
Research Article
Austin Tuberc Res Treat. 2017; 2(1): 1006.
Isoniazid Drug Resistance: Computational Study to Understand the Mechanism of Over Expressed UDPGalactopyranose Mutase Enzyme in Causing Drug Resistance in Tuberculosis
Nayak T, Jena L and Harinath BC*
JB Tropical Disease Research Centre, Mahatma Gandhi Institute of Medical Sciences, India
*Corresponding author: Harinath BC, JB Tropical Disease Research Centre, Mahatma Gandhi Institute of Medical Sciences, Sevagram-442 102 (Wardha), Maharashtra, India
Received: April 17, 2017; Accepted: June 12, 2017; Published: June 20, 2017
Abstract
Isoniazid (INH), one of the effective anti-tuberculosis drugs, has been extensively used in TB control but several clinical strains of Mycobacterium tuberculosis (MTB) have shown resistance to INH. Besides, mutation in different MTB enzymes such as KatG, NAT, InhA, KasA, AhpC etc, over expression of few enzymes are also reported to be involved in causing INH resistance. The over expression of MTB UDP-galactopyranose mutase (Glf) enzyme encoded by Rv3809c is reported to contribute to INH resistance by binding to NADH and reducing its level required for INH to form INH-NAD adduct and showing its sensitivity. In this study, we employed molecular docking technique using Auto Dock 4.2 to observe the binding affinity of Glf with FADH2 (the primary cofactor of Glf) and NADH. Further, we also performed molecular docking of Ndh enzyme with NADH and compared its binding affinity with Glf-NADH complex. The docking results revealed that, both FADH2 and NADH are showing similar binding affinity (-11.3 kcal/mol and -11.1 kcal/mol respectively) with Glf whereas; Ndh binds with NADH with binding energy of -5.0 kcal/mol. Thus, the over expression of Glf may lead to NADH binding and as a result, it may not be available to Ndh for forming NAD+, required for INH-NAD adduct formation and thus leading to INH resistance.
Keywords: Mycobacterium tuberculosis; Drug resistance; Glf; FADH2; Bioinformatics
Abbreviations
FAD: Flavin Adenine Dinucleotide; INH: Isonicotinylhydrazide (Isoniazid); MDR: Multi Drug Resistant; MTB: Mycobacterium tuberculosis; NAD: Nicotinamide Adenine Dinucleotide; NADP: Nicotinamide Adenine Dinucleotide Phosphate; NAT: Arylamine N-Acetyltransferase; UDP: Uridine Diphosphate; UDP-Galf: UDPGalactofuranose; UDP-Galp: UDP-Galactopyranose; UGM: UDPGalactopyranose Mutase
Introduction
Mycobacterium tuberculosis (MTB) has existed for millennia and remains a major global health problem with about 10.4 million new cases of tuberculosis and 1.4 million deaths reported in 2015 and an additional 0.4 million deaths resulting from TB disease among HIVpositive people [1]. Increasing emergence of Multidrug-Resistant (MDR) and Extensively Drug-Resistant (XDR) TB in some region is becoming an increasing risk of spreading TB [2-4]. Most of MTB drugs-resistant clinical strains are resistant to Isoniazid (INH), the most widely used first-line anti-TB drug [5,6]. INH also known as isonicotinyl hydrazine is an organic compound with simple structure containing two essential components (i.e. a pyridine ring and a hydrazide group) [6,7]. Different studies have meticulously explored the mode of action of INH. It is reported to generate a variety of highly reactive compounds such as reactive organic species like isonicotinic acyl radical or anion, reactive oxygen species like superoxide, peroxide and hydroxyl radical, nitric oxide, and certain electrophilic species, which then attack various MTB enzymes. As a prodrug, INH requires activation and after passively diffuses through the mycobacterial envelope, is activated by MnCl2 and the catalase– peroxidase KatG, possibly into an isonicotinoyl radical or anion, which then forms adduct in the presence of NADH/NADPH which inhibits different enzymes of MTB besides InhA (2-trans-enoyl-acyl carrier protein reductase) of MTB, thus blocking the synthesis of mycolic acid, a major lipid of the mycobacterial cell wall [6,7,12].
