Research Article
Austin J Public Health Epidemiol. 2014;1(2): 1006.
Early Diagnosis of Neonatal Sepsis Caused by Yeast Infection
Mokhtar E1, El-Shereef A2, Abdel-Kader A3, Al-Tounisy A4 and Karam El-Din A1*
1Departments of Microbiology, Ain-Shams University, Egypt
2Department of Microbiology and Immunology, Ain-Shams University, Egypt
3Department of Clinical Pathology, Ain-Shams University, Egypt
4Department of Pediatrics, Al-Azhar University, Egypt
*Corresponding author: Al-Zahraa A Karam El-Din, Department of Microbiology, Ain-Shams University, Abasia Cairo, Egypt
Received: June 20, 2014; Accepted: July 18, 2014; Published: July 21, 2014
Abstract
Early diagnosis of neonatal sepsis is lifesaving to the neonates. The present study was conducted to clarify the rate of incidence of neonatal early and late onset sepsis caused by yeast infection. Conventional methods, Buffy coat examination and molecular (PCR) methods were adopted and compared for the isolation of etiologic agents. In addition, other tests were also carried out to complete our data. One hundred and twenty neonates suspected of having sepsis were identified. One hundred and ten cases were found positive using blood cultures and PCR, and 20 neonates were negative. Torch antibodies were detected for the negative cases confirming viral septicemia. The remaining 100 positive cases were classified as bacterial (88%) and yeast infections (12%). Candida albicans was isolated from 11 cases (91.7%) while Debaryomyces hansenii was isolated from one case only, representing 8.3% of the positive yeast isolates. The analysis of neonatal cases showed that there is a perfect correlation between molecular and microbiological data. PCR for the 12 cases having yeast infection gave positive results at 615 bp, and that for C. albicans was observed at 156 bp. In conclusion PCR is the best and rapid method for early detection of neonatal yeast infection.
Keywords: Neonatal sepsis; Candidemia; Laboratory diagnosis
Introduction
Neonatal infections remain a major cause of morbidity and mortality and death in newborn infants [1]. Septicemia is a systemic illness caused by spread of microbes or their toxins via the blood stream [2]. The reported incidence of neonatal sepsis varies from 7.1 to 38 per 1000 live births in Asia [3,4], from 6.5 to 23 per 1000 live births in Africa [5,6] and from 3.5 to 8.4 per 1000 live births in South America and the Caribbean [7,8]. By comparison, rates reported in the United States and Australia range from 6-9 per 1000 [9,10]. Neonatal sepsis classified according to the time of onset of the disease: early onset (EOS) and late onset (LOS). The distinction has clinical relevance, as "EOS" disease is mainly due to pathogens acquired before and during delivery, and "LOS" disease to pathogen acquired after delivery (nosocomial or community source). Some reports distinguish between early onset (within 24 hours), "EOS" (24 to 6 days), and "LOS" (more than 6 days) sepsis [4,11]. Clinical diagnosis of sepsis is not easy, because, symptoms and signs are not specific and dramatic deterioration of clinical conditions can supervene rapidly long before blood cultures results are available even in asymptomatic newborn infants [12]. Predisposing risk factors associated with neonatal sepsis include neonatal group B streptococcal infection (GBS) colonization, premature rupture of membrane (PROM), and chorioamionitis12. Neonatal candidiasis can be subdivided into two categories, catheter related candidemia and disseminated or invasive candidiasis [13]. Candida spp are the common cause of nosocomial infections in neonatal intensive care units (ICUs). Although C. albicans has historically been the most frequently species isolated, infections caused by other species of Candida have been diagnosed with increased frequency [14,15]. Diagnosis of sepsis is difficult and there are no laboratory tests with 100% specificity and sensitivity, (with the exception of blood cultures which needs at least 48-72 hours). PCR methodology has been used to diagnose different infections, and the possibility of amplifying the DNA region common for all microorganisms could represent an optimal method for the diagnosis of sepsis [9,10]. PCR also allows rapid onset of treatment [1,15].
