Short Communication
Recently a very powerful web-server has been developed for predicting the subcellular localization of Gram-negative bacterial proteins purely according to their sequences information for the multi-label systems [1], in which a same protein may appear or move between two or more location sites and hence its ID (Identification) needs two or more labels for distinction, namely the “multi-label mark” [2].
The web-server is called as “pLoc_bal-mGneg”, where “bal” means that the predictor has been treated by balancing or quasibalancing out the training dataset [3-9], and “m” means that the predictor is with the capacity to study the multi-label systems. How the web-server is working can be clearly seen via the showcase below.
Clicking the link at http://www.jci-bioinfo.cn/pLoc_bal-mGneg/, you will see the top page of the pLoc_bal-mGneg web-server prompted on your computer’s screen Figure 1. Then, just simply following the commands given in the Step 2 and Step 3 of [4], you will see Figure 2 on the screen of your computer. The corresponding predicted reports were given [4]. As you can see from there: nearly all the success rates achieved by the web-server for the Gram-negative bacterial proteins in each of the 8 subcellular locations are within the range of 98-100%, which is far beyond the reach of any of its counterparts.
Figure 1: A semi screenshot for the top page of pLoc_bal-mGneg (Adapted
from [4] with permission).
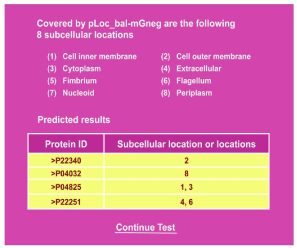
Figure 2: A semi screenshot for the webpage obtained by following Step 3 of
Section 3.5 (Adapted from [4] with permission).
In addition to the advantages of high accuracy and easy to use, the web-server has been constructed by strictly observing the “Chou’s 5-steps rule” and hence bears the following fantastic merits as agreed and supported by many investigators (see, e.g., [10-91] as well as three comprehensive review papers [2,92,93]): (1) crystal clear in logic development, (2) completely transparent in operation, (3) easily to repeat the reported results by other investigators, (4) with high potential in stimulating other sequence-analyzing methods, and (5) very convenient to be used by the majority of experimental scientists.
Besides, the approach [94-96] of PseAAC (Pseudo Amino Acid Composition) has also been applied during developing the webserver predictor. It is a very powerful approach for representing the protein samples by catching their special or key features, as done by many scientists as well [97-222].
Furthermore, the IHTS (Inserting Hypothetical Training Samples) treatment has also been applied to balance or quasi-balance out the training dataset [57,60,84].
For the amazing and awesome roles of the “5-steps rule” and general “PseAAC” in driving proteome, genome analyses and drug development, see a series of recent papers [2,93,223-232] where both the “5-steps rule” and the general “PseAAC” as well as their huge impacts have been very impressively recollected at different angles or from varieties of aspects.
References
- Chou KC, Shen HB. Recent progresses in protein subcellular location prediction. Analytical Biochemistry. 2007; 370: 1-16.
- Chou KC. Advance in predicting subcellular localization of multi-label proteins and its implication for developing multi-target drugs. Current Medicinal Chemistry. 2019; 26: 4918-4943.
- Xiao X, Cheng X, Chen G, Mao Q, Chou KC. pLoc_bal-mVirus: Predict Subcellular Localization of Multi-Label Virus Proteins by Chou’s General PseAAC and IHTS Treatment to Balance Training Dataset. Med Chem. 2019; 15: 496-509.
- Xiao X, Cheng X, Chen G, Mao Q, Chou KC. pLoc_bal-mGpos: predict subcellular localization of Gram-positive bacterial proteins by quasibalancing training dataset and PseAAC. Genomics. 2019; 111: 886-892.
- Chou KC, Cheng X, Xiao X. pLoc_bal-mEuk: predict subcellular localization of eukaryotic proteins by general PseAAC and quasi-balancing training dataset. Med Chem. 2019; 15: 472-485.
- Chou KC, Cheng X, Xiao X. pLoc_bal-mHum: predict subcellular localization of human proteins by PseAAC and quasi-balancing training dataset Genomics. 2019; 111: 1274-1282.
- Cheng X, Lin WZ, Xiao X, Chou KC. pLoc_bal-mAnimal: predict subcellular localization of animal proteins by balancing training dataset and PseAAC. Bioinformatics. 2019; 35: 398-406.
- Cheng X, Xiao X, Chou KC. pLoc_bal-mPlant: predict subcellular localization of plant proteins by general PseAAC and balancing training dataset Curr Pharm Des. 2018; 24: 4013-4022.
- Cheng X, Xiao X, Chou KC. pLoc_bal-mGneg: predict subcellular localization of Gram-negative bacterial proteins by quasi-balancing training dataset and general PseAAC. Journal of Theoretical Biology. 2018; 458: 92-102.
- Butt AH, Khan YD. Prediction of S-Sulfenylation Sites Using Statistical Moments Based Features via Chou’s 5-Step Rule. International Journal of Peptide Research and Therapeutics (IJPRT). 2018.
- Awais M, Hussain W, Khan YD, Rasool N, Khan SA, Chou KC. iPhosHPseAAC: Identify phosphohistidine sites in proteins by blending statistical moments and position relative features according to the Chou’s 5-step rule and general pseudo amino acid composition. IEEE/ACM Trans Comput Biol Bioinform. 2019.
- Barukab O, Khan YD, Khan SA, Chou KC. iSulfoTyr-PseAAC: Identify tyrosine sulfation sites by incorporating statistical moments via Chou’s 5-steps rule and pseudo components Current Genomics. 2019.
- Butt AH, Khan YD. Prediction of S-Sulfenylation Sites Using Statistical Moments Based Features via Chou’s 5-Step Rule. International Journal of Peptide Research and Therapeutics (IJPRT). 2019.
- Chen Y, Fan X. Use of Chou’s 5-Steps Rule to Reveal Active Compound and Mechanism of Shuangshen Pingfei San on Idiopathic Pulmonary Fibrosis. Current Molecular Medicine. 2019.
- Du X, Diao Y, Liu H, Li S. MsDBP: Exploring DNA-binding Proteins by Integrating Multi-scale Sequence Information via Chou’s 5-steps Rule. Journal of Proteome Research. 2019; 18; 3119-3132.
- Ehsan A, Mahmood MK, Khan YD, Barukab OM, Khan SA, Chou KC. iHyd-PseAAC (EPSV): Identify hydroxylation sites in proteins by extracting enhanced position and sequence variant feature via Chou’s 5-step rule and general pseudo amino acid composition. Current Genomics. 2019; 20: 124- 133.