Of late, several clinical strains of MTB have shown resistance to INH due to mutation in enzymes such as KatG, NAT, InhA, MabA, KasA, AhpC, NudC etc [7-13]. INH is reported to interact with around 117 enzymes of MTB [14]. These enzymes are involved in different metabolic pathways critical for growth and survival of the pathogen. Out of 117 enzymes, mutations in 82 were reported to associate with INH resistance [15,16], 15 enzymes were reported to be induced by INH [17]. Further, 18 enzymes were reported to be reactive with INH-NAD (P) adduct [7,18] possibly involving in inhibitory effect of INH.
Miesel et al. (1998) also reported that defective NADH Dehydrogenase (Ndh) enzyme is associated with INH resistance in Mycobacterium smegmatis [8]. Lee et al. (2001) identified a novel mutation Arg268His (R268H) in Ndh from INH resistance clinical isolates and reported that this mutation causing some defect in the enzyme activity thus could not utilize NADH for adduct formation, causing resistance to both Isoniazid and ethionamide [8,19]. Further, over expression of certain MTB enzymes such as Glf (Rv3809c), MmpL7 (Rv2942), Acn (Rv1475c), EfpA (Rv2846c), Mmr (Rv3065), integral membrane transport protein (Rv1258c) were reported to be involved in INH resistance [20-23].
The MTB Glf (Rv3809c) gene encoding UDP-galactopyranose mutase catalyzes the conversion of UDP-galactopyranose into UDP-Galactofuranose (Figure 1), a key building block for cell wall construction and is essential to the linking of the peptidoglycan and mycolic acid cell wall layers in MTB through a 2-keto intermediate [24]. Further, Glf has both FAD and NAD binding sites [20] and it requires reduced FAD (FADH2) and either NADH or NADPH for activity [24]. Glf catalyzes the reversible inter-conversion of UDPgalactopyranose (UDP-Galp) and UDP-Galactofuranose (UDP-Galf) [25,26].
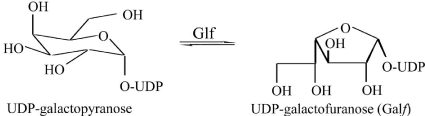
Figure 1: Inter-conversion of UDP-Galp and UDP-Galf is catalyzed by Glf.
Chen & Bishai (1998), observed in their wet laboratory experiment that the Minimum Inhibitory Concentration (MIC) of INH increased up to 50% in Mycobacterium bovis BCG strain in which they over expressed the glf gene and reported that the over expression of Glf enzyme might contribute to INH resistance, by binding with NADH which is required for formation of INH-NAD adduct for INH activity [20].
In this study, we employed molecular docking techniques to observe the binding affinity of Glf with FADH2 (the primary cofactor of Glf [27] and NADH. Further, as Ndh plays an important role in the oxidation of NADH to generate NAD+ [28], we also performed molecular docking of Ndh with NADH to compare the binding affinity of NADH with Glf and Ndh.
Materials and Methods
Hardware and software
Dell workstation with Windows operating system, having hard disc of 500 GB and 6 GB RAM was used for this in silico study. Further, computational analysis was performed using bioinformatics tools, such as Modeller, AutoDock Tools 1.5.4 [29], AutoDock Vina 1.1.2 [30], PyMol molecular visualization packages and online resources.
Sequence retrieval
The amino acid sequences of two MTB enzymes such as Glf (NCBI Accession No. - NP_218326.1) and Ndh (NCBI Accession No. - NP_216370.1) were retrieved from NCBI (https://www.ncbi.nlm. nih.gov) in FASTA format for further analyses.
Homology modeling of Glf and Ndh
Homology modeling technique was used to predict the three dimensional (3D) structure of both Glf and Ndh using Modeller9v14 [31] software. As the experimentally determined structures of Glf (PDB ID: 4RPG) [32] enzyme has some missing residues, its 3D structure was predicted using its amino acid sequence and 4RPG as template. Further, the crystal structure of bacterial type II NADH dehydrogenase from Caldalkalibacillus thermarum at 2.5A resolution (PDB ID: 4NWZ) was used as a template to predict the 3D model of Ndh enzyme. Both the 3D structures were subjected for structure validation using ProSA-web [33], ProQ [34] and RCSB validation server [35].
