Research Article
Austin Virol and Retrovirology. 2014;1(2): 4.
Molecular Characterization of the Porcine Epidemic Diarrhea Virus TW4/2014 in Taiwan
Ming-Tang Chiou1,2, Chien-Ho Yu3, Chih-Cheng Chang4, Wen-Bin Chung1, Hung-Yi Wu5, Chuen-Fu Lin4 and Chao-Nan Lin1,2*
1Department of Veterinary Medicine, National Pingtung University of Science and Technology, Taiwan
2Veterinary Hospital, National Pingtung University of Science and Technology, Taiwan
3Program of Agriculture Science, Veterinary Medicine, National Chiayi University, Taiwan
4Department of Veterinary Medicine, National Chiayi University, Taiwan
5Graduate Institute of Veterinary Pathobiology, College of Veterinary Medicine, National Chung-Hsing University, Taiwan
*Corresponding author: Chao-Nan Lin, Department of Veterinary Medicine, College of Veterinary Medicine, National Pingtung University of Science and Technology, No. 1, Hseuh Fu Rd. Nei Pu, Pingtung, 91201, Taiwan
Received: November 02, 2014; Accepted: December 03, 2014; Published: December 08, 2014
Abstract
Infections by Porcine Epidemic Diarrhea Virus (PEDV) have been shown to be significantly correlated with fatal diarrhea in suckling piglets. Although PEDV was first identified in Europe, since late 2010, it has become increasingly problematic on several continents, including Asia, North America, and South America. Since late 2013, several outbreaks of PEDV have emerged in Taiwan. Analysis of the partial PEDV S gene sequences in these strains revealed that these outbreaks were in the same clade as US strains of PEDV. To elucidate the molecular characterization of the Taiwanese and reference strains in greater detail, the full length of a local isolate (TW4/2014) that was derived from a suckling piglet was sequenced using next-generation sequencing techniques and compared to other worldwide strains. The complete genome size of TW4 (27,966 nucleotides excluding the 5’ leader sequence) appears to be almost identical (99.9%) to several of the 2013 US strains. Our results indicate that the recent PEDV isolates from Taiwan share a common evolutionary origin with US-like PEDV strain from several countries (South Korea, Canada, Mexico, and Peru) and US strains.
Keywords: PEDV; Porcine epidemic diarrhea; Complete genome; Next-generation sequencing
Abbreviation
PEDV: Porcine Epidemic Diarrhea Virus; HCoV: Human Coronavirus; TGEV: Transmissible Gastroenteritis Virus; ORFs: Open Reading Frames; nts: Nucleotides; PCV2: Porcine Circovirus Type 2.
Introduction
Porcine Epidemic Diarrhea Virus (PEDV) is an enveloped virus with a large, capped and polyadenylated RNA gnome of approximately 28,000 nucleotides [1]. PEDV belongs the genus Alphacoronavirus, family Coronaviridae, order Nidovirales. Other members of this subgroup include Human Coronavirus (HCoV) 229E, HCoV NL63, and bats coronavirus 512/05 [2]. Although PEDV was first identified in Europe, since 2010, it has become increasingly problematic on several continents, including Asia, North America, and South America [1,3-5].
Starting in April 2013, PEDV was first identified in the United States. All of the affected swine farms experienced explosive epidemics of diarrhea and vomiting affecting pigs of all ages, with 90-95% mortality in suckling piglets [4]. Since that time, US strain-like PEDV variants have become prevalent in several countries, including South Korea [5,6], Canada [7], Mexico [8], and Peru [9]. Whole-genome sequencing of the 2013 US PEDV revealed the highest identity with the Chinese strain AH2012 [10].
Since late 2013, several outbreaks of PEDV infection have emerged in Taiwan. Suckling piglets under 2 weeks of age showed severe vomiting and watery yellowish diarrhea with high morbidity and mortality. Analysis of the partial PEDV S gene sequence revealed that these outbreaks were in the same clade as the US strains of PEDV [3]. However, the full length of a Taiwanese PEDV strain remains to be analyzed.
Materials and Methods
Animal
PEDV TW4 was isolated in January 2014 from a one-day-old piglet with naturally occurring PED. Within 24 hours of birth, this piglet developed watery yellow diarrhea, weight loss and dehydration. Thinned and distended small intestine walls with watery yellowish contents were recorded during necropsy. The clinical specimens were negative for rotavirus and Transmissible Gastroenteritis Virus (TGEV) and positive for the partial PEDV S gene using reverse transcription polymerase chain reaction [3].
Isolation of viral RNA and sequencing for complete genome analysis
Total nucleic acid was extracted from the piglet’s intestine using the MagNA Pure LC 2.0 (Roche Diagnostics, Mannheim, Germany) following the manufacturer’s protocol. The nucleic acid templates were then sequenced for the whole genome using the MiSeq sequencing system (Illumina Inc, San Diego, CA, USA).