Materials and Methods
Patients
This study was carried out at the neonatal intensive care unit (during 2004) of Al-Azhar University Hospital, Cairo, Egypt on 120 neonates of an age group ranging from one day to four weeks suspected to have neonatal sepsis.
Sample collection and pre-analytical preparation
Two ml. Of venous blood were drawn in sterile EDTA-treated tubes (Bekton Dickinson Vaccutainer system, Europe, UK). One ml. of venous blood was used for blood culture and one ml. was used for molecular analysis. The blood for molecular analysis was stored at -20OC until use. A complete blood count was carried out using Hemat 8 (Radium Group) and the detection of C-reactive protein level. A Gram stained smear of the plasma buffy coat layer, obtained by centrifuging anti-coagulated capillary was examined and a rapid diagnosis of bacteremia in neonates was carried out [16].
Identification of the isolates
Venous blood samples were inoculated on Sabouraud dextrose agar (SDA) and Sabouraud-Brain Heart Infusion (BHI) broth and incubated for 2 weeks at 25°C prior to the identification of fungal isolates. The isolates were then identified using two techniques, ApI 20C Aux (Bio Mérieux SA, Marcy-L'Eliale, France) and the Micro-scan 4-Hour Rapid Yeast Identification panel (YIP, Baxter- Microscan, W. Sacramento, Calif.).
Polymerase chain reaction (PCR)
DNA Extraction from whole blood was carried out using "QIAamp DNA blood kit" (Quiagen, Inc.), this was followed by detection of yeast DNA using the universal primers "F-5'-GCETATCAATAAGCGGAGGAAAA-'3" and "R-5'- GGTCCGTGTTTCAGAAG-'3" that amplifies the V3 region of the large subunit of the ribosomal DNA [17]. For identification of Candida albicans, the following specific PCR primers were used "F-5'-TTGGAGCGGCAGGATAATCG-'3" and "R-5'GGTCCGTGTTTCAAGACG-'3" [18]. Detection of the amplified.
PCR product was carried out using gel electrophoresis. The fragments were separated in 2% agarose gel stained with ethidium bromide and visualized using ultraviolet transillumination.
Statistical analysis
The data of the study were analyzed using "SPSS" version 12 and this data was represented in a descriptive and analytical form. Appropriate statistical tests were chosen for each table and figure, depending on the type of data present, either Chi square (X2) and Fisher exact test (if cells are less than 5). The accepted levels of significance were 0.05 or less.
Ethical issues
The Ethics Committees of Al-Azhar University Hospital Cairo, Egypt approved the study. All patient information and test results were kept confidential.
Results
In the present study, one hundred neonates were positive for blood cultures and PCR, Table 1 shows the number of cases and percentage of each etiologic agent.
Causal agent
No.
%Total cases
% Positive cases
Bacteria
Viruses
Yeasts
88
20
12
73.33
16.67
10.00
88.00
-
12.00
Total
130
100
100
Table 1: Number and percentage of the etiologic agents causing sepsis.
Two yeast species were isolated from blood of 12 neonates. Candida albicans was the predominant species, and isolated from 11 neonates representing 91.7% of the total yeast cases, while D.hansenii (C. famata) was isolated from one case only, representing 8.3% of the total yeast isolates (Table 2).
Yeast isolates
No = 12
%
Candida albicans
Debaryomyces hansenii
11
1
91.7
8.3
Total
12
100
Table 2: The isolated yeast species from blood of neonates.
The different methods adopted for the identification of yeast infections were compared. There was no difference between the sensitivity of blood cultures and PCR results (Table 3). It was observed that male neonates weighing less than 2.5kg are highly affected as in Table 4.
Method
No = 12
%
Blood culture
PCR
Buffy Coat
12
12
8
100
100
66.7
Table 3: Sensitivity comparison between the different methods used in the identification of yeast isolates.
