (ii) zebrafish can absorb compounds solubilized in water, making drug administration simple and feasible; (iii) zebrafish develop rapidly, allowing for assays of drug toxicities on organ development; (iv) the maintenance cost for zebrafish is less expensive than for mammals, decreasing the cost associated with animal husbandry [6]; (v) zebrafish and human share high molecular and genetic homologies, especially for enzymes and cell surface receptors [7]; (vi) zebrafish embryos are as accessible and proliferative as cell culture systems and thus lend themselves to being as applicable as in vitro systems; and (vii) multiple cancer models have been generated in zebrafish and proven similar to their human counterparts molecularly and pathologically, providing excellent tools for anti-cancer drug discovery through large-scale screens, candidate drug testing, and target identification [8,9]. Taken together, these features indicate that the zebrafish is a simple, cost-effective, and faithful model for both drug discovery and toxicological studies.
Zebrafish Models of Human Cancer
Zebrafish cancer models induced by chemicals
While maintaining zebrafish in laboratory conditions, researchers observed diseases developing in adult fish, including cancer. Later studies clarified that after exposure of certain mutagens, zebrafish spontaneously developed almost any tumor type known from humans with similar morphology and comparable signaling pathways. The most common locations for this spontaneous neoplasia to arise include gut, thyroid, and liver. Lower levels of spontaneous neoplasia occur in blood vessels, brains, and gills. In light of spontaneous tumor acquisition, detailed chemical approaches to induce cancer have been developed [10]. To chemically induce cancer, zebrafish are soaked in water dissolved with carcinogens for varied periods of time. Advantageously, zebrafish can endure treatments at a variety of chemical concentrations and durations. For instance, smaller doses, from 5 mM or less can be applied for up to 24 hours, while doses greater than 20 mM are to be applied for 8 hours or less [11]. The treatment of zebrafish with the mutagen 7,12-dimethylbenz(a) anthracene induces the broadest range of tumors, from epithelial tumors in intestines to mesenchymal tumors in blood vessels and lymphoid malignancies (Table 1) [12]. Treatment with N-nitrosodiethylamine is reported to induce pancreatic and liver carcinomas, while N-nitrosodimethylamine specifically induces liver tumors (Table 1) [13].
Zebrafish cancer models resulted from tumor suppressor inactivation
Reverse genetics aims to discover the function of a gene by characterizing phenotypic changes upon gene inactivation. Targeting, and subsequent inactivation of specific tumor suppressor genes has led to zebrafish cancer models of Malignant Plural Nerve Sheath Tumor (MPNST), ocular, liver, and intestinal cancer [14,15]. These cancer types are modeled through silencing of the p53, ptenb, apc, or nf1tumor suppressor genes respectively (Table 1) [15-19]. Targeted in activation of tumor suppressor genesis done by taking advantage of: site-directed mutagenesis, recombination mediated by Cre or Flp recombinases, Targeted Induced Local Lesions In Genomes (TILLING), Zinc Finger Nucleases (ZFN), Transcription Activator-Like Effector Nucleases (TALENs), or Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated (CRISPR/ Cas) technologies [12].
In forward genetic screens, mutations are introduced to the adult zebrafish’s genome through chemical, viral, or transposon-based mutagenesis. The progeny of these mutagenized adult zebrafish are screened for abnormal phenotypes. Genes that harbor genetic mutations are then identified through gene mapping, sequence analysis, and phenotype validation. Using zebrafish in forward genetic screens provides a powerful approach to identify cancer susceptible or novel modifier genes in a specific oncogenic cascade based on their ability to accelerate or suppress tumor phenotypes [20].It has been found that mutations in genes encoding ribosomal proteins have led to development of MPNST, as well as mutations affecting genomic instability that have also been associatedwith MPNST in zebrafish [21,22]. Shephard et al. have discovered that the separase and bmybloss-of-function mutants are susceptible to liver and testicular cancers respectively, through forward genetic screens (Table 1) [23,24].
Transgenic zebrafish cancer models
Transgenic zebrafish expressing mammalian oncogenes provide a convenient platform for modeling human cancers through the misexpression of wild-type or constitutively active form of oncogenes under a zebrafish tissue-specific promoter. To generate transgenic zebrafish models, exogenous DNA is microinjected into one-cell-stage zebrafish embryos. Traditionally, linear or circular DNA plasmids, or artificial bacterial chromosomes are injected into fertilized zebrafish eggs. A large number of eggs need to be injected and screened to compensate for low germline transmission of the transgene of interest to the F1 generation. As transgenic lines are passed on through generations, repetitive DNA becomes susceptible to methylation, leading to the silencing of transgenes [4,5]. Modifications of these early transgenic techniques have led to the development of transposon- or I-SceI meganuclease-mediated transgenesis approaches that significantly improved germline transmission rates in zebrafish [25,26].With these improved techniques, modeling human cancers in zebrafish through transgenesis becomes much easier. There are multiple types of cancers in zebrafish developed through the use of transgenesis (Table 1) [27-44]. MYC-induced T-cell Acute Lymphoblastic Leukemia (T-ALL) and melanoma models in particular have not only been used extensively to gain mechanistic insights into disease pathogenesis, but also in small molecule screens to successfully identify candidate therapeutics for human cancers (Table 2) [25,29,30,32-37,39,40,45-48].