- Hussain W, Khan SD, Rasool N, Khan SA, Chou KC. SPalmitoylC-PseAAC: A sequence-based model developed via Chou’s 5-steps rule and general PseAAC for identifying S-palmitoylation sites in proteins. Anal Biochem. 2019; 568: 14-23.
- Hussain W, Khan YD, Rasool N, Khan SA, Chou KC. SPrenylC-PseAAC: A sequence-based model developed via Chou’s 5-steps rule and general PseAAC for identifying S-prenylation sites in proteins. J Theor Biol. 2019; 468: 1-11.
- Ju Z, Wang SY. Prediction of lysine formylation sites using the composition of k-spaced amino acid pairs via Chou’s 5-steps rule and general pseudo components. Genomics. 2019.
- Kabir M, Ahmad S, Iqbal M, Hayat M. iNR-2L: A two-level sequence-based predictor developed via Chou’s 5-steps rule and general PseAAC for identifying nuclear receptors and their families. Genomics. 2019.
- Khan ZU, Ali F, Khan IA, Hussain Y, Pi D. iRSpot-SPI: Deep learningbased recombination spots prediction byincorporating secondary sequence information coupled withphysio-chemical properties via Chou’s 5-step rule and pseudo components. Chemometrics and Intelligent Laboratory Systems (CHEMOLAB). 2019; 189: 169-180.
- Lan J, Liu J, Liao C, Merkler DJ, Han Q, Li J. A Study for Therapeutic Treatment against Parkinson’s Disease via Chou’s 5-steps Rule. Current Topics in Medicinal Chemistry. 2019.
- Le NQK. iN6-methylat (5-step): identifying DNA N(6)-methyladenine sites in rice genome using continuous bag of nucleobases via Chou’s 5-step rule. Mol Genet Genomics. 2019; 294: 1173-1182.
- Le NQK, Yapp EKY, Ho QT, Nagasundaram N, Ou YY, Yeh HY. iEnhancer- 5Step: Identifying enhancers using hidden information of DNA sequences via Chou’s 5-step rule and word embedding. Anal Biochem. 2019; 571: 53- 61.
- Le NQK, Yapp EKY, Ou YY, Yeh HY. iMotor-CNN: Identifying molecular functions of cytoskeleton motor proteins using 2D convolutional neural network via Chou’s 5-step rule. Anal Biochem. 2019; 575: 17-26.
- Liang R, Xie J, Zhang C, Zhang M, Huang H, Huo H, et al. Identifying Cancer Targets Based on Machine Learning Methods via Chou’s 5-steps Rule and General Pseudo Components. Current Topics in Medicnal Chemistry. 2019.
- Liang Y, Zhang S. Identifying DNase I hypersensitive sites using multifeatures fusion and F-score features selection via Chou’s 5-steps rule. Biophys Chem. 2019; 253: 106227.
- Malebary SJ, Rehman MSU, Khan YD. iCrotoK-PseAAC: Identify lysine crotonylation sites by blending position relative statistical features according to the Chou’s 5-step rule. PLoS One. 2019; 14: e0223993.
- Nazari M Tahir, Tayari H, Chong KT. iN6-Methyl (5-step): Identifying RNA N6-methyladenosine sites using deep learning mode via Chou’s 5-step rules and Chou’s general PseKNC. Chemometrics and Intelligent Laboratory Systems (CHEMOLAB). 2019.
- Ning Q, Ma Z, Zhao X. dForml(KNN)-PseAAC: Detecting formylation sites from protein sequences using K-nearest neighbor algorithm via Chou’s 5-step rule and pseudo components. J Theor Biol. 2019; 470: 43-49.
- Salman M Khan, Iqbal N, Hussain T, Afzal S, Chou KC. A two-level computation model based on deep learning algorithm for identification of piRNA and their functions via Chou’s 5-steps rule. International Journal of Peptide Research and Therapeutics (IJPRT). 2019.
- Tahir M, Tayara H, Chong KT. iDNA6mA (5-step rule): Identification of DNA N6-methyladenine sites in the rice genome by intelligent computational model via Chou’s 5-step rule. CHEMOLAB. 2019; 189: 96-101.
- Vishnoi S, Garg P, Arora P. Physicochemical n-Grams Tool: A tool for protein physicochemical descriptor generation via Chou’s 5-steps rule. Chem Biol Drug Des. 2019.
- Wiktorowicz A, Wit A, Dziewierz A, Rzeszutko L, Dudek D, Kleczynski P. Calcium Pattern Assessment in Patients with Severe Aortic Stenosis Via the Chou’s 5-Steps Rule. Current Pharmaceutical Design. 2019.
- Yang L, Lv Y, Wang S, Zhang Q, Pan Y, Su D, et al. Identifying FL11 subtype by characterizing tumor immune microenvironment in prostate adenocarcinoma via Chou’s 5-steps rule. Genomics. 2019.
- Yang L, Lv Y, Wang S, Zhang Q, Pan Y, Su D, et al. Zuo, Identifying FL11 subtype by characterizing tumor immune microenvironment in prostate adenocarcinoma via Chou’s 5-steps rule. Genomics. 2019.
- Khan YD, Amin N, Hussain W, Rasool N, Khan SA, Chou KC. iProtease- PseAAC(2L): A two-layer predictor for identifying proteases and their types using Chou’s 5-step-rule and general PseAAC. Anal Biochem. 2020; 588: 113477.
- Xu Y, Ding J, Wu LY, Chou KC. iSNO-PseAAC: Predict cysteine S-nitrosylation sites in proteins by incorporating position specific amino acid propensity into pseudo amino acid composition PLoS ONE. 2013; 8 e55844.
- Xu Y, Shao XJ, Wu LY, Deng NY, Chou KC. iSNO-AAPair: incorporating amino acid pairwise coupling into PseAAC for predicting cysteine S-nitrosylation sites in proteins. PeerJ. 2013; 1: e171.
- Xu Y, Wen X, Shao XJ, Deng NY, Chou KC. iHyd-PseAAC: Predicting hydroxyproline and hydroxylysine in proteins by incorporating dipeptide position-specific propensity into pseudo amino acid composition. International Journal of Molecular Sciences (IJMS). 2014; 15: 7594-7610.
- Xu Y, Wen X, Wen LS, Wu LY, Deng NY, Chou KC. iNitro-Tyr: Prediction of nitrotyrosine sites in proteins with general pseudo amino acid composition. PLoS ONE. 2014; 9: e105018.