Risk factors
C. albicans
D. hansenii
Total
X2
P
Onset sepsis
Early
8
-
8
0.14
0.71
Late
3
1
4
Birth weight
≤ 2.5
7
1
8
0.14
0.71
≥ 2.5
4
-
4
Gestational ages (weeks)
Preterm(37)
3
1
4
0.14
0.71
Term >37
8
-
8
Gender
Males
10
-
11
2.48
0.11
Females
1
1
1
Table 4: Relation between different risk factors and the isolated yeast species.
Complete blood count revealed that, 80% of the positive neonates had normal HB level, platelets, HCT and MCHC, 50% had normal WBCs count, 70% normal RBCs while 40% suffered from leukocytosis, 70% had high MCH, 60% had high MCV and most of the studied neonates had thrombocytopenia.
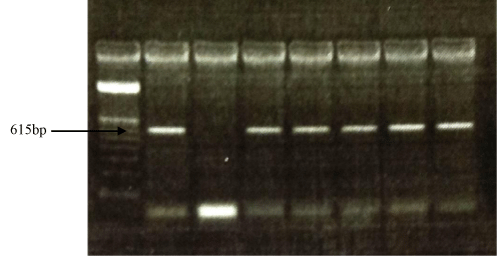
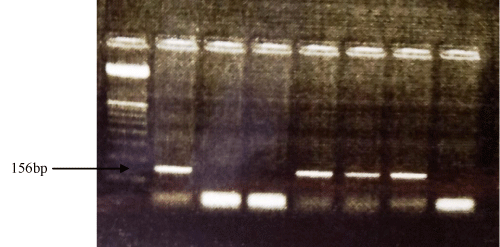
The multiplex PCR performed for universal yeast DNA showed positive results of the test sampler with the positive control at 615 bp. Also PCR for C. albicans showed positive result of the test samples with positive control at 156 bp (Figure 1 & 2).
Figure 1 : PCR amplification of yeast isolates DNA using universal primers that amplifies the V3 region of the large subunit rDNA. Lane 1: DNA marker (OX Hae III, Promega), lane 2: positive control, lane 3: negative control, lanes 4 - 8 yeast isolates.
Figure 2: PCR amplification of Candida albicans DNA using specific PCR primers. Lane 1: DNA marker (OX Hae III, Promega), lane 2: positive control, lane 3: negative control, lanes 4 - 8 yeast isolates.
Discussion
The number of neonatal deaths according to WHO [19] was3.3 million in 2009, but 98% occurred in less developed countries [20]. Sepsis remains the main cause of morbidity and mortality in neonatal intensive care units [21,22]. Host immunodeficiency, increasing microbial resistance to antibiotics, and late detection of early onset of infection are the major factors altering the course of infections [23,24]. Neonates suffering from candidiasis usually have serious morbid conditions. The epidemiology of candidemia is a marker for with other serious medical problems [25].
In the present study, systemic candidiasis occurred in 12% of the total positive test samples. C. albicans was isolated in11 cases (91.7%). These results are in agreement with other investigators who reported that candidemia accounted for 10.9% [26] and 15% [27] among infants in the neonatal intensive care units. C. albicans and C. parapsilosis, were isolated in 93% of cases [26] while Feja et al., [27] recorded that C. albicans, caused 62% of candidemia, C. parapsilosis (31%), C. glabrata, C. stellatoidea and C. lusitaniae caused 2% each [27]. D. hansenii (C. famata) was isolated from one case representing 8.3% of the yeast infection.
D. hansenii gained the status of emerging pathogen as being implicated in human infections [28,29]. However its incidence during candidemia is low based on data surveillance implemented by the Centers for Disease Control and Prevention, whereas one review on world wide-collected isolates states that D. hansenii accounts for 0.08 to 0.5% of isolates recovered during invasive candidiasis [30], But Desons-Olivier et al. [31], stated that D. hansenii has been repeatedly associated with catheter-related blood stream infection and rarely with other infections. In a recent study by Kumar et al. [32] on neonatal sepsis caused by Candida species, Candidemia was observed in 66 out of 442 neonates representing 14.9%. C. albicans was predominant isolate followed by C. glabrata (19.7%), C. tropicalis (6.1%) [33].