Cancer Type
Zebrafish Model
Compound Library Name
Small Molecule Identified
References
Leukemia
lck:EGFP
ChemBridge DIVERSet
Lenaldekar (LDK)
[73]
T-ALL
rag2:MYC-ER;mitf1a:mitf1a;
rag2:dsRed2
Prestwick Chemical Library
Spectrum Collection
Phenothiazine
[74]
T-ALL
rag2:EGFP-mMyc;CG2
Compounds obtained from Sigma Aldrich
Vincristine and Cyclophosphamide
[75]
AML
Xenotransplantation of K562, K562-R, JURKAT, NB4 human leukemia cell lines into wild-type zebrafish
Imatinib, all-trans retinoic, mafosamide, cyclopjosphamide,
4EGI-1
Imatinib
[76]
ALDH+ myelogenous leukemia
Xenotransplantation of ALDH+ and ALDH- leukemia cells into fli1:EGFP;Casper
imatinib, dasatinib, parthenolide, TDZD-8, arsenic trioxide, niclosamide, salinomycin, and thioridazine
Imatinib
[77]
AML
Hsp70:AML1-ETO
SPECTRUM library
Microsource Discovery Systems
Nimesulide
[48]
Melanoma
mitfa:BRAF(V600E); p53 -/-
SPECTRUM library
Microsource Discovery Systems
Leflunomide
[46]
Pancreatic Cancer
Morpholinos and chemical screens
selective RAR antagonists
RAR antagonist Ro-41-5253
[93]
Human Carcinoma
Xenotransplantation of YD10B and HSC-2 human oral squamous carcinoma cell lines; DLD-1 and HCT116 human colorectal carcinoma cell lines into wild-type zebrafish
Tagged triazine molecules
BII-B9
Paclitaxel
[94]
Table 2: Small Molecule Screens with Zebrafish Cancer Models.
Zebrafish xenograft models of human cancer
An additional methodology to establish cancer models involves the transplantation of human cancer cells into zebrafish embryos. Zebrafish lack an innate immune system until 72 hours postfertilization (hpf) and a mature adaptive immune response until 4 weeks of life [49]. Therefore, human cancer cell lines, purified subpopulations of cancer cells or primary patient cells can be directly injected into zebrafish embryos to study many aspects of tumor biology, such as vasculature remodeling, cancer invasion, and metastasis [50,51]. So far, multiple types of human cancer cell lines and primary patient samples, including gastrointestinal, neuroendocrine, leukemic, and melanoma clinical tumor samples, have been successfully transplanted into 48-hpf zebrafish embryos and demonstrated their usefulness in studying cancer pathogenesis as well as novel drug screening and therapeutic testing of candidate cancer drugs. Invasiveness and micrometastasis of primary human tumors occurs within 24 hours of transplantation [52]. These zebrafish xenograft models of human cancer are especially useful in drug screens allowing for the simultaneous examination of in vivo efficacy and toxicity of candidate drugs [53]. Finally, the advent of pigmentless Casper adult fish has enabled visualization of tumor cell proliferation and dissemination in transplanted recipients beyond zebrafish embryonic stages. Adult zebrafish have three distinct classes of pigment cells: black melanophores, reflective iridophores, and yellow xanothophores. Nacre mutant zebrafish lack melanocytes, while roy orbison zebrafish lack iridophores. Casper zebrafish are double mutant for nacre and roy lacking both melanocytoes and iridophores throughout embryogenesis and adulthood. Casper permits all organs to be seen with stereomicroscopy [40].
Basic Design of Small Molecule Screens using Zebrafish
The ultimate goal of small molecule library screens utilizing zebrafish is to record the greatest number of small molecules that can modulate the activity of zebrafish proteins. Thus, the best molecular library should include as much chemical diversity as possible with a variety of core chemical structures. Moreover, it is of best interest to develop a chemical library that has a high percentage of compounds with similar and consistent physiochemical properties. Chemical treatments can occur at any point during development allowing study of gene inactivation effects by small molecule inhibitors throughout fish development. Moreover, the chemical dosage can be controlled which is advantageous when studying essential functions of tissue specificity [54,55].
The most common chemical library applied in zebrafish screens is the DIVERSet E from Chembridge. This chemical library contains 50,000 compounds occupying a broad pharmacophore space, while excluding non-drug like compounds with undesirable chemical groups and structures. Using the DIVERSet E, Peterson et al., Sternson et al., and Murphey et al. have evaluated phenotypic changes in the central nervous system, cardiovascular system, pigmentation, and organogenesis respectively [56-58]. Phenotypic changes in hematopoiesis are evaluated after application of the NINDS custom collection from the NIH/NINDS, the Spectrum collection from Microsource, and the ICCB collection of known bioactive compounds from BioMol [59]. Changes in embryogenesis and angiogenesis are evaluated through application of 1,3-dioxane library from the Schreiber lab, trisubstituted triazines from the Chang lab, and the LOPAC1280 chemical library from Sigma Aldrich [60-62].
In general, drug screens can be divided into target- or phenotypebased approaches [54,63]. In target-based approaches, the aim is to discover novel probes for defined biological pathways or molecular targets. When this approach is imparted in screening, a greater quality of hits can be generated instead of quantity. A well-designed screen should be able to inform not only the activity of the compounds directed against a particular target, but also other properties of the compounds, such as their permeability, specificity, and off-target effects [55]. Furthermore, target-based chemical screens can either be engineered to report the activity of a specific pathway or they can be formatted to identify compounds that phenocopy a known pathway disruption. Although target-based approaches can lead to the discovery of compounds directed against a specific protein or its related pathways, compounds identified in this manner may not significantly modify the disease of interest [64].