- Xu Y, Chou KC. Recent progress in predicting posttranslational modification sites in proteins. Curr Top Med Chem. 2016; 16: 591-603.
- Liu LM, Xu Y, Chou KC. iPGK-PseAAC: identify lysine phosphoglycerylation sites in proteins by incorporating four different tiers of amino acid pairwise coupling information into the general PseAAC. Med Chem. 2017; 13: 552- 559.
- Xu Y, Li C, Chou KC. iPreny-PseAAC: identify C-terminal cysteine prenylation sites in proteins by incorporating two tiers of sequence couplings into PseAAC. Med Chem. 2017; 13: 544-551.
- Cai L, Wan CL, He L, Jong S, Chou KC. Gestational influenza increases the risk of psychosis in adults. Medicinal Chemistry. 2015; 11: 676-682.
- Liu J, Song J, Wang MY, He L, Cai L, Chou KC. Association of EGF rs4444903 and XPD rs13181 polymorphisms with cutaneous melanoma in Caucasians. Medicinal Chemistry. 2015; 11: 551-559.
- Cai L, Yang YH, He L, Chou KC. Modulation of cytokine network in the comorbidity of schizophrenia and tuberculosis. Curr Top Med Chem. 2016; 16: 655-665.
- Cai L, Yuan W, Zhang Z, He L, Chou KC. In-depth comparison of somatic point mutation callers based on different tumor next-generation sequencing depth data Scientific Reports. 2016; 6: 36540.
- Zhu Y, Cong QW, Liu Y, Wan CL, Yu T, He G, et al. Antithrombin is an importantly inhibitory role against blood clots. Curr Top Med Chem. 2016; 16: 666-674.
- Zhang ZD, Liang K, Li K, Wang GQ, Zhang KW, Cai L, et al. Chlorella vulgaris induces apoptosis of human non-small cell lung carcinoma (NSCLC) cells. Med Chem. 2017; 13: 560-568.
- Cai L, Huang T, Su J, Zhang X, Chen W, Zhang F, et al. Implications of newly identified brain eQTL genes and their interactors in Schizophrenia. Molecular Therapy - Nucleic Acids. 2018; 12: 433-442.
- Niu B, Zhang M, Du P, Jiang L, R Qin, Su Q, et al. Chou, Small molecular floribundiquinone B derived from medicinal plants inhibits acetylcholinesterase activity. Oncotarget. 2017; 8: 57149-57162.
- Su Q, Lu W, Du D, Chen F, Niu B, Chou KC. Prediction of the aquatic toxicity of aromatic compounds to tetrahymena pyriformis through support vector regression. Oncotarget. 2017; 8: 49359-49369.
- Lu Y, Wang S, Wang J, Zhou G, Zhang Q, Zhou X, et al. An Epidemic Avian Influenza Prediction Model Based on Google Trends. Letters in Organic Chemistry. 2019; 16: 303-310.
- Niu B, Liang C, Lu Y, Zhao M, Chen Q, Zhang Y, et al. Glioma stages prediction based on machine learning algorithm combined with proteinprotein interaction networks. Genomics. 2019.
- Jia J, Liu Z, Xiao X, Liu B, Chou KC. Identification of protein-protein binding sites by incorporating the physicochemical properties and stationary wavelet transforms into pseudo amino acid composition (iPPBS-PseAAC). J Biomol Struct Dyn (JBSD). 2016; 34: 1946-1961.
- Jia J, Liu Z, Xiao X, Liu B, Chou KC. iSuc-PseOpt: Identifying lysine succinylation sites in proteins by incorporating sequence-coupling effects into pseudo components and optimizing imbalanced training dataset. Anal Biochem. 2016; 497: 48-56.
- Jia J, Liu Z, Xiao X, Liu B, Chou KC. pSuc-Lys: Predict lysine succinylation sites in proteins with PseAAC and ensemble random forest approach. Journal of Theoretical Biology. 2016; 394: 223-230.
- Jia J, Liu Z, Xiao X, Liu B, Chou KC. iCar-PseCp: identify carbonylation sites in proteins by Monto Carlo sampling and incorporating sequence coupled effects into general PseAAC. Oncotarget. 2016; 7: 34558-34570.
- Jia J, Liu Z, Xiao X, Liu B, Chou KC. iPPBS-Opt: A Sequence-Based Ensemble Classifier for Identifying Protein-Protein Binding Sites by Optimizing Imbalanced Training Datasets. Molecules. 2016; 21: E95.
- Jia J, Zhang L, Liu Z, Xiao X, Chou KC. pSumo-CD: Predicting sumoylation sites in proteins with covariance discriminant algorithm by incorporating sequence-coupled effects into general PseAAC. Bioinformatics. 2016; 32: 3133-3141.
- Liu Z, Xiao X, Yu DJ, Jia J, Qiu WR, Chou KC. pRNAm-PC: Predicting N-methyladenosine sites in RNA sequences via physical-chemical properties. Anal Biochem. 2016; 497: 60-67.
- Xiao X, Ye HX, Liu Z, Jia JH, Chou KC. iROS-gPseKNC: predicting replication origin sites in DNA by incorporating dinucleotide position-specific propensity into general pseudo nucleotide composition. Oncotarget. 2016; 7: 34180-34189.
- Qiu WR, Sun BQ, Xiao X, Xu ZC, Jia JH, Chou KC. iKcr-PseEns: Identify lysine crotonylation sites in histone proteins with pseudo components and ensemble classifier. Genomics. 2018; 110: 239-246.
- Jia J, Li X, Qiu W, Xiao X, Chou KC. iPPI-PseAAC(CGR): Identify proteinprotein interactions by incorporating chaos game representation into PseAAC. Journal of Theoretical Biology. 2019; 460: 195-203.
- Chen W, Ding H, Feng P, Lin H, Chou KC. iACP: a sequence-based tool for identifying anticancer peptides. Oncotarget. 2016; 7: 16895-16909.
- Chen W, Feng P, Ding H, Lin H, Chou KC. Using deformation energy to analyze nucleosome positioning in genomes. Genomics. 2016; 107: 69-75.
- Chen W, Tang H, Ye J, Lin H, Chou KC. iRNA-PseU: Identifying RNA pseudouridine sites Molecular Therapy - Nucleic Acids. 2016; 5: e332.
- Zhang CJ, Tang H, Li WC, Lin H, Chen W, Chou KC. iOri-Human: identify human origin of replication by incorporating dinucleotide physicochemical properties into pseudo nucleotide composition. Oncotarget. 2016; 7: 69783- 69793.