Cahanand Deville [34] reported in a study that lasted for four years that neonatal candidiasis has risen with a mortality rate of 35%. The mortality rate in developing countries among neonates was 23-52% [11]. Concerning the risk factors, yeast infection among neonatal males represented 91.7 (11 cases) and a single female case represented 8.3%. This male predominance is apparent in almost all studies of sepsis in newborn infants [35,36]. Klein and Marcy [37] stated that this might be due to a gene located on the X-chromosome and involved with the function of the thymus, or with synthesis of immunoglobulins. Premature infants represent 33.34% of the test group, which is lower than other studies [38]. Early onset sepsis represented 66.7% of the yeast infection. While late onset sepsis represented 33.3% and C. albicans was the predominant species in both cases. In a study performed by Shin et al., [39] the estimated incidence rate of neonatal sepsis was 30.5 per 1000 live births for clinical sepsis and 6.1 per 1000 live births for sepsis with positive culture, with case-fatality rates of 4.7% and 2.2%, respectively. When only early-onset sepsis considered, the incidence and fatality rates were 2.1 per 1000 live births and 6.1% for clinical sepsis, and 4.1 per 1000 live births and 2.5% for culture confirmed sepsis, respectively. The incidence of candidiasis is 3-4% in low birth weight infants [36,40], while Friedman et al [41], found that the incidence in very low birth weight infants was 2-9% with mortality rate 37%. In the present study, infection in low birth weight infants was 66.6%. According to published data, 0.004% to 1.5% of all patients in NICU, 2.6 to 3.1% of very low birth weight infants (birth weight < 1500 gm), and 5.5% to 10% of extremely low birth weight infants (birth weight <1000 gm) develop candidemia [36,42,43].
The analysis of neonatal yeast infection in this study showed that there is a perfect correlation between molecular and microbiological data (100% sensitivity). PCR is more sensitive than blood culture, since some of the neonates at risk for invasive yeast infection, whose blood cultures was negative for yeast, tested positive in PCR amplification [37]. C-reactive protein is the most accessible and widely used as a marker for detecting infection [44]. This test (C-RP) is considered of low diagnostic value because of its low specificity (66.7%), where in this study it recorded 66.7% in detecting yeast infection compared with PCR and blood culture (100%). It may be helpful in excluding infections in some cases of sepsis, if normal levels are obtained 24-48 hours after the onset of sepsis.
In conclusion, the incidence of neonatal yeast infection was 10% of the total test group. The identification of the isolated yeast species using PCR method matched the routine blood culture (100% specificity). PCR is a rapid method that saves time and medical cost of treatment and hospital accommodation and prevents the use of antibiotics for nonseptic cases. Preventive strategies for blood stream infection among neonates in intensive care unit should continue to focus on the possible risk factors leading to neonatal species.
Recommendation
It is important to publish this data in order to carry out a future study to find out the changing pattern of fungemia in cases of neonatal sepsis.
References
- Laforgi N, Coppola, B, Crpone R, Grassi A, Mautine A, Iolascon A. Rapid detection of neonatal sepsis using polymerase chain reaction. Acta Pediatr. 1997; 86: 1097-1099.
- Totoff Sp. Infections of neonatal infant. In: Nelson Textbook of Pediatrics, Behrman, Req. Kliegman RM, Arvin AM, editors. 16th Ed. IWB Saunders. 2000.
- Lim NL, Wong YH, Boo NY. Bcteraemic infections in a neonatal intensive care unit: a nine months survey. Med. J. Malaysia.1995; 50: 59-63.
- Tallur SS, Kasturi AV, Nadgir SD, Krishna BV. Clinico-bacteriological study of neonatal septicemia in Hubli. Indian J Pediatr. 2000; 67: 169-174.
- Airede AI. Neonatal septicaemia in an African city of high altitude. J Trop Pediatr. 1992; 38: 189-191.
- [No authors listed]. Clinical prediction of serious bacterial infections in young infants in developing countries. The WHO Young Infants Study Group. Pediatr Infect Dis J. 1999; 18: S23-31.