Phenotype-based approaches possess unique advantages in discovering compounds that either elicit a disease phenotype or conversely, rescue a disease phenotype. Novel or well-characterized pharmacological probes are screened in order to identify compounds that induce or modulate a specific phenotype. In their simplest forms, phenotype-based screens might involve searching for an agent that suppresses a cell cycle defect in embryos or rescues mutant phenotype affecting cancer genes. Screens for reversal or prevention of disease phenotypes in an organism are powerful because it allows for the identification of promising chemotherapeutics without bias about which targets are involved in disease pathogenesis. Moreover, many potential targets can be assessed in one screen, and bioactive compounds identified would be of high quality given that the screen output is a suppressor of a disease phenotype in a whole organism [65].
Throughput assays are another basic design of small molecule screens that consist of a small lot of well-characterized pharmacological compounds with known bioactivities. This enables generation of biological connections and hypotheses to be tested quickly since background information about the relevant subset of pharmacological compounds is available [66]. Finally, novel modifier screens can be employed to either identify or rescue disease-related phenotypes. These drug screens involve large libraries of uncharacterized compounds followed by significant efforts to ascertain the mechanisms of action of the identified hits in order to pursue clinical testing [67,68].
Application of Zebrafish in Cancer Drug Discovery
Suitability of zebrafish embryos for anti-cancer drug screens
A remarkable range of biological and disease processes can be studied in the early stages of zebrafish development, but may be limited when using adult zebrafish. Importantly, researchers made the connection between abnormal embryonic phenotypes and increased cancer susceptibility in adulthood, allowing chemical screens to be performed during embryogenesis in order to identify potential compounds and targets that could influence carcinogenesis. To rapidly estimate the therapeutic efficacy of small molecule compounds, wild-type, transgenic, or mutant zebrafish embryos are incubated with the compounds of interest in water (Figure 1). Because the embryos are optically transparent, it is possible to detect functional and morphological changes in internal organs without having to kill and dissect the organism post-compound treatment, a significant advantage over the use of other vertebrate models. Quantitative analysis using microplates can also facilitate comparisons between zebrafish embryos incubated with compounds and vehicles. Microplate analysis of embryos is similar to cell based assays, and is used for primary compound screening at high throughput cores. The small size of zebrafish embryos enables them to be arrayed in a variety of plate formats (12, 24, or 96-wells) and bathed in water containing the compound of interest (Figure 1) [65].
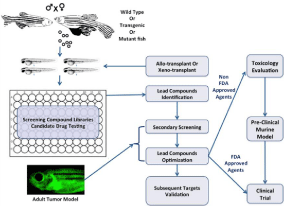
Figure 1: Integrating zebrafish into the cancer drug discovery platform. Zebrafish embryos from wild-type, transgenic, or mutant fish are arrayed into a plate
with candidate agents or compound library to screen for small molecules that elicit desired phenotypes in embryos. Prior to drug treatment, embryos can also
be transplanted with either zebrafish or human cancer cells to determine a compound’s effect in killing tumor cells or inhibiting their migration properties. Adult
zebrafish with cancer can be treated with identified or modified compounds to validate and prioritize the lead compounds for preclinical and clinical testing. Anticancer
agents of unknown biological properties must undergo a toxicological and pre-clinical evaluation prior to clinical tests, while FDA-approved drugs can directly
enter human clinical trials.
The use of multiple transgenic lines with fluorescent reporter molecules further enables detailed analysis of functional and morphological changes [69]. For instance, when compounds are fluorescently-labeled, it facilitates direct visualization of absorption into transparent embryos. Compound excretion following treatment can be observed and measured based on fluorescence properties as well [40]. In the early 2000s, forward chemical screens using zebrafish embryos led to the identification of compounds affecting vertebrate development. These embryo screens narrowed down the number of molecules for future testing in adult zebrafish models of tumor and mammals (Figure 1) [51]. As multiple zebrafish xenograft models of cancer are established, researchers have demonstrated their efficacy in small molecule screens. This is accomplished by soaking xenografted embryos in a compound solution or treating cancer cells with small molecules before implantation into transparent embryos (Figure 1) [45].
Fortunately, many toxic responses have been conserved between zebrafish and human toxicological studies.Therefore, zebrafish embryos are valuable in the assessment of system toxicity, which can be evaluated using overall embryonic mortality as a metric from which the working range of a compound can be determined. To do this, embryos are arrayed in a multi-well plate and exposed to drugs with a dose gradient to determine the maximum tolerable dose. Preliminary hits from screening can all be tested for toxicity as a means of prioritizing compounds [70,71].
Adult zebrafish for cancer drug validation
Adult zebrafish are often limited in their capacity to aid in drug screenings as adult zebrafish are relatively larger and less transparent. With the availability of multiple zebrafish models of cancer (Table 1), investigators can treat adult fish with tumors to validate the antineoplastic efficacy of candidate agents. Dissemination of tumors throughout the zebrafish can occur at varying rates, typically from 5 to 10 days post transplantation. Moreover, the appearance of these disseminated tumor cells may not occur until 2-3 weeks post transplantation. The innovation of the transparent Casper zebrafish has improved opacity in adult zebrafish [72], and provided a tool for exploration of tumor dissemination mechanism and subsequent metastatic growth throughout fish development. Advantages of using the Casper fish, in combination with syngeneic transplantation, include in vivo monitoring of tumor engraftment and migration following transplantation.