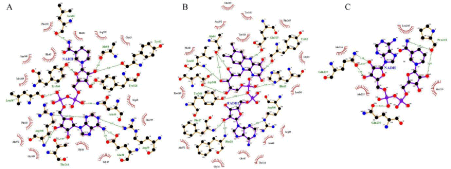
Figure 4: Docking interaction of Glf with (A) NADH, (B) FADH2 (C) Docking interaction of Ndh with NADH.
Identification of FAD & NAD binding site
The Glf, also known as flavoenzyme UDP-Galactopyranose Mutase (UGM) has both NAD and FAD binding sites. NAD binder server was used to obtain NAD binding site amino acid residues of Glf proteins in MTB [36] whereas the FAD binding site amino acid residues were identified from UniProt Knowledge Base (ID: P9WIQ1).
Ligand preparation
Two compounds such as NADH (PubChem ID: 439153), and FADH2 (ChemSpider ID: 393487) were selected in this study and their 3D structures (Figure 2A,B) were retrieved from PubChem [37,38] & ChemSpider [39] databases. All the pre-processing steps for ligands were performed using the AutoDock Tools 1.5.4 program (ADT) [29] and all ligands were converted to Protein Data Bank Extension File Format (PDBQT), an extended PDB format, used for coordinate files, which include atomic partial charges and atom types.
Figure 2: 2D structure of (A) NADH; (B) FADH2.
Receptor preparation and protein-ligand docking
AutoDock Tool 1.5.4 program (ADT) was used to prepare receptor molecule (Glf and Ndh) by adding all hydrogen atoms into the carbon atoms of the receptor. Kollman charges were also assigned and the entire receptor molecules were converted to Protein Data Bank Extension File Format (PDBQT). Receptor (Glf) - Ligand (FADH2, NADH) and Receptor (Ndh) with Ligand (NADH) docking studies were performed using the Autodock vina 1.1.2.
Visualization
The visualization of structure files was done using PyMol molecular graphics system (www.pymol.org) and the graphical interface of the ADT tool.
Results and Discussion
M. tuberculosis Glf (UDP-galactopyranose mutase) is a flavoenzyme containing 399 amino acids in its protein sequence, which catalyzes the conversion of UDP-galactopyranose to UDPGalactofuranose. Sanders et al, (2001) reported that Uridine Diphosphogalactofuranose (UDP-Galf) is the precursor of the D-Galactofuranose (Galf) residues found in the MTB cell walls [40]. Glf is also involved in different pathways such as Galactose metabolism, amino sugar and nucleotide sugar metabolism, UDPGalactofuranose biosynthesis pathway, mycolyl-arabinogalactanpeptidoglycan complex biosynthesis pathway, and Super pathway of UDP-glucose-derived O-antigen building blocks biosynthesis [41-43]. Further, Glf is very essential for mycobacterial viability [44] and is reported to be a suitable drug target because there are no comparable enzymes present in humans [45].
Homology modeling and structure validation
The stereochemistry of Glf model (Figure 3A) (Procheck analysis) revealed that 94.8% of residues were situated in the most favorable region and 4.6% were in additional allowed region whereas 0.6% of the residue fell in the generously allowed region of the Ramachandran plot (Figure 3B). ProSA-web evaluation of Glf model revealed a compatible Z score value of -10.1 (Figure 3C) and LG score of 5.013 as predicted by the Protein Quality Predictor (ProQ).
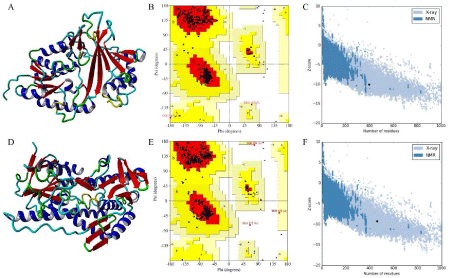
Figure 3: (A) 3D model of Glf, (B) Ramchandran plot of Glf, (C) Z plot of Glf, (D) 3D model of Ndh, (E) Ramchandran plot of Ndh, (F) Z plot of Ndh.
The procheck analysis of Ndh model (Figure 3D) revealed that 94.0% of residue was situated in the most favorable region and 5.3% were in allowed region, whereas 0.3% and 0.5% of the residue fell in the generously allowed and disallowed region of the Ramachandran plot (Figure 3E) respectively. Further, ProSA-web evaluation of Ndh model revealed a compatible Z score value of -9.32 (Figure 3F) and LG score of 4.013 as predicted by the Protein Quality Predictor (ProQ).