- Chen W, Feng P, Yang H, Ding H, Lin H, Chou KC. iRNA-AI: identifying the adenosine to inosine editing sites in RNA sequences. Oncotarget. 2017; 8: 4208-4217.
- Feng P, Ding H, Yang H, Chen W, Lin H, Chou KC. iRNA-PseColl: Identifying the occurrence sites of different RNA modifications by incorporating collective effects of nucleotides into PseKNC. Molecular Therapy - Nucleic Acids. 2017; 7: 155-163.
- Chen W, Ding H, Zhou X, Lin H, Chou KC. iRNA(m6A)-PseDNC: Identifying N6-methyladenosine sites using pseudo dinucleotide composition. Analytical Biochemistry. 2018; 561-562: 59-65.
- Chen W, Feng P, Yang H, Ding H, Lin H, Chou KC. iRNA-3typeA: identifying 3-types of modification at RNA’s adenosine sites. Molecular Therapy: Nucleic Acid. 2018; 11: 468-474.
- Su ZD, Huang Y, Zhang ZY, Zhao YW, Wang D, Chen W, et al. iLoc-lncRNA: predict the subcellular location of lncRNAs by incorporating octamer composition into general PseKNC. Bioinformatics. 2018; 34: 4196-4204.
- Yang H, Qiu WR, Liu G, Guo FB, Chen W, Chou KC, et al. iRSpot- Pse6NC: Identifying recombination spots in Saccharomyces cerevisiae by incorporating hexamer composition into general PseKNC International Journal of Biological Sciences. 2018; 14: 883-891.
- Feng P, Yang H, Ding H, Lin H, Chen W, Chou KC. iDNA6mA-PseKNC: Identifying DNA N(6)-methyladenosine sites by incorporating nucleotide physicochemical properties into PseKNC. Genomics. 2019; 111: 96-102.
- Du QS, Wang SQ, Xie NZ, Wang QY, Huang RB, Chou KC. 2L-PCA: A two-level principal component analyzer for quantitative drug design and its applications. Oncotarget. 2017; 8: 70564-70578.
- Liu B, Fang L, Long R, Lan X, Chou KC. iEnhancer-2L: a two-layer predictor for identifying enhancers and their strength by pseudo k-tuple nucleotide composition. Bioinformatics. 2016; 32: 362-369.
- Liu B, Long R, Chou KC. iDHS-EL: Identifying DNase I hypersensi-tivesites by fusing three different modes of pseudo nucleotide composition into an ensemble learning framework. Bioinformatics. 2016; 32: 2411-2418.
- Liu B, Wang S, Long R, Chou KC. iRSpot-EL: identify recombination spots with an ensemble learning approach. Bioinformatics. 2017; 33: 35-41.
- Liu B, Yang F, Chou KC. 2L-piRNA: A two-layer ensemble classifier for identifying piwi-interacting RNAs and their function. Molecular Therapy - Nucleic Acids. 2017; 7: 267-277.
- Liu B, Li K, Huang DS, Chou KC. iEnhancer-EL: Identifying enhancers and their strength with ensemble learning approach. Bioinformatics. 2018; 343: 835-3842.
- Liu B, Weng F, Huang DS, Chou KC. iRO-3wPseKNC: Identify DNA replication origins by three-window-based PseKNC. Bioinformatics. 2018; 34: 3086-3093.
- Liu B, Yang F, Huang DS, Chou KC. iPromoter-2L: a two-layer predictor for identifying promoters and their types by multi-window-based PseKNC. Bioinformatics. 2018; 34: 33-40.
- Qiu WR, Sun BQ, Xiao X, Xu ZC, Chou KC. iHyd-PseCp: Identify hydroxyproline and hydroxylysine in proteins by incorporating sequencecoupled effects into general PseAAC. Oncotarget. 2016; 7; 44310-44321.
- Qiu WR, Sun BQ, Xiao X, Xu ZC, Chou KC. iPTM-mLys: identifying multiple lysine PTM sites and their different types. Bioinformatics. 2016; 32: 3116- 3123.
- Qiu WR, Xiao X, Xu ZC, Chou KC. iPhos-PseEn: identifying phosphorylation sites in proteins by fusing different pseudo components into an ensemble classifier. Oncotarget. 2016; 7: 51270-51283.
- Qiu WR, Jiang SY, Sun BQ, Xiao X, Cheng X, Chou KC. iRNA-2methyl: identify RNA 2'-O-methylation sites by incorporating sequence-coupled effects into general PseKNC and ensemble classifier. Medicinal Chemistry. 2017; 13: 734-743.
- Qiu WR, Jiang SY, Xu ZC, Xiao X, Chou KC. iRNAm5C-PseDNC: identifying RNA 5-methylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition. Oncotarget. 2017; 8: 41178-41188.
- Qiu WR, Sun BQ, Xiao X, Xu D, Chou KC. iPhos-PseEvo: Identifying human phosphorylated proteins by incorporating evolutionary information into general PseAAC via grey system theory. Molecular Informatics. 2017; 36: UNSP 1600010.
- Zhai X, Chen M, Lu W. Accelerated search for perovskite materials with higher Curie temperature based on the machine learning methods. Computational Materials Science. 2018; 151: 41-48.
- Chou KC. Some remarks on protein attribute prediction and pseudo amino acid composition (50th Anniversary Year Review, 5-steps rule). Journal of Theoretical Biology. 2011; 273; 236-247.
- Chou KC. Impacts of pseudo amino acid components and 5-steps rule to proteomics and proteome analysis. Current Topics in Medicinak Chemistry (CTMC). 2019.
- Chou KC. Prediction of protein cellular attributes using pseudo amino acid composition. PROTEINS: Structure, Function, and Genetics. 2001; 43: 246- 255.
- Chou KC. Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics. 2005; 21: 10-19.
- Chou KC. Pseudo amino acid composition and its applications in bioinformatics, proteomics and system biology. Current Proteomics. 2009; 626: 2-274.
- Ding YS, Zhang TL. Using Chou’s pseudo amino acid composition to predict subcellular localization of apoptosis proteins: an approach with immune genetic algorithm-based ensemble classifier. Pattern Recognition Letters. 2008; 29: 1887-1892.
- Fang Y, Guo Y, Feng Y, Li M. Predicting DNA-binding proteins: approached from Chou’s pseudo amino acid composition and other specific sequence features. Amino Acids. 2008; 34: 103-109.
- Jiang X, Wei R, Zhang TL, Gu Q. Using the concept of Chou’s pseudo amino acid composition to predict apoptosis proteins subcellular location: an approach by approximate entropy. Protein & Peptide Letters. 2008; 15: 392-396.