Methods for modeling zebrafish cancers
Chemicals or genetic alterations
Cancer types
References
Chemical Treatment
N-2-Flurenylacetamide
N-nitrosodiethylamine
N-dimethylnitrosamine
N-methyl-N'-nitro-N-nitrosoguanidine
7,12-dimethylbenz[a]anthracene
Liver
Cholangiolar tumors
Testicular germ cell tumor
Liver, Intestinal
Testis, Germ cell tumor, Vascular
[13]
[91]
[92]
[12]
[23]
Reverse genetics
p53
ptena
ptenb
apc
nf1a
nf1b
MPNST
Ocular
Hemangiosarcoma
Liver, intestinal
Gliomas
[15]
[16]
[16,17]
[18]
[19]
Forward genetics
Ribosomal protein gene
separase
bmyb
Genomic instability mutations
MPNST
Epithelial tumors
Liver, Intestinal
MPNST/others
Testicular germ cell tumor, Vascular tumor
[21]
[23]
[24]
[22]
[12]
Transgenesis
TEL-AML1
Myc
MYC
Akt2
NOTCH1
MY5T3-NCOA2
NUP98-HOXA9
KRASG12D
BRAF-V600E
HRASG12V
HRASG12V
MYCN
Xmrk, kras, Myc
B-ALL
T-ALL
T-ALL
T-ALL
T-ALL
AML
MPN
Rhabdomyosarcoma
Melanoma
Melanoma, liver cancer
Melanoma
Neuroendocrine tumor, Neuroblastoma, MPN
Liver cancer
[27]
[28-30]
[31]
[31]
[32]
[33]
[44]
[34,35]
[36]
[37,38]
[39]
[40-42]
[43]
Table 1: Zebrafish Cancer Models.
Advances using zebrafish in cancer drug discovery
Collectively, the use of embryo screens and drug validation in adult zebrafish models of cancer represents the ideal means to identify and prioritize candidate agents for further testing in mammals (Figure 1). Researchers have taken advantage of the zebrafish cancer paradigm and have successfully identified candidate compounds for cancer treatment. A novel high-content in vivo screen was conducted by Ridges et al. using genetically engineered T-cell reporting zebrafish larvae (lck:EGFP) (Table 2) [68]. By exploiting the developmental similarities between immature and malignant T-cells, the activity of small molecule libraries was assayed against immature T-cells with a corresponding visual readout in zebrafish larvae. It was found that the compound, Lehaldekar, was able to abolish immature T-cells in developing zebrafish without affecting cell cycle in other cell variants. Moreover, Lehaldekar was tolerated in murine models. Notably, Myc-driven T-ALL in adult zebrafish was induced into remission upon Lehaldekar treatment [73].
Gutierrez et al. designed a fluorescence-based small molecule screen to identify compounds that were selectively cytotoxic to MYC-overexpressing thymocytes in a MYC-induced (activated by tamoxifen) T-ALL background (Table 2) [74]. At three days post fertilization, FDA-approved drugs were added to embryos. Four days later, dsRed2 fluorescence expression in thymocytes was imaged via microscopy. Screening for the decreased dsRed2 expression in thymocytes lead to the identification of perphenazine, an antipsychotic drug, with antileukemic activity. Importantly, perphenazine was also found as an antileukemic agent in a parallel cell-based screen, and validated to induce apoptosis in fish, mouse and human T-ALL cells [74].
Mizgrev et al. aimed to validate the efficacy and faithfulness of zebrafish embryos in drug discovery. T-ALL was induced in zebrafish by mosaic expression of EGFP-fused mMYC transgene under a lymphocyte-specific promoter rag2 (Table 2) [75]. Subsequently, primary tumor cells were transplanted into recipient zebrafish larvae that were treated with vincristine and cyclophosphamide five days post leukemia engraftment. Drug efficacy and relevance toembryonic development was determined based on a compound’s ability to increase larvae lifespan [75].
Pruvot et al. injected human leukemia cell lines or blasts from patients with acute myelogenous leukemia into zebrafish embryos at 48 hpf (Table 2). Compounds that demonstrated no toxicity in normal zebrafish embryos and decreased leukemia burdens in xenografted zebrafish were ideal, ultimately validating anti-leukemic efficacy of imatinib and oxaphorines [76]. Similarly, Zhang et al. injected purified human leukemic stem cells into zebrafish embryos based on kusabriaorange fluorescence [77]. Post-transplanted fish were treated with selected therapeutic agents (e.g. imatinib, dasatinib, parthenolide, etc.). Cell proliferation and migration were subsequently evaluated using high-content imaging, and recapitulated the ability of the drugs’ to inhibit LSCs in both in vitro and murine studies – thus validating their methodology for future anti-LSC drug discovery (Table 2) [77].
Yeh et al. created an assay with zebrafish embryos that faithfully recapitulated the effects of the oncogene AML1-ETO to block cell differentiation in multipotent progenitor cells (Tables 1 and 2). A chemical screen of 200 bioactive chemicals was then performed in an effort to find a compound that would suppress the differentiation blockade of oncogenic AML1-ETO on hematopoietic progenitor cells. The Cox2 inhibitor nimesulide was identified as the lead hit of the screen. Their follow-up study demonstrated a previously unknown role for COX-2 and β-catenin in AML1-ETO-mediated hematopoietic differentiation [48].