Docking analysis
Docking of Glf with FADH2 was performed around the FAD binding site of Glf such as PHE18, Glu38, Asn46, Leu66, Arg360 retrieved from UniprotKB [27,46]. Further, The Glf-NADH docking was performed around NAD binding site residues of Glf such as Gly16, Phe18, Glu38, Arg360 predicted by NAD binder server [36].
From the docking analysis, it was observed that Glf binds with FADH2 with binding energy of -11.3 kcal/mol where as in case of NADH the binding energy was -11.1 kcal/mol. There were 14 hydrogen bond interactions were observed in Glf-FADH2 complex (Figure 4B) where as 13 hydrogen bonds interactions were observed in Glf-NADH complex (Figure 4A) respectively. FADH2 from hydrogen bond with binding site residue such as Gly16, Phe17, Phe18, Arg39, Asn46, Tyr62, Ala64, His65, Leu66, Glu315, Arg360, Tyr366, Leu367, Asp368 (Figure 4B) where as NADH from hydrogen bond with binding site residue such as Gly16, Glu38, Arg39, Arg40, Asn46, Tyr62, Ala64, Leu66, Thr244, Tyr328, Arg360, Tyr366, Leu367 of Glf protein (Figure 4A).
Docking of Ndh enzyme with NADH were performed around NAD binding site residue such as Ala178, Gly179, Pro180, Thr181, Glu184, predicted by NAD Binder Server. From docking analysis, it was observed that NADH bind with Ndh with a binding energy of -5.0 kcal/mol and it forms 3 hydrogen bonds with binding site residues such as Pro218, Gln237, Gln230 of Ndh protein (Figure 4C).
Glf is a both NAD and FAD binding enzyme [47] and it requires reduced Flavin Adenine Dinucleotide (FAD) and either NADH or NADPH for activity [24]. Further, it’s over expression is reported to involve in Isoniazid resistance by reducing the levels of NAD+ that required for formation of INH-NAD adduct or by binding of active form Isoniazid [13,20]. As, NAD(P)H is also required for FAD reduction [48,49], the over expression of Glf may depletes FADH2 as hydrogen donor and thus more molecules of NADH are required to reduce FAD. Thus, there may be decrease in concentration of reduced NAD and as a result it may not be available for forming INH-NAD adduct, thus reducing the effectivity of Isoniazid as inhibitor. From our binding energy and hydrogen bond interactions study, it was also revealed that Glf has similar binding affinity towards FADH2 and NADH. Thus, in case of Glf over expression in MTB, it might be possible that NADH may also donate hydrogen for increased Glf activity and reducing NADH level, required for adduct formation [50]. Further, the NADH Dehydrogenase (Ndh) enzyme also utilizes NADH and produces NAD+ and mutation in Ndh gene is reported to cause INH resistance by influencing the NADH/NAD+ ratio [8,19]. In our docking study, the binding affinity of NADH towards Glf was observed to be more as compared to Ndh and thus it may be possible that Glf over expression may require more NADH, limiting NADH for INH activity resulting drug resistance.
Conclusion
Drug resistance in tuberculosis is an important problem around the world. The most effective anti tuberculosis drug Isoniazid is widely reported to become resistant in TB treatment. The INH is associated with various enzymes of MTB and beside mutations in different enzymes. Over expression of certain MTB enzymes are reported to be causing INH resistance. Thus it is important to know the molecular mechanism of drug resistance. The Glf is an essential enzymes of MTB and it’s over expression is reported to cause INH resistance by reducing the levels of NADH that required for the formation of the INH-NAD adduct. In order to know the molecular mechanism of INH resistance, we performed molecular docking study of Glf with FADH2 and NADH and found that both having similar binding affinity towards Glf. Thus, it’s over expression may utilize more NADH for either reducing FAD, and/or directly binding with Glf, necessary for Glf activity. In both cases, the concentration of NADH is decreased, resulting in decreased INH-NAD adduct formation causing INH resistance.
References
- World Health Organization, WHO Global Tuberculosis Report 2016, World Health Organization, Geneva, Switzerland. 2016.
