
Research Article
Ann Agric Crop Sci. 2025; 10(1): 1177.
Identification of BRI1-Associated Receptor Kinase 1 (BAK1) - Interacting Proteins in Arabidopsis Thaliana
Ere Choi†, Hyun-Ji Seo† and Man-Ho Oh*
Department of Biological Sciences, College of Biological Sciences and Biotechnology, Chungnam National University, Daejeon 34134, Republic of Korea †These Authors Contributed Equally to this Work
*Corresponding author: Man-Ho Oh, Department of Biological Sciences, College of Biological Sciences and Biotechnology, Chungnam National University, Daejeon 34134, Republic of Korea Tel: 010-3001-6367; Email: manhooh@cnu.ac.kr
Received: February 20, 2025; Accepted: March 12, 2025; Published: March 17, 2025;
Abstract
Plants utilize receptor-like kinases (RLKs) like BAK1 to mediate complex immune responses and adapt to environmental stress. BAK1 is a co-receptor involved in signal transduction triggered by microbe-associated molecular patterns (MAMPs) and damage-associated molecular patterns (DAMPs). These patterns active plant immunity, including pathogen-associated molecular patterntriggered immunity (PTI) and responses to environmental challenges. Catalase families, NOT9A, and IOS1 are essential in regulating stress responses and enhancing plant resilience. This study investigates the interaction of BAK1 with these proteins in Arabidopsis thaliana, aiming to uncover mechanisms underlying plant immune signaling and stress adaption.
The study examined the effects of ABA, flg22, chitin, and pep1 treatments on the expression of Catalase1, Catalase2, Catalase3, NOT9A, and IOS1 in Arabidopsis thaliana. ABA treatment increased the expression of CAT families and IOS1, indicating their role in mitigating oxidative stress during environmental stress. Chitin treatment downregulated CAT families and NOT9A, suggesting ROS accumulation as a signaling mechanism to enhance immune responses. Both flg22 and pep1 increased the expression of CAT families, NOT9A, and IOS1, highlighting their involvement in maintaining oxidative balance during immune responses. These finding reveals the coordination between oxidative stress mitigation and immune activation in plants. The BiFC assay confirmed BAK1-Catalase2 interaction in vivo. Y2H assays showed BAK1 interacts with Catalase1, NOT9A, and IOS1, but BiFC showed weak fluorescence for NOT9A and none for Catalase1 or IOS1, possibly due to low protein stability or expression.
Keywords: Arabidopsis thaliana; BAK1; Immune signaling; Catalase1; Catalase2; Catalase3; NOT9A; IOS1
Introduction
Plants are sessile organisms, meaning they are fixed in place. To respond to various factors arising from the external environment, they possess complex signaling mechanisms related to immunity. Peptides play a critical role in these processes, performing diverse physiological and biochemical functions. In the field of biological research, the widely used model organisms Arabidopsis thaliana has over 1,000 genes encoding peptides. Most signaling processes are initiated by the interaction between these peptides and kinases [1].
Receptor-like kinases (RLKs) are receptors located on the plasma membrane of plant cells. Depending on which amino acid residues they phosphorylate, they can be classified as serine/threonine protein kinase or tyrosine kinases. These proteins not only function as receptors but also act as kinases. When ligands bind to the extracellular domain of RLKs, phosphorylation occurs in the intracellular domain, mediating signal transduction [2].
BAK1 (BRI1-Associated Receptor Kinase 1) is a gene located on chromosome 4 of Arabidopsis thaliana. BAK1 consists of various domain that regulates its structure and function. The extracellular domain is characterized by a repetitive arrangement of leucine residues, classifying it as a Leucine-rich repeat receptor-like kinase (LRR-RLK). A transmembrane domain (TMD) spans the phospholipid bilayer to transmit external signals into the cell. Inside the cell, it features a juxta membrane domain (JMD) that regulates kinase activity, a kinase domain (KD) responsible for phosphorylation-dependent responses, and a carboxy-terminus region involved in protein stability and interactions [3].
The interaction between RLKs and LRR-RLKs is essential for plant growth, development, and adaptation to various environmental conditions, including pathogen defense. They play a key role in perceiving and transmitting external signals. Therefore, transcriptional and post-translational regulation of their activity is pivotal for receptor signaling in plants. Recognition of microbe-associated molecular patterns (MAMPs) by pattern recognition receptors (PRRs) triggers the induction of various proteins such as Flagellin Sensing 2 (FLS2) and Elongation Factor Tu (EFR), which is the first line of inducible defense against invading pathogens. Also, the CERK1 receptor, which recognize chitin as a ligand, and PEPR1 and PEPR2, which recognize pep1 as ligand, also play a role in plant immune signaling [2].