- Jiang X, Wei R, Zhao Y, Zhang T. Using Chou’s pseudo amino acid composition based on approximate entropy and an ensemble of AdaBoost classifiers to predict protein subnuclear location. Amino Acids. 2008; 34: 669-675.
- Li FM, Li QZ. Predicting protein subcellular location using Chou’s pseudo amino acid composition and improved hybrid approach. Protein & Peptide Letters. 2008; 15: 612-616.
- Lin H. The modified Mahalanobis discriminant for predicting outer membrane proteins by using Chou’s pseudo amino acid composition. Journal of Theoretical Biology. 2008; 252: 350-356.
- Lin H, Ding H, Feng-Biao Guo FB, Zhang AY, Huang J. Predicting subcellular localization of mycobacterial proteins by using Chou’s pseudo amino acid composition. Protein & Peptide Letters. 2008; 15: 739-744.
- Nanni L, Lumini A. Genetic programming for creating Chou’s pseudo amino acid based features for submitochondria localization. Amino Acids. 2008; 34: 653-660.
- Zhang GY, Li HC, Gao JQ, Fang BS. Predicting lipase types by improved Chou’s pseudo amino acid composition. Protein & Peptide Letters. 2008; 15: 1132-1137.
- Zhang SW, Chen W, Yang F, Pan Q. Using Chou’s pseudo amino acid composition to predict protein quaternary structure: a sequence-segmented PseAAC approach. Amino Acids. 2008; 35: 591-598.
- Zhang SW, Zhang YL, Yang HF, Zhao CH, Pan Q. Using the concept of Chou’s pseudo amino acid composition to predict protein subcellular localization: an approach by incorporating evolutionary information and von Neumann entropies. Amino Acids. 2008; 34: 565-572.
- Chen C, Chen L, Zou X, Cai P. Prediction of protein secondary structure content by using the concept of Chou’s pseudo amino acid composition and support vector machine. Protein & Peptide Letters. 2009; 16: 27-31.
- Georgiou DN, Karakasidis TE, Nieto JJ, Torres A. Use of fuzzy clustering technique and matrices to classify amino acids and its impact to Chou’s pseudo amino acid composition. Journal of Theoretical Biology. 2009; 257: 17-26.
- Li ZC, Zhou XB, Dai Z, Zou XY. Prediction of protein structural classes by Chou’s pseudo amino acid composition: approached using continuous wavelet transform and principal component analysis. Amino Acids. 2009; 37: 415-425.
- Lin H, Wang H, Ding H, Chen YL, Li QZ. Prediction of Subcellular Localization of Apoptosis Protein Using Chou’s Pseudo Amino Acid Composition. Acta Biotheoretica. 2009; 57: 321-330.
- Qiu JD, Huang JH, Liang RP, Lu XQ. Prediction of G-protein-coupled receptor classes based on the concept of Chou’s pseudo amino acid composition: an approach from discrete wavelet transform. Analytical Biochemistry. 2009; 390: 68-73.
- Zeng YH, Guo YZ, Xiao RQ, Yang L, Yu LZ, Li ML. Using the augmented Chou’s pseudo amino acid composition for predicting protein submitochondria locations based on auto covariance approach. Journal of Theoretical Biology. 2009; 259: 366-372.
- Esmaeili M, Mohabatkar H, Mohsenzadeh S. Using the concept of Chou’s pseudo amino acid composition for risk type prediction of human papillomaviruses. Journal of Theoretical Biology. 2010; 263: 203-209.
- Gu Q, Ding YS, Zhang TL. Prediction of G-Protein-Coupled Receptor Classes in Low Homology Using Chou’s Pseudo Amino Acid Composition with Approximate Entropy and Hydrophobicity Patterns. Protein & Peptide Letters. 2010; 17: 559-567.
- Mohabatkar H. Prediction of cyclin proteins using Chou’s pseudo amino acid composition. Protein & Peptide Letters. 2010; 17: 1207-1214.
- Qiu JD, Huang JH, Shi SP, Liang RP. Using the concept of Chou’s pseudo amino acid composition to predict enzyme family classes: an approach with support vector machine based on discrete wavelet transform. Protein & Peptide Letters. 2010; 17: 715-722.
- Sahu SS, Panda G. A novel feature representation method based on Chou’s pseudo amino acid composition for protein structural class prediction. Computational Biology and Chemistry. 2010; 34: 320-327.
- Yu L, Guo Y, Li Y, Li G, Li M, Luo J, et al. SecretP: Identifying bacterial secreted proteins by fusing new features into Chou’s pseudo amino acid composition. Journal of Theoretical Biology. 2010; 267: 1-6.
- Guo J, Rao N, Liu G, Yang Y, Wang G. Predicting protein folding rates using the concept of Chou’s pseudo amino acid composition. Journal of Computational Chemistry. 2011; 32: 1612-1617.
- Lin J, Wang Y. Using a novel AdaBoost algorithm and Chou’s pseudo amino acid composition for predicting protein subcellular localization. Protein & Peptide Letters. 2011; 18: 1219-1225.
- Lin J, Wang Y, Xu X. A novel ensemble and composite approach for classifying proteins based on Chou’s pseudo amino acid composition. African Journal of Biotechnology. 2011; 10: 16963-16968.
- Mohabatkar H, Mohammad Beigi M, Esmaeili A. Prediction of GABA(A) receptor proteins using the concept of Chou’s pseudo amino acid composition and support vector machine. Journal of Theoretical Biology. 2011; 281: 18-23.
- Mohammad BM, Behjati M, Mohabatkar H. Prediction of metalloproteinase family based on the concept of Chou’s pseudo amino acid composition using a machine learning approach. Journal of Structural and Functional Genomics. 2011; 12: 191-197.
- Qiu JD, Suo SB, Sun XY, Shi SP, Liang RP. OligoPred: A web-server for predicting homo-oligomeric proteins by incorporating discrete wavelet transform into Chou’s pseudo amino acid composition. Journal of Molecular Graphics & Modelling. 2011; 30: 129-134.
- Zou D, He Z, He J, Xia Y. Supersecondary structure prediction using Chou’s pseudo amino acid composition. Journal of Computational Chemistry. 2011; 32: 271-278.
- Cao JZ, Liu WQ, Gu H. Predicting Viral Protein Subcellular Localization with Chou’s Pseudo Amino Acid Composition and Imbalance-Weighted Multi- Label K-Nearest Neighbor Algorithm. Protein and Peptide Letters. 2012; 19: 1163-1169.