- Shah NS, Wright A, Bai GH, Barrera L, Boulahbal F, Martín-Casabona N, et al. Worldwide emergence of extensively drug resistant tuberculosis. Emerg Infect Dis. 2007; 13: 380-387.
- Almeida Da, Silva PE, Palomino JC. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother. 2011; 66: 1417-1430.
- Unissa AN, Subbian S, Hanna LE, Selvakumar N. Overview on mechanisms of isoniazid action and resistance in Mycobacterium tuberculosis. Infect Genet Evol. 2016; 45: 474-492.
- Marrakchi H, Lanéelle G, Quémard A. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology. 2000; 146: 289-296.
- Sarathy JP, Dartois V, Lee EJ. The role of transport mechanisms in mycobacterium tuberculosis drug resistance and tolerance. Pharmaceuticals (Basel). 2012; 5: 1210-1235.
- Timmins GS, Deretic V. Mechanisms of action of isoniazid. Mol Microbiol. 2006; 62: 1220-1227.
- Miesel L, Weisbrod TR, Marcinkeviciene JA, Bittman R, Jacobs WR Jr. NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J Bacteriol. 1998; 180: 2459-2467.
- Sandy J, Mushtaq A, Kawamura A, Sinclair J, Sim E, Noble M, et al. The structure of arylamine N-acetyltransferase from Mycobacterium smegmatis- -an enzyme which inactivates the anti-tubercular drug, isoniazid. J Mol Biol. 2002; 318: 1071-1083.
- Sholto-Douglas-Vernon C, Sandy J, Victor TC, Sim E, Helden PD. Mutational and expression analysis of tbnat and its response to isoniazid. J Med Microbiol. 2005; 54: 1189-1197.
- Müller B, Streicher EM, Hoek KG, Tait M, Trollip A, Bosman ME, et al. inhA promoter mutations: a gateway to extensively drug-resistant tuberculosis in South Africa? Int J Tuberc Lung Dis. 2011; 15: 344-351.
- Jena L, Waghmare P, Kashikar S, Kumar S, Harinath BC. Computational approach to understanding the mechanism of action of isoniazid, an anti-TB drug. Int J Mycobacteriol. 2014; 3: 276-282.
- Vilchèze C, Jacobs WR Jr. Resistance to Isoniazid and Ethionamide in Mycobacterium tuberculosis: Genes, Mutations, and Causalities. Microbiol Spectr. 2014; 2: MGM2-0014-2013.
- Jena L, Nayak T, Deshmukh S, Wankhade G, Waghmare P. Isoniazid with Multiple Mode of Action on Various Mycobacterial Enzymes Resulting in Drug Resistance. J Infect Dis Ther. 2016; 4: 297.
- Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM. Tuberculosis drug resistance mutation database. PLoS Med. 2009; 10: e2.
- Shekar S, Yeo ZX, Wong JC, Chan MK, Ong DC. Detecting novel genetic variants associated with isoniazid-resistant Mycobacterium tuberculosis. PLoS One. 2014; 15: e102383.
- Wilson M, DeRisi J, Kristensen HH, Imboden P, Rane S, Brown PO, et al. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA. 1999; 96: 12833-12838.
- Argyrou A, Jin L, Siconilfi-Baez L, Angeletti RH, Blanchard JS. Proteomewide profiling of isoniazid targets in Mycobacterium tuberculosis. Biochemistry. 2006; 45: 13947-13953.
- Lee AS, Teo AS, Wong SY. Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2001; 45: 2157-2159.
- Chen P, Bishai WR. Novel selection for isoniazid (INH) resistance genes supports a role for NAD+-binding proteins in mycobacterial INH resistance. Infect Immun. 1998; 66: 5099-5106.
- Pasca MR, Guglierame P, De Rossi E, Zara F, Riccardi G. mmpL7 gene of Mycobacterium tuberculosis is responsible for isoniazid efflux in Mycobacterium smegmatis. Antimicrob Agents Chemother. 2005; 49: 4775- 4777.
- Pandey R, Rodriguez GM. A ferritin mutant of Mycobacterium tuberculosis is highly susceptible to killing by antibiotics and is unable to establish a chronic infection in mice. Infect Immun. 2012; 80: 3650-3659.