Abscisic acid (ABA) is a plant hormone involved in stress responses, such as drought and cold. It signals through PYR/PYL/ PCAR receptors, which interact with PP2C proteins to activate SnRK2 kinases, leading to stress adaptations [4]. Chitin, a fungal cell wall component, is recognized by CERK1 and CEBiP receptors, activating immune responses like ROS production and PR gene expression [5]. Flg22, derived from flagellin, binds FLS2 and recruits BAK1 to trigger plant immunity, including callose deposition. Pep1, a DAMP, is recognized by PEPR1/2, leading to immune signaling and root growth inhibition, with BAK1 involvement [6].
Cells near the site of infection in plants rapidly synthesize toxic compounds formed by the reduction of oxygen molecules. These compounds include superoxide anion, H2O2, and hydroxyl radicals. NADPH-dependent oxidase, located in the plasma membrane, produces superoxide anions, which are subsequently converted into H2O2 and hydroxyl radicals. Among these, hydroxyl radicals are the most potent oxidants and can cause lipid peroxidation, enzyme inactivation, and nucleic acid degradation. Thus, reactive oxygen species (ROS) function as part of the hypersensitive responses, contributing to host cell death or directly killing pathogens [7]. ROS also act as signaling molecules. Abiotic stress applied to one part of a plant generates signals that are transmitted to other parts, inducing systemic acquired resistance (SAR). This leads to systemic acquired acclimation (SAA), where acclimation occurs event in unstressed regions. These responses are mediated by ROS waves generated by RBOHD (Respiratory Burst Oxidase Homolog D).
When ROS accumulate, they must be removed by antioxidants that accept electrons from ROS. Plants have evolved antioxidant enzymes to active ROS scavenging more efficiently. Superoxide dismutase (SOD) converts two molecules of superoxide anion by combining them with hydrogen ions into oxygen and hydrogen peroxide (H2O2). Ascorbate peroxidase uses ascorbate as a reducing agent to add hydrogen ions to H2O2, converting it into monodehydroascorbate and H2O. These two antioxidant enzymes function in chloroplasts, peroxisomes, mitochondria, apoplast, and the cytoplasm. Catalase plays a role in detoxifying H2O2 into H2O and oxygen in the peroxisomes [8].
Arabidopsis thaliana has three types of Catalases. Catalase1 is located on chromosome 1 of Arabidopsis thaliana. It is an enzyme that uses a heme group as a cofactor to catalyze the reduction of hydrogen peroxide. It is mainly expressed in pollen and seeds. Catalase2 is located on chromosome 4 of Arabidopsis thaliana. It is primarily expressed in photosynthetically active tissues and interacts with NCA1 (No Catalase Activity 1) to enhance the plant’s resistance to abiotic stress. Catalase3 is located on chromosome 1 of Arabidopsis thaliana. It is mainly expressed in vascular tissues and aging leaves. It is activated by phosphorylation through CPK8 (Calcium-dependent Protein Kinase 8) in response to drought stress [9].
Almost all eukaryotic mRNA molecules possess poly(A) tails at the 3’ end. The length of a poly(A) tail is important for post transcriptional regulation because it infnces mRNA stability and/or translational efficiency. Poly(A) tail have a critical role in enhancing the translational efficiency of some transcripts, especially in certain developmental stages [10,11]. Various factors in the nucleus and cytoplasm are involved in determining the length of a poly(A) tail. One such cytoplasmic factor is deadenylase, a poly(A)-specific ribonuclease. Carbon catabolite repressor 4 (CCR4) and CCR4- associated factor 1 (CAF1) are widely conserved deadenylases in eukaryotes. CCR4 is a major cytoplasmic deadenylase [12,13]. NOT proteins serve as core components in the regulation of multiple levels of gene expression. The CCR4-NOT (Carbon Catabolite Repressor 4 – Negative On TATA) complex is a highly conserved and essential protein complex. It consists of at least six core subunits organized in a specific molecular structure. This complex plays a crucial role in various aspects of gene expression regulation [14]. The CCR4-NOT complex is built around a scaffold protein called NOT1/CNOT1. Other subunits, which have specific functions, attach to this scaffold to form the complete complex. Another highly conserved subunit of the complex is NOT9 (also called Required for Cell Differentiation 1 [RQCD1], CAF40, CNOT9), which spans the bridge between the RNA-induced silencing complex (RISC) and CCR4-NOT [15,16].