- Chen C, Shen ZB, Zou XY. Dual-Layer Wavelet SVM for Predicting Protein Structural Class Via the General Form of Chou’s Pseudo Amino Acid Composition. Protein & Peptide Letters. 2012; 19: 422-429.
- Du P, Wang X, Xu C, Gao Y. PseAAC-Builder: A cross-platform standalone program for generating various special Chou’s pseudo amino acid compositions. Analytical Biochemistry. 2012; 425: 117-119.
- Fan GL, Li QZ. Predict mycobacterial proteins subcellular locations by incorporating pseudo-average chemical shift into the general form of Chou’s pseudo amino acid composition. Journal of Theoretical Biology. 2012; 304: 88-95.
- Fan GL, Li QZ. Predicting protein submitochondria locations by combining different descriptors into the general form of Chou’s pseudo amino acid composition. Amino Acids. 2012; 43: 545-555.
- Hayat M, Khan A. Discriminating Outer Membrane Proteins with Fuzzy K-Nearest Neighbor Algorithms Based on the General Form of Chou’s PseAAC. Protein & Peptide Letters. 2012; 19: 411-421.
- Li LQ, Zhang Y, Zou LY, Zhou Y, Zheng XQ. Prediction of Protein Subcellular Multi-Localization Based on the General form of Chou’s Pseudo Amino Acid Composition. Protein & Peptide Letters. 2012; 19: 375-387.
- Liao B, Xiang Q, Li D. Incorporating Secondary Features into the General form of Chou’s PseAAC for Predicting Protein Structural Class. Protein & Peptide Letters. 2012; 19: 1133-1138.
- Liu L, Hu XZ, Liu XX, Wang Y, Li SB. Predicting Protein Fold Types by the General Form of Chou’s Pseudo Amino Acid Composition: Approached from Optimal Feature Extractions. Protein & Peptide Letters. 2012; 19: 439-449.
- Mei S. Multi-kernel transfer learning based on Chou’s PseAAC formulation for protein submitochondria localization. Journal of Theoretical Biology. 2012; 293: 121-130.
- Mei S. Predicting plant protein subcellular multi-localization by Chou’s PseAAC formulation based multi-label homolog knowledge transfer learning. Journal of Theoretical Biology. 2012; 310: 80-87.
- Nanni L, Brahnam S, Lumini A. Wavelet images and Chou’s pseudo amino acid composition for protein classification. Amino Acids. 2012; 43: 657-665.
- Nanni L, Lumini A, Gupta D, Garg A. Identifying bacterial virulent proteins by fusing a set of classifiers based on variants of Chou’s pseudo amino acid composition and on evolutionary information. IEEE-ACM Transaction on Computational Biolology and Bioinformatics. 2012; 9: 467-475.
- Niu XH, Hu XH, Shi F, Xia JB. Predicting Protein Solubility by the General Form of Chou’s Pseudo Amino Acid Composition: Approached from Chaos Game Representation and Fractal Dimension. Protein & Peptide Letters. 2012; 19: 940-948.
- Qin YF, Wang CH, Yu XQ, Zhu J, Liu TG, Zheng XQ. Predicting Protein Structural Class by Incorporating Patterns of Over- Represented k-mers into the General form of Chou’s PseAAC. Protein & Peptide Letters. 2012; 19: 388-397.
- Ren LY, Zhang YS, Gutman I. Predicting the Classification of Transcription Factors by Incorporating their Binding Site Properties into a Novel Mode of Chou’s Pseudo Amino Acid Composition. Protein & Peptide Letters. 2012; 191: 170-1176.
- Sun XY, Shi SP, Qiu JD, Suo SB, Huang SY, Liang RP. Identifying protein quaternary structural attributes by incorporating physicochemical properties into the general form of Chou’s PseAAC via discrete wavelet transform. Molecular BioSystems. 2012; 8: 3178-3184.
- Zhao XW, Ma ZQ, Yin MH. Predicting protein-protein interactions by combing various sequence- derived features into the general form of Chou’s Pseudo amino acid composition. Protein & Peptide Letters. 2012; 19: 492-500.
- Zia-ur-Rehman, Khan A. Identifying GPCRs and their Types with Chou’s Pseudo Amino Acid Composition: An Approach from Multi-scale Energy Representation and Position Specific Scoring Matrix. Protein & Peptide Letters. 2012; 19: 890-903.
- Cao DS, Xu QS, Liang YZ. propy: a tool to generate various modes of Chou’s PseAAC. Bioinformatics. 2013; 29: 960-962.
- Chang TH, Wu LC, Lee TY, Chen SP, Huang HD, Horng JT. EuLoc: a webserver for accurately predict protein subcellular localization in eukaryotes by incorporating various features of sequence segments into the general form of Chou’s PseAAC. Journal of Computer-Aided Molecular Design. 2013; 27: 91-103.
- Chen YK, Li KB. Predicting membrane protein types by incorporating protein topology, domains, signal peptides, and physicochemical properties into the general form of Chou’s pseudo amino acid composition. Journal of Theoretical Biology. 2013; 318; 1-12.
- Fan GL, Li QZ, Zuo YC. Predicting acidic and alkaline enzymes by incorporating the average chemical shift and gene ontology informations into the general form of Chou’s PseAAC. Pocess Biochemistry. 2013; 48: 1048-1053.
- Fan GL, Li QZ. Discriminating bioluminescent proteins by incorporating average chemical shift and evolutionary information into the general form of Chou’s pseudo amino acid composition. Journal of Theoretical Biology. 2013; 334: 45-51.
- Georgiou DN, Karakasidis TE, Megaritis AC. A short survey on genetic sequences, Chou’s pseudo amino acid composition and its combination with fuzzy set theory. The Open Bioinformatics Journal. 2013; 7: 41-48.
- Gupta MK, Niyogi R, Misra M. An alignment-free method to find similarity among protein sequences via the general form of Chou’s pseudo amino acid composition. SAR QSAR Environ Res. 2013; 24: 597-609.
- Huang C, Yuan J. Using radial basis function on the general form of Chou’s pseudo amino acid composition and PSSM to predict subcellular locations of proteins with both single and multiple sites. Biosystems. 2013; 113: 50-57.
- Huang C, Yuan JQ. A multilabel model based on Chou’s pseudo amino acid composition for identifying membrane proteins with both single and multiple functional types. J Membr Biol. 2013; 246: 327-334.
- Huang C, Yuan JQ. Predicting protein subchloroplast locations with both single and multiple sites https://www.ncbi.nlm.nih.gov/pubmed/22894156 three different modes of Chou’s pseudo amino acid compositions. Journal of Theoretical Biology. 2013; 335: 205-212.