Recently, White et al. preformed a small molecule screen that identified an inhibitor of Dihydroorotate Dehydrogenase (DHODH), leflunomide, which diminished the self-renewal of mammalian neural crest stem cells (Table 2) [46]. This chemical screen was performed using transgenic mitfa:BRAF(V600E) zebrafish embryos that had defective p53 activity. The inhibition of DHODH through leflunomide alone or in combination with an oncogenic inhibitor of BRAF (V600E) was able to successfully suppress melanoma growth in vitro and in murine models [46].
The Zebrafish as a Model for Studying Tumor Metastasis
Tumor metastasis is an extremely complex, multistep cascade [78]. In total, there are at least four major steps involved: the local invasion and intravasation; the survival of tumor cells in circulation; the extravasation (the exit of the tumor cells from vasculature); and the distant proliferation and colonization [79]. By coupling optical transparency with the differential fluorescent labeling of tumor cells, inflammatory cells, and vasculature, zebrafish enable noninvasive, real-time visualization of metastatic events in vivo, distinguishing among each of the cell types and their dynamic interactions leading to intravasation and metastasis [80]. In particular, zebrafish xenograft models of cancer faithfully recapitulate the metastatic potentials of human cancer cells as in xenografted mouse models and in patients [81,82]. As such the zebrafish is particularly well suited for realtime monitoring of metastatic events in vivo [80]. The generation of fli1:EGFP and flk1:mCherry fish, in which the vasculature expresses EGFP or mCherry fluorescent protein, provides unprecedented high-resolution imaging of complex intravasation and extravasation processes [83,84].
Stoletov et al. combined the use of fli1:EGFP transgenic fish and fluorescently-labeled human cancer cellsin high-resolution confocal microscopy studies, demonstrating the role of RhoC and VEGF in intravasation [85]. Using a similar methodology, they subsequently visualized extravasation dynamics of metastatic tumor cells in zebrafish [86]. Their findings show that different from intravasation, extravasation does not require damaged vessels or vascular leakage, but instead is induced by local vessel remodeling (i.e., clustering of endothelial cells and cell-cell junctions). The availability of transparent Casper fish makes it feasible to study tumor cell intravasation and metastasis in adult fish. Using Casper; fli1:EGFP fish, Feng et al. demonstrated that elevated S1P1 levels inhibit lymphoma cell intravasation, whereas AKT activation or an S1P1 antagonist can overcome this blockade [87]. Ignatius et al. showed that in embryonal rhabdomyosarcoma (ERMS), myf5+ ERMS-propagating cells do not intravasate, while the more differentiated myogenin+ ERMS cells can readily invade the vasculature [88]. The ability of studying metastatic events in zebrafish coupled with the high fecundity of zebrafish suit this organism for small molecule screens to identify novel antimetastatic agents, especially repurposing the FDA-approved drugs.
The Future Perspectives
Due to their genetic similarities to humans, the zebrafish serve as a relevant in vivo system to complement mammalian systems. The optical clarity of zebrafish embryos and mutant Casper adult zebrafish has provided a foundation for assessment and imaging of tumor phenotypes through application of novel or known compounds. The embryonic zebrafish serves as the jumping off point for lead compound identification, while validation in adult zebrafish allows for longitudinal observation of compound impact on tumor cell growth, dissemination, and metastasis in an anatomically relevant system. Additionally, zebrafish allow for the assessment of absorption, distribution, metabolism, excretion, and toxicology properties of thousands of chemical compounds. These assessments are done at much quicker speeds, lower costs, and greater scale and efficiency. Although only being applied in small molecule screens for a few years, zebrafish have already demonstrated their great potential in cancer drug screens and novel compound identification.
The establishment of zebrafish cancer cell lines, especially for the ones with known and relevant oncogenes,would be advantageous for functional cell-based studies and would also enable the prescreening genetic and chemical modifiers of cancer. Without a doubt, anti-cancer screens in zebrafish will be rapidly improved as robots and more readout automation are implemented in the procedures. An automated micro-injector allows a greater number of fish to be xenografted with high accuracy, improving the standardization of the screens and allow a large-scale screen to be performed in a shorter time frame [89]. Furthermore, automated imaging system (or an ImageJ plugin) optimized to detect or quantify a specific phenotypic feature of interest will drastically improve the readout quality and lower extensive labor necessary to perform such screens and identify candidates molecules [90]. With automation and standardization of these procedures, zebrafish can contribute to personalized medicine. For instance, ex vivo screening of cancer cells from an individual patient’s could be conducted in a zebrafish xenograft model for the testing of multiple drugs and drug combinations, and the readout would inform clinicians on measures of both drug potency and toxicity, improving therapy selection on a case-by-case basis.
While we continue to maximize the use of zebrafish in highthroughput screens, we must keep in mind that the anatomical and molecular differences of zebrafish with humans may cause the elimination of a fraction of the hits generated. Thus, the human relevancy of compounds identified through the use of the zebrafish models must be carefully investigated. Because it still remains unclear to what degree the molecules discovered in zebrafish screens impart similar effects in humans, the predictive toxicity in zebrafish may not equate to that of human toxicity. Fortunately, so far, tested compounds (e.g., ones in regards to cell cycle affecters) showed a 50- 70% similarity in effect between mammalian and zebrafish cells [3]. An even greater degree of drug conservation of 95% was observed in treating zebrafish and human with cardiac modulators [4]. The high degree of conservation between fish and man further establishes the usefulness of the zebrafish as an important tool to human cancer drug discovery.