Plants generally possess resistance to most pathogens, and the recognition of MAMPs triggers a complex set of responses known as PAMP-triggered immunity (PTI), which activates defense mechanisms to resist pathogen attacks [17]. Additionally, infection by microbial pathogens often triggers UPR (Unfolded Protein Response) in plants, which plays a crucial role in regulating immune responses [18]. In this context, IOS1 (Impaired Oomycete Susceptibility 1) is known to regulate ER stress response, including UPR [15]. IOS1 is a member of a subfamily of about 50 RLKs in Arabidopsis thaliana. These RLKs have an extracellular region that contains a malectinlike domain (MLD) along with two to three leucine-rich repeats [19]. The protein sequence and domain organization of IOS1 show strong similarities to legumes, which are key regulators of fungal and bacterial symbiont accommodation [20].
The research aims to identify the specific conditions under which the relative expression levels of Catalase1, Catalase2, Catalase3, NOT9A, and IOS1 are altered in the BAK1-Flag line upon treatment with various ligands, including ABA, chitin, flg22, and pep1.
Through this approach, they study seeks to elucidate the relationship between each ligand and ROS metabolism in the signal transduction pathway induced by BAK1. Furthermore, a critical objective is to confirm whether BAK1 interacts with Catalase1, Catalase2, NOT9A, and IOS1 in vivo. While BAK1 functions as a signaling receptor involved in immune responses, its interaction with Catalase1, Catalase2, which are responsible for ROS removal, could provide pivotal insights into regulatory mechanisms of immune responses. Additionally, BAK1’s interaction with NOT9A, which is responsible for RNA stability and gene expression regulation, could provide insights into the crosstalk between immune signaling and stress responses. Furthermore, the interaction between IOS1, which regulates ER stress during plant infection, can provide important insights into understanding plant stress responses and the balance between immunity and growth during pathogen attacks.
In summary, this study contributes to understanding the relationship between BAK1-mediated immune signaling, ROS metabolism, regulation of gene expression, regulation of PAMPtriggered immunity (PTI) and ER stress response. Potentially offering foundational knowledge for stress response mechanism, immune response mechanism in plants.
Materials and Methods
Seed sterilization, Liquid Culture, Hormone Treatment
Sterilized seeds were placed in a 1.5mL ep tube. Then, 700uL of 80% ethanol was added, and the tube was reacted in a shaking machine for 5 minutes. After removing the 80% ethanol, 500uL of seed sterilization solution (7.6mL Clorox, 17.5mL 0.05% Triton, 25mL Sterile distilled water) was added and reacted in the shaking machine for 20 minutes. The tube was the spun down in a clean bench, and the seed sterilization solution was removed. Sterile distilled water was added, inverted, and spun down; this process was repeated more than five times. Finally, 1mL of sterile distilled water was added, and vernalization treatment was perform at 4°C for two days before planting the seeds on 1/2 MS media. Shaking incubation set at 23°C and 130 RPM. On the 9th day, the 1/2 MS media was changed, and on the 10th day, hormones were applied. ABA, chitin, flg22, and pep1 were applied at a final dilution of 10-6, and each hormone was treated for 2 hours.
RNA Extraction, cDNA Synthesis
For total RNA extraction from plants, samples that were harvested and frozen were places in a mortar, and liquid nitrogen was added before grinding. The EZTM Total RNA Miniprep Kit (Cat. No. EP30150N) from Enzynomics was used, and the integrity of the RNA was checked through gel electrophoresis. For cDNA synthesis, the RNA extraction product was measured using a Nanodrop, and the concentration was adjusted to 1,000ng/uL for use. The First Strand cDNA Synthesis Kit ReverTra Ace-a- (Code: FSK-101) from TOYOBA was used. The reaction was carried out in a SimpliAmp thermal cycler at 42°C for 20 minutes and 99°C for 5 minutes.
Quantitative PCR
Using the synthesized cDNA, a reaction mixture was prepared for each well of the BioFACTTM 0.1mL qPCR 8 Strip Tubes with 1uL of cDNA, 2uL of primers, 2uL of distilled water, and 5uL of RbTaqTM SYBR Green qPCR PreMIX (Cat. #RT5305), making a total volume of 10uL. After sinning down the tubes, the reaction was carried out using the CFX-8-Connect Real-Time PCR system (Cat. No. BR1855200, BIORAD, California, U.S.A). The PCR program consisted of denaturation at 95°C for 15 minutes, followed by 45 cycles of denaturation at 95°C for 10 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 30 seconds. A melting curve was measured from 55°C to 95°C with a 0.5°C interval for 5 seconds. The ΔCq values of each gene were normalized against the ΔCq value of ACTIN to obtain the ΔΔCq values, which were used to analyze gene expression levels (Table 1).