- Khosravian M, Faramarzi FK, Beigi MM, Behbahani M, Mohabatkar H. Predicting Antibacterial Peptides by the Concept of Chou’s Pseudo amino Acid Composition and Machine Learning Methods. Protein & Peptide Letters. 2013; 20: 180-186.
- Lin H, Ding C, Yuan LF, Chen W, Ding H, Li ZQ, et al. Predicting subchloroplast locations of proteins based on the general form of Chou’s pseudo amino acid composition: Approached from optimal tripeptide composition. International Journal of Biomethmatics. 2013; 6: 1350003.
- Liu B, Wang X, Zou Q, Dong Q, Chen Q. Protein remote homology detection by combining Chou’s pseudo amino acid composition and profile-based protein representation. Molecular Informatics. 2013; 32: 775-782.
- Mohabatkar H, Beigi MM, Abdolahi K, Mohsenzadeh S. Prediction of Allergenic Proteins by Means of the Concept of Chou’s Pseudo Amino Acid Composition and a Machine Learning Approach. Medicinal Chemistry. 2013; 9: 133-137.
- Pacharawongsakda E, Theeramunkong T. Predict Subcellular Locations of Singleplex and Multiplex Proteins by Semi-Supervised Learning and Dimension-Reducing General Mode of Chou’s PseAAC. IEEE Transactions on Nanobioscience. 2013; 12: 311-320.
- Qin YF, Zheng L, Huang J. Locating apoptosis proteins by incorporating the signal peptide cleavage sites into the general form of Chou’s Pseudo amino acid composition. International Journal of Quantum Chemistry. 2013; 113: 1660-1667.
- Sarangi AN, Lohani M, Aggarwal R. Prediction of Essential Proteins in Prokaryotes by Incorporating Various Physico-chemical Features into the General form of Chou’s Pseudo Amino Acid Composition. Protein Pept Lett. 2013; 20: 781-795.
- Wan S, Mak MW, Kung SY. GOASVM: A subcellular location predictor by incorporating term-frequency gene ontology into the general form of Chou’s pseudo amino acid composition. Journal of Theoretical Biology. 2013; 323: 40-48.
- Wang X, Li GZ, Lu WC. Virus-ECC-mPLoc: a multi-label predictor for predicting the subcellular localization of virus proteins with both single and multiple sites based on a general form of Chou’s pseudo amino acid composition. Protein & Peptide Letters. 2013; 20: 309-317.
- Xiaohui N, Nana L, Jingbo X, Dingyan C, Yuehua P, Yang X, et al. Using the concept of Chou’s pseudo amino acid composition to predict protein solubility: An approach with entropies in information theory. Journal of Theoretical Biology. 2013; 332: 211-217.
- Xie HL, Fu L, Nie XD. Using ensemble SVM to identify human GPCRs N-linked glycosylation sites based on the general form of Chou’s PseAAC. Protein Eng Des Sel. 2013; 26: 735-742.
- Du P, Gu S, Jiao Y. PseAAC-General: Fast building various modes of general form of Chou’s pseudo amino acid composition for large-scale protein datasets. International Journal of Molecular Sciences. 2014; 15: 3495-3506.
- Hajisharifi Z, Piryaiee M, Mohammad Beigi M, Behbahani M, Mohabatkar H. Predicting anticancer peptides with Chou’s pseudo amino acid composition and investigating their mutagenicity via Ames test. Journal of Theoretical Biology. 2014; 341: 34-40.
- Han GS, Yu ZG, Anh V. A two-stage SVM method to predict membrane protein types by incorporating amino acid classifications and physicochemical properties into a general form of Chou’s PseAAC. J Theor Biol. 2014; 344: 31-9.
- Jia C, Lin X, Wang Z. Prediction of Protein S-Nitrosylation Sites Based on Adapted Normal Distribution Bi-Profile Bayes and Chou’s Pseudo Amino Acid Composition. Int J Mol Sci. 2014; 15: 10410-104123.
- Kong L, Zhang L, Lv J. Accurate prediction of protein structural classes by incorporating predicted secondary structure information into the general form of Chou’s pseudo amino acid composition. J Theor Biol. 2014; 344; 12-18.
- Li L, Yu S, Xiao W, Li Y, Li M, Huang L, et al. Prediction of bacterial protein subcellular localization by incorporating various features into Chou’s PseAAC and a backward feature selection approach. Biochimie. 2014; 104: 100-107.
- Nanni L, Brahnam S, Lumini A. Prediction of protein structure classes by incorporating different protein descriptors into general Chou’s pseudo amino acid composition. J Theor Biol. 2014; 360: 109-116.
- Zhang J, Sun P, Zhao X, Ma Z. PECM: Prediction of extracellular matrix proteins using the concept of Chou’s pseudo amino acid composition. Journal of Theoretical Biology. 2014; 363; 412-418.
- Zhang J, Zhao X, Sun P, Ma Z. PSNO: Predicting Cysteine S-Nitrosylation Sites by Incorporating Various Sequence-Derived Features into the General Form of Chou’s PseAAC. Int J Mol Sci. 2014; 15: 11204-11219.
- Zhang L, Zhao X, Kong L. Predict protein structural class for low-similarity sequences by evolutionary difference information into the general form of Chou’s pseudo amino acid composition. J Theor Biol. 2014; 355: 105-110.
- Zuo YC, Peng Y, Liu L, Chen W, Yang L, Fan GL. Predicting peroxidase subcellular location by hybridizing different descriptors of Chou’s pseudo amino acid patterns. Anal Biochem. 2014; 458: 14-19.
- Ali F, Hayat M. Classification of membrane protein types using Voting Feature Interval in combination with Chou’s Pseudo Amino Acid Composition. J Theor Biol. 2015; 384: 78-83.
- Fan GL, Zhang XY, Liu YL, Nang Y, Wang H. DSPMP: Discriminating secretory proteins of malaria parasite by hybridizing different descriptors of Chou’s pseudo amino acid patterns. J Comput Chem. 2015; 36: 2317-2327.
- Huang C, Yuan JQ. Simultaneously Identify Three Different Attributes of Proteins by Fusing their Three Different Modes of Chou’s Pseudo Amino Acid Compositions. Protein Pept Lett. 2015; 22: 547-556.
- Khan ZU, Hayat M, Khan MA. Discrimination of acidic and alkaline enzyme using Chou’s pseudo amino acid composition in conjunction with probabilistic neural network model. J Theor Biol. 2015; 365: 197-203.