Acknowledgements
The authors thank Jay Baxter, Baker Logan, and Julia Warren for helpful comments and suggestions. L.N.H is supported by a training grant (T32GM008541) from the National Institutes of Health. H.F. is supported by a grant (R00CA134743) from the National Institutes of Health, a career development grant from the St. Baldrick’s Foundation, a Karin Grunebaum Faculty Fellowship from the Karin Grunebaum Cancer Foundation, a Ralph Edwards Career Development Professorship from Boston University, and an Institutional Research Grant (IRG –72-001-36-IRG) from the American Cancer Society.
References
- Hughes JP, Rees S, Kalindjian SB, Philpott KL. Principles of early drug discovery. Br J Pharmacol. 2011; 162: 1239-1249.
- Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010; 9: 203-214.
- Ablain J, Zon LI. Of fish and men: using zebrafish to fight human diseases. Trends Cell Biol. 2013; 23: 584-586.
- Goldsmith JR, Jobin C. Think small: zebrafish as a model system of human pathology. J Biomed Biotechnol. 2012; 2012: 817341.
- Shin JT, Fishman MC. From Zebrafish to human: modular medical models. Annu Rev Genomics Hum Genet. 2002; 3: 311-340.
- Goessling W, North TE, Zon LI. New waves of discovery: modeling cancer in zebrafish. J Clin Oncol. 2007; 25: 2473-2479.
- Barbazuk WB, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bedell JA. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000; 10: 1351-1358.
- Blackburn JS, Langenau DM. Zebrafish as a model to assess cancer heterogeneity, progression and relapse. Dis Model Mech. 2014; 7: 755-762.
- Phillips JB, Westerfield M. Zebrafish models in translational research: tipping the scales toward advancements in human health. Dis Model Mech. 2014; 7: 739-743.
- Feitsma H, Cuppen E. Zebrafish as a cancer model. Mol Cancer Res. 2008; 6: 685-694.
- Pliss GB, Khudoley VV. Tumor induction by carcinogenic agents in aquarium fish. J Natl Cancer Inst. 1975; 55: 129-136.
- Neumann JC, Dovey JS, Chandler GL, Carbajal L, Amatruda JF. Identification of a heritable model of testicular germ cell tumor in the zebrafish. Zebrafish. 2009; 6: 319-327.
- Lam SH, Wu YL, Vega VB, Miller LD, Spitsbergen J, Tong Y, et al. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nature biotechnology. 2006; 24: 73-75.
- Skromne I, Prince VE. Current perspectives in zebrafish reverse genetics: moving forward. Developmental dynamics: an official publication of the American Association of Anatomists. 2008; 237: 861-882.
- Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proceedings of the National Academy of Sciences of the United States of America. 2005; 102: 407-412.
- Faucherre A, Taylor GS, Overvoorde J, Dixon JE, Hertog Jd. Zebrafish pten genes have overlapping and non-redundant functions in tumorigenesis and embryonic development. Oncogene. 2008; 27: 1079-1086.
- Choorapoikayil S, Kuiper RV, de Bruin A, den Hertog J. Haploinsufficiency of the genes encoding the tumor suppressor Pten predisposes zebrafish to hemangiosarcoma. Dis Model Mech. 2012; 5: 241-247.
- Haramis AP, Hurlstone A, van der Velden Y, Begthel H, van den Born M, Offerhaus GJ, et al. Adenomatous polyposis coli-deficient zebrafish are susceptible to digestive tract neoplasia. EMBO Rep. 2006; 7: 444-449.
- Shin J, Padmanabhan A, de Groh ED, Lee JS, Haidar S, Dahlberg S, et al. Zebrafish neurofibromatosis type 1 genes have redundant functions in tumorigenesis and embryonic development. Disease models & mechanisms. 2012; 5: 881-894.
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996; 123: 37-46.
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004; 2: E139.
- Moore JL, Rush LM, Breneman C, Mohideen MA, Cheng KC. Zebrafish genomic instability mutants and cancer susceptibility. Genetics. 2006; 174: 585-600.
- Shepard JL, Amatruda JF, Finkelstein D, Ziai J, Finley KR, Stern HM, et al. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes & development. 2007; 21: 55-59.
- Shepard JL, Amatruda JF, Stern HM, Subramanian A, Finkelstein D, Ziai J, et al. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proceedings of the National Academy of Sciences of the United States of America. 2005; 102: 13194-13199.
- Kawakami K1. Transposon tools and methods in zebrafish. Dev Dyn. 2005; 234: 244-254.
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002; 118: 91-98.
- Sabaawy HE, Azuma M, Embree LJ, Tsai HJ, Starost MF, Hickstein DD. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2006; 103: 15166-15171.
- Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, Lin S. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003; 299: 887-890.
- Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2005; 102: 6068-6073.
- Feng H, Langenau DM, Madge JA, Quinkertz A, Gutierrez A, Neuberg DS, et al. Heat-shock induction of T-cell lymphoma/leukaemia in conditional Cre/lox-regulated transgenic zebrafish. Br J Haematol. 2007; 138: 169-175.
- Gutierrez A, Grebliunaite R, Feng H, Kozakewich E, Zhu S, Guo F, et al. Pten mediates Myc oncogene dependence in a conditional zebrafish model of T cell acute lymphoblastic leukemia. The Journal of experimental medicine. 2011; 208: 1595-1603.