Name of Primer
Primer sequence
Catalase1_F
5’– TGC CTT TAC AAC CTG TTG GTC –3’
Catalase1_R
5’– CTG AAT AGT GGA TGC CTG GAA –3’
Catalase2 _F
5’– GGA AAC CAA CTT GTG GAG TCA –3’
Catalase2 _R
5’– CAT TCA GGG TAG TTT CCA GCA –3’
Catalase3 _F
5’– CAA CAC TCC GGT GTT CTT CAT –3’
Catalase3 _R
5’– AGC ACC ATG TGA GCA AAC TCT –3’
NOT9A_F
5’– CTG ACT CCT GCT CAG TCC AAC –3’
NOT9A_R
5’– TAG AAC GGG ATA TGA GCC TTG –3’
IOS1_F
5’– TTG CAG CAT CAC TTG TTT CAG –3’
IOS1_R
5’–ACG GTG TTG TAC CAG TTC CTG –3’
Table 1: Quantitative PCR.
Gene Cloning
In this study, cDNA obtained from seeds transformed with BAK1-Flag, based on Arabidopsis thaliana (WS-2), was used for the experiments. cDNA PCR was performed using Pfu DNA polymerase (Agilent Technologies, Santa Clara, CA, United States). PCR amplification of Catalase1, Catalase2, NOT9A, and IOS1 was confirmed through gel electrophoresis, and gel extraction was performed using the HigeneTM Gel & PCR Purification System Kit. The product was then cloned into the pENTR/D-TOPO vector and transformed in to E. coli. To verify insertion, the bacteria were plated on kanamycin-resistant media and incubated at 37°C for 16 hours. Plasmid DNA was extracted using the Biomedic Plasmid DNA Miniprep Kit (Cat. No. BM-K110B), and sequencing results were checked to confirm that the insert was in the correct position and orientation (Table 2).
Name of Primer
Primer sequence
Catalase1_F
5’– CAC CAT GGA TCC ATA CAG GGT TC –3’
Catalase1_R
5’– TCA GAA GTT TGG CCT CAC GTT AAG AC –3’
Catalase2 _F
5’– CAC CAT GGA TCC TTA CAA GTA TCG TCC –3’
Catalase2 _R
5’– TTA GAT GCT TGG TCT CAC GTT CAG AC –3’
NOT9A_F
5’– CAC CAT GGC GAA TCT ACC TTC TTC –3’
NOT9A_R
5’– TCA AAG CAT GTG CTC AAA TCC TCC –3’
IOS1_F
5’– CAC CAT GGC GTT TTC TTC TTG TTT TCT C –3’
IOS1_R
5’– TCT AGC TCC TGG ATT AAG CTC TGT TG –3’
Table 2: Gene Cloning.
BiFC Assay
Reverse reaction (LR) recombinations of appropriate open reading frame (AtCAT1, AtCAT2, AtNOT9A, AtIOS1) in pENTR/DTOPO were performed with the split-YFP destination vectors pDESTGWVYNE and pDEST-GWVYCE to generate N- or C-terminal fusions with the N- and C-terminal yellow fluorescent proteins (YFP) moieties, respectively [21]. Recombined vectors were transformed into Agrobacterium strain GV3101. Six-week-old Nicotiana benthamiana leaves were agro-filtrated as previously described [22]. After 48 hours, YFP fluorescence was visualized using a Super Resolution Confocal Laser Scanning Microscope (LSM 880 with Airyscan, Zeiss, Jena, Germany).
Results
In plants, hormones act as chemical substances that can induce various responses. ABA, chitin, flg22, and pep1 induce defense responses. Catalase families play a role in braking down reactive oxygen species produced by plants under stress conditions. NOT9A regulates the expression of genes related to immune responses and plays a role in managing cellular stress responses. IOS1 plays a role in regulating ER stress responses, including UPR. To examine the expression levels of Catalase1, Catalase2, Catalase3, NOT9A, and IOS1 after 2 hours of hormone treatment, quantitative PCR was performed.
In these experiments, the BiFC (Biomolecular Fluorescent Complementation) was performed using the Gateway vector system. First, the DNA fragments of insert, including the coding sequences of Catalase1, Catalase2, NOT9A, and IOS1, were amplified through PCR. For each gene, the expected PCR product sizes were confirmed, with the coding sequences of Catalase 1 and Catalase 2 being 1,479bp, NOT9A being 950bp, and IOS1 being 2,685bp. The amplified DNA was then extracted from the electrophoresis gel after verification of the PCR results. Second, after inserting the Catalase1, Catalase2, NOT9A, and IOS1 fragments into pENTR/D-TOPO, the constructs were cultured in E. coli, followed by miniprep. The resulting plasmids were again confirmed by electrophoresis at the expected size. Subsequently, an LR reaction was performed to transfer the DNA into the Gateway destination vector. Throughout the process, antibiotic markers were used to ensure proper insertion, and gel electrophoresis was performed to confirm the correct size of the fragments. Finally, sequencing was conducted to verify that the DNA was inserted in the correct orientation and at the desired location without inversion.