Sequence analysis
The complete sequences of PEDV TW4 were then compared with reference strains, and the results are summarized in Table 1. Multiple alignments of nucleic acid sequences were performed using the Clustal W methods within the MegAlign program (DNASTAR Inc., WI, USA). The phylogenetic analyses were conducted using the maximum likelihood method within MEGA 5, version 5.05.
Isolation
Virus
Year
Country
Sequence identity
GenBank accession no.
PEDV CV777
1978
Belgium
96.9
NC003436
PEDV DR13
1999
South Korea
97.7
JQ023161
PEDV LZC
2006
China
96.6
EF185992
PEDV JS2008
2008
China
97.4
KC210146
PEDV SM98
2011
South Korea
96.7
GU937797
PEDV CHGD-01
2011
China
98.3
JX261936
PEDV CH/FJND-3
2011
China
99.0
JQ282909
PEDV AH2012
2012
China
99.6
KC210145
PEDV CH/YNKM-8
2013
China
99.0
KF761675
PEDV IA2
2013
USA
99.9
KF468754
PEDV Iowa/18984
2013
USA
99.9
KF804028
PEDV MN
2013
USA
99.9
KF468752
PEDV MEX/104
2013
Mexico
99.8
KJ645708
PEDV VN/KCHY-310113
2013
Vietnam
98.1
KJ960180
PEDV OH851
2014
USA
99.1
KJ399978
PEDV K14JB01
2014
South Korea
99.8
KJ623926
PEDV ON-018
2014
Canada
99.9
KM189367
Table 1: Sequence identity of PEDV TW4 and reference strains.
Results
Genomic sequence of PEDV TW4
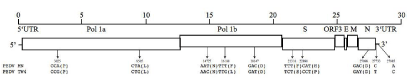
The full genomic RNA sequence of PEDV TW4 comprises 27,966 nucleotides (nts), excluding the 5’ leader sequences. Sequence analysis revealed that PEDV TW4 contains several conserved open reading frames (ORFs) with an overall genome organization similar to known PEDV strains (Table 2). The overall nucleotide composition is as follows: A, 24.8%; C, 19.1%; G, 22.7%; and T, 33.4%. The G+C content is 41.8 %.
ORFs
Start-end (nucleotide position)
No. of nucleotides
No. of amino acids
5’ UTR
1-220
220
-
ORF 1a
221-12,529
12,309
4103
ORF 1ab
221-20,565
(shift at 12,610)
20,345
6781
S
20,562-24,722
4161
1387
ORF 3
24,722-25,396
675
225
E
25,377-25,607
231
77
M
25,615-26,295
681
227
N
26,307-27,632
1326
442
3’ UTR
27,633-27,966
334
-
Table 2: Coding potentials and gene size in PEDV TW4/2014.
5’, 3’ Untranslated regions (UTRs) and ORFs
The 5’ UTR of PEDV TW4 comprises 220 nts, identical to other known PEDV strains; the 3’ UTR of our virus comprises 334 nts, which is also identical to other PEDV strains. The 5’ two-thirds of the genome contain the 1a (nt 221-12,529) and 1ab (nt 221-20,565) genes that encode the nonstructural polyproteins. A typical coronavirus ‘‘slip site,’’ 5’-UUUAAAC-3’ (nt 12,610–12,616), is located within this gene. These genes are followed by genes encoding the four structural proteins: spike (nt 20,562-24,722), envelope (nt 25,377-25,607), membrane (nt 25,615-26,295), and nucleocapsid (26,307-27,632) (Table 2). The accessory gene (ORF3: nt 24,722-25,396) identified in all of the known PEDV strains was also found in PEDV TW4 (Table 2).
Genetic comparison and phylogenetic analysis with reference PEDV strains
The overall sequence comparison revealed that PEDV TW4 was more closely related to the known subgroup 1b CoV but not 1a (TGEV Purdue and Feline coronavirus NTU156) within the alphacoronaviruses (Figure 1). PEDV TW4 was not clustered with the prototypical PEDV CV777. In contrast to the low nucleotide sequence similarity between Chinese strain AH2012 (99.6%) and recent Taiwanese strains, the homology levels between this PEDV TW4 isolate and US strains (99.9%) appeared to be much higher (Table 1 and Figure 2).
Figure 1: Phylogenetic relationships constructed using the complete genome sequences of PEDV TW4/2014 and reference strains. The analysis was performed using the maximum likelihood method based on 1,000 replicates within the MEGA 5 software. Bootstrap support values greater than 75 are shown. The complete genome sequence of transmissible gastroenteritis virus (TGEV) and feline coronavirus (FCoV) were included as an outgroup in this study.
Figure 2: Schematic comparison of the complete genome between PEDV MN and PEDV MN. The arrows indicate the different features on the sequence. Brackets enclose the amino acids. Numerical position on the genome of PEDV MN, accession number KF468752.