- Moreno MT, Vargas S, Poveda R, Sáez-Llorens X. Neonatal sepsis and meningitis in a developing Latin American country. Pediatr Infect Dis J. 1994; 13: 516-520.
- Robillard PY, Nabeth P, Hulsey TC, Sergent MP, Périanin J, Janky E, et al. Neonatal bacterial septicemia in a tropical area. Four-year experience in Guadeloupe (French West Indies). Acta Paediatr. 1993; 82: 687-689.
- Hyde TB, Hilger TM, Reingold A, Farley MM, O'Brien KL, Schuchat A; Active Bacterial Core surveillance (ABCs) of the Emerging Infections Program Network. Trends in incidence and antimicrobial resistance of early-onset sepsis: population-based surveillance in San Francisco and Atlanta. Pediatrics. 2002; 110: 690-695.
- Isaacs D, Royale JA. Intrapartum antibiotics and early-onset sepsis caused by group B Streptococcus and other organisms in Australia. Pediatri. Infect. Dis.J. 1999; 18: 524-528.
- Rodrigo I. Changing pattern of neonatal sepsis. SriLanka Journal of Child Health. 2002; 31: 3-8.
- Belling LB, Bryan LO, Scott M. Neonatal sepsis. E Medicine Online. 2003.
- Butler KM, Rench MA, Baker CJ. Amphotericin B as a single agent in the treatment of systemic candidiasis in neonates. Pediatr Infect Dis J. 1990; 9: 51-56.
- Kossoff EH, Buescher ES, Karlowicz MG. Candidemia in a neonatal intensive care unit: trends during fifteen years and clinical features of 111 cases. Pediatr Infect Dis J. 1998; 17: 504-508.
- Roilides E, Farmaki E, Evdoridou J, Francesconi A, Kasai M, Filioti J, et al. Candida tropicalis in a neonatal intensive care unit: epidemiologic and molecular analysis of an outbreak of infection with an uncommon neonatal pathogen. J Clin Microbiol. 2003; 41: 735-741.
- Cheesbrough M. District Laboratory Practice in Tropical Countries. Part 2. Cambridge University Press. 2000; 127.
- Haynes KA, Westerneng TJ, Fell JW, Moens W. Rapid detection and identification of pathogenic fungi by polymerase chain reaction amplification of large subunit ribosomal DNA. J Med Vet Mycol. 1995; 33: 319-325.
- Cruikshank A. Practical Medical Microbiology. 14thEd. College JG and Fraser AG, Williams and Wilkins. 1996; 43-207, 245-261 & 263-274).
- World Health Organization (WHO). Neonatal and prenatal mortality: Country, regional and global estimates. Geneva: World Health Organization. 2009.
- World Health Organization (WHO). 4th Ed. Prenatal mortality. A listing of available information. Geneva: WHO, Maternal Health and safe Motherhood program. 1996.
- Proulx F, Fayon M, Farrell CA, Lacroix J, Gauthier M. Epidemiology of sepsis and multiple organ dysfunction syndrome in children. Chest. 1996; 109: 1033-1037.
- Carvalho PR, Feldens L, Seitz EE, Rocha TS, Soledade MA, Trotta EA, et al. [Prevalence of systemic inflammatory syndromes at a tertiary pediatric intensive care unit]. J Pediatr (Rio J). 2005; 81: 143-148.
- Madhi SA, Petersen K, Khoosal M, Klugman KP. Increased disease burden and antibiotic resistance of bacteria causing sever community-acquired lower respiratory tract infections in human immunodeficiency. Virus type 1-infected children. Clin. Infect. Dis. 2000; 31: 170-176.
- Kang CL, Kim SH, Park WB, Lee KD, Kim HB, Kim EC, et al. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob. Agents Chemother. 2005; 49: 760-766.
- Tiraboschi IN, Bennett JE, Kauffman CA, Rex JH, Girmenia C, Sobel JD, et al. Deep Candida infections in the neutropenic and non-neutropenic host: an ISHAM symposium. Med Mycol. 2000; 38 Suppl 1: 199-204.