To determine whether BAK1 physically interacts with Catalase1, Catalase2, NOT9A, and IOS1 in vivo, a BiFC assay was performed. As positive control, SOS2-YFPn and SOS3-YFPc, which are known to interact, were used. For the negative controls, combinations such as YFPn empty vector and YFPc empty vector. YFPn and CAT1-YFPc, CAT2-YFPc, NOT9A-YFPc, and IOS1-YFPc, as wells as BAK1-YFPn and YFPc empty vector, were employed. The interaction of BAK1- YFPn with CAT1-YFPc, CAT2-YFPc, NOT9A-YFPc, and IOS1-YFPc, which were the focus of this study, were also examined. Fluorescent signals were observed using a super-resolution confocal laser scanning microscope.
Discussion
This study aims to investigate the expression levels of Catalase1, Catalase2, Catalase3, NOT9A, and IOS1 under immune responses triggered by Damage-associated molecular patterns (DAMPs), such as ABA, and Microbe-associated molecular patterns (MAMPs), such as chitin, flg22, and pep1. Additionally, the study seeks to determine This study aims to investigate the expression levels of Catalase1, Catalase2, Catalase3, NOT9A, and IOS1 under immune responses triggered by Damage-associated molecular patterns (DAMPs), such as ABA, and Microbe-associated molecular patterns (MAMPs), such as chitin, flg22, and pep1. Additionally, the study seeks to determine whether the Arabidopsis thaliana co-receptor BAK1 directly interacts with Catalase1, Catalase2, NOT9A, and IOS1 in vivo. BAK1 is a coreceptor that functions as a receptor and a kinase. It participates in plant immune signaling by interacting with various receptors, such as PYR/PYL/PCAR, which binds the ligand abscisic acid (ABA); FLS2, which binds the ligand flagellin-derived peptide 22 (flg22); CERK1, which binds the ligand chitin; and PEPR1/PEPR2, which binds the ligand pep1-derived peptides [23].
Plants have Pattern recognition receptors (PRRs) that recognize Microbe-associated molecular patterns (MAMPs) to distinguish between self and non-self during pathogen infection. When the receptor recognizes a ligand, it triggers Pattern-triggered immunity (PTI), a localized basal defense response. As part of the defense response, a Reactive oxygen species (ROS) burst occurs. This burst can directly attack pathogens around the cell or inhibit them by causing toxicity. Additionally, it can activate defense signaling pathways, such as the influx of Ca2+, which leads to the expression of further immunerelated genes [24]. The study investigated the effects of ABA, chitin, flg22, and pep1 treatments on the expression of Catalase1, Catalase2, Catalase3, NOT9A, and IOS1 genes in Arabidopsis thaliana (Figure 1). The increased expression of CAT families and IOS1 following ABA treatment indicates that the plant activates oxidative stress mitigation mechanisms in response to environmental stress such as drought or salinity. ABA not only regulates stomatal behavior but also protects cells by eliminating ROS, highlighting the critical role of Catalase families and IOS1 in cellular defense mechanisms. Chitin, a component of fungal cell walls, functions as a Microbe-associated molecular patterns (MAMPs) that activates the plant’s innate immunity [25]. The observed downregulation of CAT families and NOT9A after chitin treatment suggests that the plant allow for a transient accumulation of ROS as a signaling mechanism to amplify immune responses. ROS accumulation may act as a secondary messenger to initiate defenserelated pathways, including the activation of pathogenesis-related genes and hypersensitive responses (HR). Similarly, flg22 and pep1 act as Microbe-associated molecular patterns (MAMPs) and Damageassociated molecular patterns (DAMPs), respectively, which trigger the plant’s innate immune system. The increased expression of CAT families, NOT9A, and IOS1 after flg22 and pep1 treatments suggests that these genes play a key role in mitigating oxidative imbalances during immune responses.