Discussion
PEDV has recently become an economic concern in the swine industry in Asia, North America, and South America [1,3-6,8,9]. In Taiwan, several outbreaks of PEDV infection have emerged since late 2013. Suckling piglets under 2 weeks of age show severe vomiting and watery yellowish diarrhea with high morbidity and mortality. Our previous study suggested that this outbreak of viruses clustered in the same clade as the US strains according to the partial S gene analysis [3]. This is the first report of a complete PEDV genome in Taiwan. Interestingly, comparative genome analysis of reference PEDV isolates revealed that the complete genome sequences of recent Taiwanese strains were almost identical (99.9%) to several of the 2013 US strains (PEDV IA2, Iowa/18984, and MN) (Table 1).
A previous study suggested that the US PEDV strains were most closely related to a strain isolated in 2012 in China (AH2012) [10]. Although the complete genome sequence of PEDV TW4 clustered with the US strains and AH2012, TW4 was more closely related to the US strains with high bootstrap values. Such US-like PEDV isolates have not only been observed in Taiwan [3] but also in other countries, including South Korea [5,6], Canada [7], Mexico [8], and Peru [9]. Taken together, our results indicate that recent PEDV outbreaks share a common evolutionary origin with PEDV strains throughout most of the swine industry. Those US-like PEDVs seen like highly virulent in piglets in several countries. Specific treatments of PEDV are not available [1]. Therefore, strict biosecurity measures should be established.
These US-like strains of the virus might have gained entry into different countries via unknown routes as early as late 2013. Spray- Dried Porcine Plasma (SDPP) has been suspected and tested for the presence of PEDV genome by real-time PCR [7,11]. Although feed that tested positive for the PEDV genome did not result in obvious piglet infection, contaminated feed still cannot be ruled out as a source of PEDV introduction in the field [7].
These US-like PEDV variants are highly virulent in piglets in Taiwan. Several pig farms are still facing re-emergences of these US-like PEDV strains despite several cycles of feedback with pooled homogenized intestines from suckling pigs (data not shown). The reasons for this feedback failure need to be further investigated, including infection with Porcine Circovirus Type 2 (PCV2) in sows. The clinical course of PEDV disease is markedly affected by transplacental infection with PCV2 according to previous report [12]. In addition, the key variations in the amino acid sequences need to be further studied, including in the S proteins, which plays a crucial role in receptor binding and eliciting protective immunity [13]. Similar to most RNA viruses, coronaviruses mutate at a high frequency due to the high error rate during RNA polymerization. In addition, a unique feature of coronavirus genetics is the high frequency of RNA recombination in the natural evolution of this virus [14]. Therefore, additional PEDV cases need to be investigated using continuous surveillance and complete genome analysis to gain better evolutionary insight into PEDV.
References
- Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012; 44: 167-175.
- Vijaykrishna D, Smith GJ, Zhang JX, Peiris JS, Chen H, Guan Y. Evolutionary insights into the ecology of coronaviruses. J Virol. 2007; 81: 4012-4020.
- Lin CN, Chung WB, Chang SW, Wen CC, Liu H, Chien CH, et al. US-like strain of porcine epidemic diarrhea virus outbreaks in Taiwan, 2013-2014. J Vet Med Sci. 2014; 76: 1297-1299.
- Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, et al. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 2013; 25: 649-654.
- Lee S, Lee C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg Infect Dis. 2014; 20: 1223-1226.
- Choi JC, Lee KK, Pi JH, Park SY, Song CS, Choi IS, et al. Comparative genome analysis and molecular epidemiology of the reemerging porcine epidemic diarrhea virus strains isolated in Korea. Infect Genet Evol. 2014; 26: 348-351.
- Pasick J, Berhane Y, Ojkic D, Maxie G, Embury-Hyatt C, Swekla K, et al. Investigation into the role of potentially contaminated feed as a source of the first-detected outbreaks of porcine epidemic diarrhea in Canada. Transbound Emerg Dis. 2014; 61: 397-410.
- Diosdado F, Martinez A, Socci G, Carrera E, Cordova D, Garcia A, et al. Molecular identification of the PEDV. In Proceeding of the 23rd International Pig Veterinary Society Congress; Cancun, Mexico. 2014: 306.
- More-Bayona J, Ramirez-Velasquez M, Manchego-Sayan A, Quevedo-Valle M, Rivera-Geronimo H: Molecular detection of emerging strains of PEDV in Peru. In Proceeding of the 23rd International Pig Veterinary Society Congress, Cancun, Mexico. 2014: 247.
- Huang YW, Dickerman AW, Piñeyro P, Li L, Fang L, Kiehne R, et al. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. 2013; 4: e00737-00713.
- Opriessnig T, Xiao CT, Gerber PF, Zhang J, Halbur PG. Porcine epidemic diarrhea virus RNA present in commercial spray-dried porcine plasma is not infectious to naïve pigs. PLoS One. 2014; 9: e104766.
- Jung K, Kim J, Ha Y, Choi C, Chae C. The effects of transplacental porcine circovirus type 2 infection on porcine epidemic diarrhoea virus-induced enteritis in preweaning piglets. Vet J. 2006; 171: 445-450.
- Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009; 7: 226-236.
- Lai MMC, Perlman S, Anderson LJ, Coronaviridae. In Fields Virology. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, et al, editors. Philadelphia, PA: Lippincott Williams & Wilkins; 2007: 1305-1335.