- Chen J, Jette C, Kanki JP, Aster JC, Look AT, Griffin JD. NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia. 2007; 21: 462-471.
- Zhuravleva J, Paggetti J, Martin L, Hammann A, Solary E, Bastie JN, et al. MOZ/TIF2-induced acute myeloid leukaemia in transgenic fish. Br J Haematol. 2008; 143: 378-382.
- Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007; 21: 1382-1395.
- Le X, Langenau DM, Keefe MD, Kutok JL, Neuberg DS, Zon LI. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2007; 104: 9410-9415.
- Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005; 15: 249-254.
- Anelli V, Santoriello C, Distel M, Köster RW, Ciccarelli FD, Mione M. Global repression of cancer gene expression in a zebrafish model of melanoma is linked to epigenetic regulation. Zebrafish. 2009; 6: 417-424.
- Nguyen AT, Emelyanov A, Koh CH, Spitsbergen JM, Parinov S, Gong Z. An inducible kras(V12) transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis Model Mech. 2012; 5: 63-72.
- Dovey M, White RM, Zon LI. Oncogenic NRAS cooperates with p53 loss to generate melanoma in zebrafish. Zebrafish. 2009; 6: 397-404.
- Yang HW, Kutok JL, Lee NH, Piao HY, Fletcher CD, Kanki JP, et al. Targeted expression of human MYCN selectively causes pancreatic neuroendocrine tumors in transgenic zebrafish. Cancer Res. 2004; 64: 7256-7262.
- Zhu S, Lee JS, Guo F, Shin J, Perez-Atayde AR, Kutok JL, et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell. 2012; 21: 362-373.
- Shen LJ, Chen FY, Zhang Y, Cao LF, Kuang Y, Zhong M, et al. MYCN transgenic zebrafish model with the characterization of acute myeloid leukemia and altered hematopoiesis. PLoS One. 2013; 8: e59070.
- Zheng W, Li Z, Nguyen AT, Li C, Emelyanov A, Gong Z. Xmrk, kras and myc transgenic zebrafish liver cancer models share molecular signatures with subsets of human hepatocellular carcinoma. PloS one. 2014; 9: e91179.
- Forrester AM, Grabher C, McBride ER, Boyd ER, Vigerstad MH, Edgar A, et al. NUP98-HOXA9-transgenic zebrafish develop a myeloproliferative neoplasm and provide new insight into mechanisms of myeloid leukaemogenesis. British journal of haematology. 2011; 155: 167-181.
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011; 471: 513-517.
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011; 471: 518-522.
- Yeh JR, Munson KM, Chao YL, Peterson QP, Macrae CA, Peterson RT. AML1-ETO reprograms hematopoietic cell fate by downregulating scl expression. Development. 2008; 135: 401-410.
- Yeh JR, Munson KM, Elagib KE, Goldfarb AN, Sweetser DA, Peterson RT. Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nature chemical biology. 2009; 5: 236-243.
- Novoa B, Figueras A. Zebrafish: model for the study of inflammation and the innate immune response to infectious diseases. Adv Exp Med Biol. 2012; 946: 253-275.
- Konantz M, Balci TB, Hartwig UF, Dellaire G, André MC, Berman JN, et al. Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci. 2012; 1266: 124-137.
- Veinotte CJ, Dellaire G, Berman JN. Hooking the big one: the potential of zebrafish xenotransplantation to reform cancer drug screening in the genomic era. Dis Model Mech. 2014; 7: 745-754.
- Stuart GW, McMurray JV, Westerfield M. Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development. 1988; 103: 403-412.
- Ignatius MS, Langenau DM. Fluorescent imaging of cancer in zebrafish. Methods Cell Biol. 2011; 105: 437-459.
- Murphey RD, Zon LI. Small molecule screening in the zebrafish. Methods. 2006; 39: 255-261.
- Tamplin OJ, White RM, Jing L, Kaufman CK, Lacadie SA, Li P, et al. Small molecule screening in zebrafish: swimming in potential drug therapies. Wiley interdisciplinary reviews Developmental biology. 2012; 1: 459-468.
- Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proceedings of the National Academy of Sciences of the United States of America. 2000; 97: 12965-12969.
- Sternson SM, Louca JB, Wong JC, Schreiber SL. Split--pool synthesis of, 3-dioxanes leading to arrayed stock solutions of single compounds sufficient for multiple phenotypic and protein-binding assays. J Am Chem Soc. 2001; 123: 1740-1747.
- Murphey RD, Stern HM, Straub CT, Zon LI. A chemical genetic screen for cell cycle inhibitors in zebrafish embryos. Chem Biol Drug Des. 2006; 68: 213-219.
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007; 447: 1007-1011.
- Spring DR, Krishnan S, Blackwell HE, Schreiber SL. Diversity-oriented synthesis of biaryl-containing medium rings using a one bead/one stock solution platform. J Am Chem Soc. 2002; 124: 1354-1363.
- Jung DW, Williams D, Khersonsky SM, Kang TW, Heidary N, Chang YT, et al. Identification of the F1F0 mitochondrial ATPase as a target for modulating skin pigmentation by screening a tagged triazine library in zebrafish. Mol Biosyst. 2005; 1: 85-92.
- Tran TC, Sneed B, Haider J, Blavo D, White A, Aiyejorun T, et al. Automated, quantitative screening assay for antiangiogenic compounds using transgenic zebrafish. Cancer research. 2007; 67: 11386-11392.