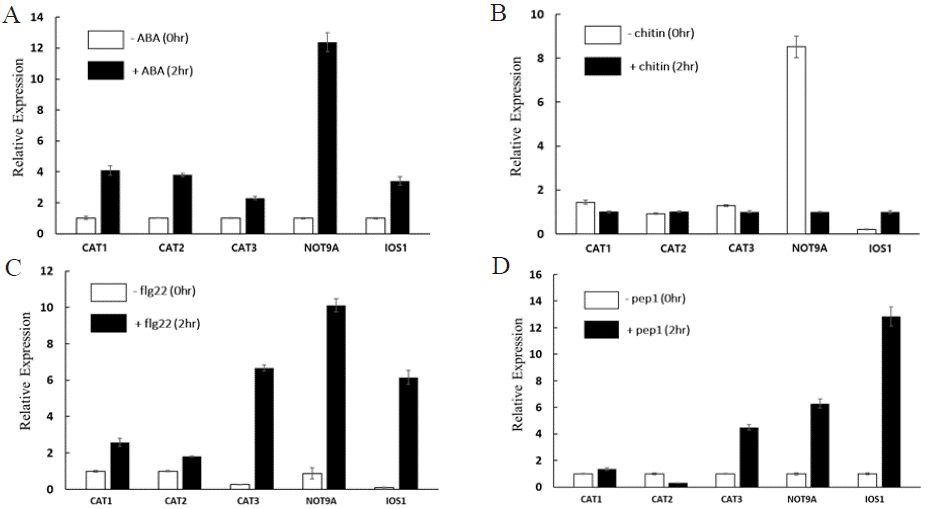
Figure 1: Relative expression of CAT1, CAT2, CAT3, NOT9A, and IOS1 after ABA, chitin, flg22, and pep1 treatment.
Each figure represents the relative expression of CAT1, CAT2, CAT3, NOT9A, and IOS1 when treated with A ABA, B chitin, C flg22, and D pep1. The relative
expression levels were normalized against ACTIN. Three independent experiments were conducted, and error bars represent the standard error.
The regulation of CAT families by ABA, chitin, flg22, and pep1 treatments demonstrates the complex crosstalk between defense responses and oxidative stress mitigation. These findings suggest that hormone signaling may interact with immune responses, providing an integrated strategy for plants to cope with environmental stress and pathogen attacks. Further studies are necessary to clarify the individual roles of Catalase1, Catalase2, and Catalase3 in oxidative stress and defense mechanism. The qPCR results following ABA, flg22, chitin, and pep1 treatments highlight the significant roles of the CAT families in oxidative stress mitigation and immune responses. The increase in NOT9A gene expression following ABA, flg22, and pep1 treatments suggests that NOT9A plays a crucial regulatory role in plant immune and stress responses by regulating gene expression, helping plants more effectively cope with environmental stress and pathogen attacks. Furthermore, the increase in IOS1 gene expression following ABA, flg22, chitin, and pep1 treatments suggests that IOS1 plays a crucial role in regulating plant immune responses. It indicates that IOS1 may contribute to stabilizing cellular processes under stress conditions, thereby activating or enhancing immune responses. These findings provide new insights into the integrated defense mechanisms of plants and contribute to the developmental of strategies to enhance crop resistance to environmental changes and pathogen infections.
The results from the BiFC assay confirmed that BAK1 physically interacts with Catalase 2 in vivo (Figure 2B). Although not shown in the figure, results from the Y2H assay indicate that BAK1 interacts with Catalase1, NOT9A, and IOS1. However, in the BiFC experiments, weak fluorescence was observed for NOT9A (Figure 2C), while no fluorescence was detected for Catalase1 (Figure 2A) and IOS1 (Figure 2D). This lack of detectable fluorescence might be due to low protein stability, improper folding, or insufficient expression levels of the corresponding fusion proteins under the experimental conditions.
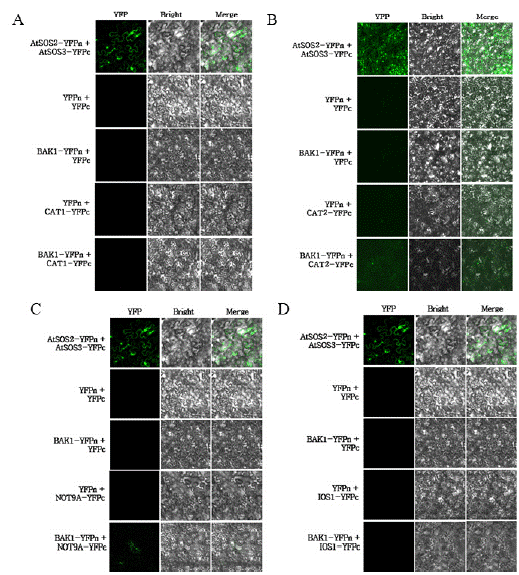
Figure 2: BiFC was used to confirm whether BAK1 interacts with CAT1, CAT2, NOT9A, and IOS1 in vivo.
To determined whether BAK1 physically interacts with A Catalase1, B Catalase2, C NOT9A, and D IOS1 in vivo, a BiFC assay was performed. As positive
control, SOS2-YFPn and SOS3-YFPc, which are known to interact, were used. For the negative controls, combinations such as YFPn empty vector and
YFPc empty vector. YFPn and CAT1-YFPc, CAT2-YFPc, NOT9A-YFPc, and IOS1-YFPc, as well as BAK1-YFPn and YFPc empty vector, were employed.