- Parsa K, Hasnain SE. Proteomics of multidrug resistant Mycobacterium tuberculosis clinical isolates: a peep show on mechanism of drug resistance & perhaps more. Indian J Med Res. 2015; 141: 8-9.
- Weston A, Stern RJ, Lee RE, Nassau PM, Monsey D, Martin SL, et al. Biosynthetic origin of mycobacterial cell wall galactofuranosyl residues. Tuber Lung Dis. 1997; 78: 123-131.
- Gruber TD, Borrok MJ, Westler WM, Forest KT, Kiessling LL. Ligand binding and substrate discrimination by UDP-galactopyranose mutase. J Mol Biol. 2009; 391: 327-340.
- Sun HG, Ruszczycky MW, Chang WC, Thibodeaux CJ, Liu HW. Nucleophilic participation of reduced flavin coenzyme in mechanism of UDPgalactopyranose mutase. J Biol Chem. 2012; 287: 4602-4608.
- Beis K, Srikannathasan V, Liu H, Fullerton SW, Bamford VA, Sanders DA, et al. Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidized state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state. J Mol Biol. 2005; 348: 971-982.
- Bald D, Koul A. Respiratory ATP synthesis: the new generation of mycobacterial drug targets? FEMS Microbiol Lett. 2010; 308: 1-7.
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Computational Chemistry. 2009; 16: 2785-2791.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010; 31: 455-461.
- Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Protein Sci. 2016; 86: 2.9.1-2.9.37.
- van Straaten KE, Kuttiyatveetil JR, Sevrain CM, Villaume SA, Jiménez- Barbero J, Linclau B, et al. Structural basis of ligand binding to UDPgalactopyranose mutase from Mycobacterium tuberculosis using substrate and tetrafluorinated substrate analogues. J Am Chem Soc. 2015; 137: 1230- 1244.
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids. 2007; 35: W407-410.
- Wallner B, Fang H, Elofsson A. Automatic consensus-based fold recognition using Pcons, ProQ, and Pmodeller. Proteins. 2003; 6: 534-541.
- Laskowski RA, MacArthur MW, Thornton JM. PROCHECK: validation of protein structure coordinates, in International Tables of Crystallography. Crystallography of Biological Macromolecules. 2001; 722-725.
- Ansari HR, Raghava GP. Identification of NAD interacting residues in proteins. BMC Bioinformatics. 2010; 11: 160.
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016; 44: D1202- 1213.
- Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH, et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009; 37: W623-633.
- Harry E. Pence. ChemSpider: An Online Chemical Information Resource. ChemSpider, Royal Society of Chemistry. 2010; 87: 123-1124.
- Sanders DA, Staines AG, McMahon SA, McNeil MR, Whitfield C, Naismith JH, et al. UDP-galactopyranose mutase has a novel structure and mechanism. Nat Struct Biol. 2001; 8: 858-863.
- Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012; 40: D742-753.
- Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016; 44: D471-480.
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45: D353-D361.
- Dykhuizen EC, Kiessling LL. Potent ligands for prokaryotic UDPgalactopyranose mutase that exploit an enzyme subsite. Org Lett. 2009; 11: 193-196.
- Dykhuizen EC, May JF, Tongpenyai A, Kiessling LL. Inhibitors of UDPgalactopyranose mutase thwart mycobacterial growth. J Am Chem Soc. 2008; 130: 6706-6707.
- Magrane M, UniProt Consortium. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford). 2011: bar009.
- Vilchèze C, Weisbrod TR, Chen B, Kremer L, Hazbón MH, Wang F, et al. Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob Agents Chemother. 2005; 49: 708-720.
- Oppenheimer M, Valenciano AL, Kizjakina K, Qi J, Sobrado P. Chemical mechanism of UDP-galactopyranose mutase from Trypanosoma cruzi: a potential drug target against Chagas’ disease. PLoS One. 2012; 7: e32918.
- Tanner JJ, Boechi L, Andrew McCammon J, Sobrado P. Structure, mechanism, and dynamics of UDP-galactopyranose mutase. Arch Biochem Biophys. 2014; 544: 128-141.
- Rawat R, Whitty A, Tonge PJ. The isoniazid-NAD adduct is a slow, tightbinding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: Adduct affinity and drug resistance. Proceedings of the National Academy of Sciences of the United States of America. 2003; 100: 13881-13886.