- MacRae CA, Peterson RT. Zebrafish-based small molecule discovery. Chem Biol. 2003; 10: 901-908.
- Peterson RT, Fishman MC. Designing zebrafish chemical screens. Methods Cell Biol. 2011; 105: 525-541.
- Laggner C, Kokel D, Setola V, Tolia A, Lin H, Irwin JJ, et al. Chemical informatics and target identification in a zebrafish phenotypic screen. Nature chemical biology. 2012; 8: 144-146.
- Delvecchio C, Tiefenbach J, Krause HM. The zebrafish: a powerful platform for in vivo, HTS drug discovery. Assay Drug Dev Technol. 2011; 9: 354-361.
- Taylor KL, Grant NJ, Temperley ND, Patton EE. Small molecule screening in zebrafish: an in vivo approach to identifying new chemical tools and drug leads. Cell Commun Signal. 2010; 8: 11.
- Basu S, Sachidanandan C. Zebrafish: a multifaceted tool for chemical biologists. Chem Rev. 2013; 113: 7952-7980.
- Terriente J, Pujades C. Use of zebrafish embryos for small molecule screening related to cancer. Dev Dyn. 2013; 242: 97-107.
- Eimon PM, Rubinstein AL. The use of in vivo zebrafish assays in drug toxicity screening. Expert Opin Drug Metab Toxicol. 2009; 5: 393-401.
- Hill AJ, Teraoka H, Heideman W, Peterson RE. Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicol Sci. 2005; 86: 6-19.
- White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008; 2: 183-189.
- Ridges S, Heaton WL, Joshi D, Choi H, Eiring A, Batchelor L, et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood. 2012; 119: 5621-5631.
- Gutierrez A, Pan L, Groen RW, Baleydier F, Kentsis A, Marineau J, et al. Phenothiazines induce PP2A-mediated apoptosis in T cell acute lymphoblastic leukemia. The Journal of clinical investigation. 2014; 124: 644-655.
- Mizgirev IV, Revskoy S. A new zebrafish model for experimental leukemia therapy. Cancer Biol Ther. 2010; 9: 895-902.
- Pruvot B, Jacquel A, Droin N, Auberger P, Bouscary D, Tamburini J, et al. Leukemic cell xenograft in zebrafish embryo for investigating drug efficacy. Haematologica. 2011; 96: 612-616.
- Zhang B, Shimada Y, Kuroyanagi J, Umemoto N, Nishimura Y, Tanaka T. Quantitative phenotyping-based in vivo chemical screening in a zebrafish model of leukemia stem cell xenotransplantation. PLoS One. 2014; 9: e85439.
- Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006; 127: 679-695.
- Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003; 3: 453-458.
- White R, Rose K, Zon L. Zebrafish cancer: the state of the art and the path forward. Nat Rev Cancer. 2013; 13: 624-636.
- Marques IJ, Weiss FU, Vlecken DH, Nitsche C, Bakkers J, Lagendijk AK, et al. Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC cancer. 2009; 9: 128.
- Teng Y, Xie X, Walker S, White DT, Mumm JS, Cowell JK. Evaluating human cancer cell metastasis in zebrafish. BMC Cancer. 2013; 13: 453.
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002; 248: 307-318.
- Gore AV, Monzo K, Cha YR, Pan W, Weinstein BM. Vascular development in the zebrafish. Cold Spring Harb Perspect Med. 2012; 2: a006684.
- Stoletov K, Montel V, Lester RD, Gonias SL, Klemke R. High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2007; 104: 17406-17411.
- Stoletov K, Kato H, Zardouzian E, Kelber J, Yang J, Shattil S, et al. Visualizing extravasation dynamics of metastatic tumor cells. J Cell Sci. 2010; 123: 2332-2341.
- Feng H, Stachura DL, White RM, Gutierrez A, Zhang L, Sanda T, et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P, and ICAM, leading to a blockade of tumor cell intravasation. Cancer Cell. 2010; 18: 353-366.
- Ignatius MS, Chen E, Elpek NM, Fuller AZ, Tenente IM, Clagg Ret al. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer cell. 2012; 21: 680-693.
- Spaink HP, Cui C, Wiweger MI, Jansen HJ, Veneman WJ, Marín-Juez R, et al. Robotic injection of zebrafish embryos for high-throughput screening in disease models. Methods. 2013; 62: 246-254.
- Veneman WJ, Stockhammer OW, de Boer L, Zaat SA, Meijer AH, Spaink HP. A zebrafish high throughput screening system used for Staphylococcus epidermidis infection marker discovery. BMC Genomics. 2013; 14: 255.
- Mizgireuv IV, Majorova IG, Gorodinskaya VM, Khudoley VV, Revskoy SY. Carcinogenic effect of N-nitrosodimethylamine on diploid and triploid zebrafish (Danio rerio). Toxicol Pathol. 2004; 32: 514-518.
- Mizgireuv IV, Revskoy SY. Transplantable tumor lines generated in clonal zebrafish. Cancer Res. 2006; 66: 3120-3125.
- Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009; 137: 2136-2145.e1-7.
- Jung DW, Oh ES, Park SH, Chang YT, Kim CH, Choi SY, Williams DR. A novel zebrafish human tumor xenograft model validated for anti-cancer drug screening. Mol Biosyst. 2012; 8: 1930-1939.