The interaction of BAK1-YFPn with CAT2-YFP c was observed, a positive reaction. Conversely, the interaction between BAK1-YFPn and NOT9A-YFPc was
observed, but it was weak. No interaction was detected between BAK1-YFPn and CAT1-YFPc or IOS1-YFPc. Fluorescent signals were observed using a superresolution
confocal laser scanning microscope.
The results from the BiFC assay confirmed that BAK1 physically interacts with Catalase 2 in vivo, providing important evidence for the functional relationship between BAK1 and the Catalase families. Previous studies have shown that BAK1 suppresses H2O2 accumulation under high light conditions, thereby regulating growth and development. In this process, BAK1 was found to phosphorylate members of the Catalase families. In plants with mutated Catalase family genes, the inhibitory effect of BAK1 under high light conditions was lost, suggesting that the interaction between BAK1 and the Catalase families play a role in regulating ROS and supporting proper plant growth and development [9]. This finding provides foundational data for understanding the functional relationship between BAK1 and the Catalase families. Such interactions are not limited to high light stress, but can also occur under various abiotic stresses such as high temperature, drought, salinity, or pathogen infection. Therefore, research to confirm the in vivo interaction between BAK1 and Catalase under diverse abiotic stress conditions using BiFC is needed. Such studies could uncover new regulatory mechanisms under abiotic stress, providing valuable insights into plant adaptation and resilience.
If the interaction between BAK1 and NOT9A is confirmed, it could lead to research highlighting their critical role in the integrated regulation of plant immune signaling pathways and stress response mechanisms. BAK1 plays an important role in activating immune responses by recognizing various microbial and damage-related signals. NOT9A functions as a link between the RISC and CCR4- NOT complexes, both of which are important for regulating gene expression. If the physical interaction between these two proteins, which perform such functions, is confirmed, future studies could provide a foundation for understanding the regulatory mechanisms of ROS production, HR, and the expression of PR genes.
Previous studies have shown that IOS1 regulates the PTI response through both BAK1-dependent and BAK1-independent PRR complexes [26]. The interaction between BAK1 and IOS1 plays a essential role in plant immune responses and stress adaptation. BAK1 is central to plant immune signaling and regulates various PTI responses. IOS1, a key protein involved in ER stress responses, particularly the UPR, helps alleviate stress and enhance immune responses during cellular stress. The interaction between BAK1 and IOS1 integrates PTI and ER stress responses, stabilizing the cellular environment and activating immune pathways. Therefore, if their physical interaction is confirmed, it will provide valuable insights into the regulatory mechanisms under various abiotic stress conditions.
After repeating the experiment and confirming the interaction, it will be necessary to identify which specific site on BAK1 plays a crucial role in its interaction with Catalase1, Catalase2, NOT9A, and IOS1. By examining the structural domains of BAK1 and selecting residues that are likely to contribute to the interaction based on known domains or functional regions, Site-directed mutagenesis (SDM) can be performed to substitute these residues with other amino acids. BiFC experiments with each mutated BAK1 protein can then be conducted to assess the interaction with Catalase1, Catalase2, NOT9A, and IOS1. Once the critical residues are identified, further structure-function analyses can be carried out to reveal the structural basis of this interaction.
Ultimately, this research aims to elucidate the physiological mechanisms plants use to adapt to changing environments, laying the foundation for the development of sustainable agricultural technologies. Modern agriculture faces various environmental stressors, such as climate change, increasing pests and diseases, and soil degradation. As such, understanding the adaptation mechanisms plants employ is essential. In this context, the interactions of key proteins such as BAK1, Catalase families, NOT9A, and IOS1 are crucial in maintaining the balance between plant immunity and ROS metabolism, facilitating stress response mechanisms, and regulating gene expression. By gaining insight into these adaptation mechanisms, it becomes possible to develop resilient crop varieties capable of withstanding extreme conditions or enhancing stress resistance through targeted genetic modifications. For instance, genetic modifications based on the interactions between BAK1 and Catalase families, NOT9A, and IOS1 could lead to the development of stress-resistant crops or strategies to improve pathogen resistance [27].
Moreover, these findings have the potential to increase productivity across a range of crops and contribute to the creation of sustainable agricultural systems. This research not only enhances our understanding of plant physiological processes but also provides the foundation for developing agricultural systems that maximize productivity while maintaining harmony with environmental factors. These insights will accelerate the development of crop varieties resistant to environmental stresses and pathogens, driving improvements in crop management and farming practices. Ultimately, this research will play a critical role in advancing agricultural biotechnology, ensuring food security, and enhancing sustainable agricultural productivity in the face of environmental challenges, such as climate change.
Acknowledgments
This work was supported in part by a grant from Chungnam National University (CNU) and Republic of Korea, and this work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01481605)” Rural Development Administration, Republic of Korea.