- Clerihew L, Lamagni TL, Brocklehurst P, McGuire W. Invasive fungal infection in very low birthweight infants; National prospective surveillance study. Arch. Dis. Child Fetal Neonatal Ed. 2006; 91: F188-F192.
- Feja KN, Wu F, Roberts K, Loughrey M, Nesin M, Larson E, et al. Risk factors for candidemia in critically ill infants: a matched case-control study. J Pediatr. 2005; 147: 156-161.
- Gupta A, Mi H, Wroe C, Jaques B, Talbot D. Fatal Candida famata peritonitis complicating sclerosing peritonitis in a peritoneal dialysis patient. Nephrol Dial Transplant. 2006; 21: 2036-2037.
- Wagner D, Sander A, Bertz H, Finke J, Kern WV. Break through invasive infection due to Debaryomyces hansenii (telomorphCandida famata) and scopulariopsis brevicaulis in a stem cell transplant patient receiving liposomal amphotericin B and caspofungin for suspected aspergillosis. Infect. 2005; 33: 397-400.
- Desnos-Ollivier M, Ragon M, Robert V, Raoux D, Gantier JC, Dromer F, et al. Debaryomyces hansenii (Candida famata), a rare human fungal pathogen often misidentified as Pichia guilliermondii (Candida guilliermondii). J Clin Microbiol. 2008; 46: 3237-3242.
- Bai FY, Liang HY, Jia JH. Taxonomic relation among the taxa in the Candida guilliermondii complex, as reveled by comparative electrophoretic karyotyping. Int. J. Syst. Evol. Microbiol. 2000; 50: 417-422.
- Diekema DJ, Messer SA, Brueggemann AB, Coffman SL, Doern GV, Herwaldt LA, et al. Epidemiology of candidemia: 3-year results from the emerging infections and the epidemiology of Iowa organisms study. J Clin Microbiol. 2002; 40: 1298-1302.
- Krcméry V, Kunová A. Candida famata fungemia in a cancer patient: case report. J Chemother. 2000; 12: 189-190.
- Cahan H, Deville JG. Outcomes of neonatal candidiasis: the impact of delayed initiation of antifungal therapy. Int J Pediatr. 2011; 2011: 813871.
- Kumar S, Vasant B, Mathur A, De M. A study of neonatal sepsis due to Candida species. Bombay Hospital Journal. 2011. 53: 524-528.
- Saiman L, Ludington E, Pfaller M, Rangel-Frausto S, Wiblin RT, Dawson J, et al. Risk factors for candidemia in Neonatal Intensive Care Unit patients. The National Epidemiology of Mycosis Survey study group. Pediatr Infect Dis J. 2000; 19: 319-324.
- Prinsloo B, Weldhagen GF, Blaine RW. Candida famata central nervous system infection. S Afr Med J. 2003; 93: 601-602.
- Klien JO, Marcy SM. Bacterial sepsis and meningitis. In: Infectious diseases of the fetus and newborn infant. 4th edn. Klein, Remington JS, editors. WB Saunders, Philadelphia. 1995; 1-19.
- Shin YJ, Ki M, Foxman B. Epidemiology of neonatal sepsis in South Korea. Pediatr Int. 2009; 51: 225-232.
- McDonnell M, Isaacs D. Neonatal systemic candidiasis. J Paediatr Child Health. 1995; 31: 490-492.
- Friedman S, Richardson SE, Jacobs SE, O'Brien K. Systemic Candida infection in extremely low birth weight infants: short term morbidity and long term neurodevelopmental outcome. Pediatr Infect Dis J. 2000; 19: 499-504.
- Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, et al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics. 2002; 110: 285-291.
- Benjamin DK Jr, Poole C, Steinbach WJ, Rowen JL, Walsh TJ. Neonatal candidemia and end-organ damage: a critical appraisal of the literature using meta-analytic techniques. Pediatrics. 2003; 112: 634-640.
- Kyr M, Fedora M, Elbl L, Kugan N, Michalek J. Modeling effect of the septic condition and trauma on C-reactive protein levels in children with sepsis: a retrospective study. Crit Care. 2007; 11: R70.