References
- Murphy E, Smith S and Smet ID. Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. The Plant cell. 2012; 24: 3198-3217.
- Singh V, Perraki A, Kim SY, Shrivastava S, Lee JH, Zhao Y, et al. Tyrosine-610 in the Receptor Kinase BAK1 Does Not Play a Major Role in Brassinosteroid Signaling or Innate Immunity. Frontiers in plant sceicne. 2017; 8: 1273.
- Zhou Q, Liu J, Wang J, Chen S, Chen L, Wang J, Wang HB and Liu B. The juxtamembrane domains of Arabidopsis CERK1, BAK1, and FLS2 play a conserved role in chitin-induced signaling. Journal of Integrative plant biology. 2010; 62: 556-562.
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, et al. In vitro reconstitution of an abscisic acid signaling pathway. Nature. 2009; 462: 660-664.
- Miya A, Albert P, Shinya T and Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. PNAS. 2007; 104: 19613- 19618.
- Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. PNAS. 2006; 103: 10098-10103.
- Peter Balint-Kurti. The plant hypersensitive response: concepts, control and consequences. Molecular plant pathology. 2019; 20: 1163-1178.
- Andréia Caverzan, Alice Casassola, and Sandra Patussi Brammer. Reactive Oxygen Species and Antioxidant Enzymes Involved in Plant Tolerance to Stress. Abiotic and biotic stress in plants – Recent Advances and Future Perspectives. 2016.
- Shan Zhang, Cheng Li, Haihua Ren, Tong Zhao, Qi Li, Shufen Wang, et al. BAK1 Mediates Light Intensity to Phosphorylate and Activate Catalases to Regulate Plant Growth and Development. International journal of molecular sciences. 2020; 21: 1437.
- Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes & Development. 2012; 26: 2724-2736.
- Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, et al. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Molecular cell. 2012; 47: 253-266.
- Chen J, Chiang YC and Denis CL. CCR4, a 3’-5’ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. The EMBO journal. 2002; 21: 1414-1426.
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D and Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. The EMBO journal. 2002; 21: 1427- 1436.
- Robert Buschauer, Yoshitaka Matsuo, Takato Sugiyama, Ying-Hsin Chen, Najwa Alhusaini, Thomas Sweet, et al. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science. 2020; 368: eaay6912.
- Chen Y, Hu D, Yabe R, Tateno H, Qin SY, Matsumoto N, et al. Role of malectin in Glc(2)Man(9)GlcNAc(2)-dependent quality control of a1-antitrypsin. Molecular biology of the cell. 2011; 22: 3559-3570.
- Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, et al. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Molecular cell. 2014; 54: 751-765.
- Boller T and Felix G. A renaissance of elicitors: perception of microbeassociated molecular patterns and danger signals by pattern-recognition receptors. Annual review of plant biology. 2009; 60: 379-406.
- Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, et al. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PloS one. 2012; 7: e31944.
- Hok S, Danchin EGJ, Allasia V, Ranabières F, Attard A and Keller H. An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant, cell & environment. 2011; 34: 1944-1957.
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002; 417: 959-962.
- Gehl C, Waadt R, Kudla J, Mendel RR, Hänsch R. New GATEWAY vectors for high throughput analyses of protein-protein interactions by biomolecular fluorescence complementation. Molecular plant. 2009; 2: 1051-1058.
- Kumar D, Kumar R, Baek D, Hyun TK, Chung WS, Yun DJ, et al. Arabidopsis thaliana RECEPTOR DEAD KINASE1 Functions as a Positive Regulator in Plant Responses to ABA. Molecular plant. 2017; 10: 223-243.
- Delphine Chinchilla, Libo Shan, Ping He, Sacco de Vries, and Birgit Kemmerling. One for all: the receptor-associated kinase BAK1. Trends in plant science. 2009; 14: 535-541.
- Gernot Sellge and Thomas A Kufer. PRR-signaling pathways: Learning from microbial tactics. Seminars in immunology. 2015; 27: 75-84.
- Dongping Lu, Shujing Wu, Xiquan Gao, Yulan Zhang, Libo Shan, and Ping He. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proceedings of the National Academy of Sciences of the United States of America. 2010; 107: 496-501.
- Yu-Hung Yeh, Dario Panzeri, Yasuhiro Kadota, Yi-Chun Huang, Pin-Yao Huang, Chia-Nan Tao, et al. The Arabidopsis Malectin-Like/LRR-RLK IOS1 Is Critical for BAK1-Dependent and BAK1-Independent Pattern-Triggered Immunity. The Plant cell. 2016; 28: 1701-1721.
- Iti Sharma and Parvaiz Ahmad. Catalase: A Versatile Antioxidant in Plants. Oxidative Damage to Plants. 2014; 131-148.