- Kumar R, Srivastava A, Kumari B, Kumar M. Prediction of beta-lactamase and its class by Chou’s pseudo amino acid composition and support vector machine. J Theor Biol. 2015; 365: 96-103.
- Liu B, Xu J, Fan S, Xu R, Jiyun Zhou J, Wang X. PseDNA-Pro: DNA-binding protein identification by combining Chou’s PseAAC and physicochemical distance transformation. Molecular Informatics. 2015; 34: 8-17.
- Mandal M, Mukhopadhyay A, Maulik U. Prediction of protein subcellular localization by incorporating multiobjective PSO-based feature subset selection into the general form of Chou’s PseAAC. Med Biol Eng Comput. 2015; 53: 331-344.
- Sanchez V, Peinado AM, Perez-Cordoba JL, Gomez AM. A new signal characterization and signal-based Chou’s PseAAC representation of protein sequences. J Bioinform Comput Biol. 2015; 13: 1550024.
- Wang X, Zhang W, Zhang Q, Li GZ. MultiP-SChlo: multi-label protein subchloroplast localization prediction with Chou’s pseudo amino acid composition and a novel multi-label classifier. Bioinformatics. 2015; 31: 2639-2645.
- Jiao YS, Du PF. Prediction of Golgi-resident protein types using general form of Chou’s pseudo amino acid compositions: Approaches with minimal redundancy maximal relevance feature selection. J Theor Biol. 2016; 402: 38-44.
- Kabir M, Hayat M. iRSpot-GAEnsC: identifing recombination spots via ensemble classifier and extending the concept of Chou’s PseAAC to formulate DNA samples. Molecular Genetics and Genomics. 2016; 291: 285-296.
- Tahir M, Hayat M. iNuc-STNC: a sequence-based predictor for identification of nucleosome positioning in genomes by extending the concept of SAAC and Chou’s PseAAC. Mol Biosyst. 2016; 12: 2587-2593.
- Tang H, Chen W, Lin H. Identification of immunoglobulins using Chou’s pseudo amino acid composition with feature selection technique. Mol Biosyst. 2016; 12: 1269-1275.
- Zou HL, Xiao X. Predicting the Functional Types of Singleplex and Multiplex Eukaryotic Membrane Proteins via Different Models of Chou’s Pseudo Amino Acid Compositions. J Membr Biol. 2016; 249: 23-29.
- Huo H, Li T, Wang S, Lv Y, Zuo Y, Yang L. Prediction of presynaptic and postsynaptic neurotoxins by combining various Chou’s pseudo components. Sci Rep. 2017; 7: 5827.
- Ju Z, He JJ. Prediction of lysine propionylation sites using biased SVM and incorporating four different sequence features into Chou’s PseAAC. J Mol Graph Model. 2017; 76: 356-363.
- Rahimi M, Bakhtiarizadeh MR, Mohammadi-Sangcheshmeh A. OOgenesis_ Pred: A sequence-based method for predicting oogenesis proteins by six different modes of Chou’s pseudo amino acid composition. J Theor Biol. 2017; 414; 128-136.
- Tripathi P, Pandey PN. A novel alignment-free method to classify protein folding types by combining spectral graph clustering with Chou’s pseudo amino acid composition. J Theor Biol. 2017; 424: 49-54.
- Yu B, Li S, Qiu WY, Chen C, Chen RX, Wang L, et al. Accurate prediction of subcellular location of apoptosis proteins combining Chou’s PseAAC and PsePSSM based on wavelet denoising. Oncotarget. 2017; 8: 107640- 107665.
- Yu B, Lou L, Li S, Zhang Y, Qiu W, Wu X, et al. Tian, Prediction of protein structural class for low-similarity sequences using Chou’s pseudo amino acid composition and wavelet denoising. J Mol Graph Model. 2017; 76: 260-273.
- Ahmad J, Hayat M. MFSC: Multi-voting based Feature Selection for Classification of Golgi Proteins by Adopting the General form of Chou’s PseAAC components. J Theor Biol. 2018; 463: 99-109.
- Akbar S, Hayat M. iMethyl-STTNC: Identification of N(6)-methyladenosine sites by extending the Idea of SAAC into Chou’s PseAAC to formulate RNA sequences. J Theor Biol. 2018; 455: 205-211.
- Al Maruf MA, Shatabda S. iRSpot-SF: Prediction of recombination hotspots by incorporating sequence based features into Chou’s Pseudo components. Genomics. 2019.
- Arif M, Hayat M, Jan Z. iMem-2LSAAC: A two-level model for discrimination of membrane proteins and their types by extending the notion of SAAC into Chou’s pseudo amino acid composition. J Theor Biol. 2018; 442: 11-21.
- Contreras-Torres E. Predicting structural classes of proteins by incorporating their global and local physicochemical and conformational properties into general Chou’s PseAAC. J Theor Biol. 2018; 454: 139-145.
- Cui X, Yu Z, Yu B, Wang M, Tian B, Ma Q. UbiSitePred: A novel method for improving the accuracy of ubiquitination sites prediction by using LASSO to select the optimal Chou’s pseudo components. Chemometrics and Intelligent Laboratory Systems (CHEMOLAB). 2018.
- Fu X, Zhu W, Liso B, Cai L, Peng L, Yang J. Improved DNA-binding protein identification by incorporating evolutionary information into the Chou’s PseAAC. IEEE Access. 2018.
- Javed F, Hayat M. Predicting subcellular localizations of multi-label proteins by incorporating the sequence features into Chou’s PseAAC. Genomics. 2018.
- Mei J, Zhao J. Prediction of HIV-1 and HIV-2 proteins by using Chou’s pseudo amino acid compositions and different classifiers. Sci Rep. 2018; 8: 2359.
- Mousavizadegan M, Mohabatkar H. Computational prediction of antifungal peptides via Chou’s PseAAC and SVM. J Bioinform Comput Biol. 2018; 1850016.
- Qiu W, Li S, Cui X, Yu Z, Wang M, Du J, et al. Predicting protein submitochondrial locations by incorporating the pseudo-position specific scoring matrix into the general Chou’s pseudo-amino acid composition. J Theor Biol. 2018; 450: 86-103.
- Zhang L, Kong L. iRSpot-ADPM: Identify recombination spots by incorporating the associated dinucleotide product model into Chou’s pseudo components. J Theor Biol. 2018; 441: 1-8.
- Zhang S, Liang Y. Predicting apoptosis protein subcellular localization by integrating auto-cross correlation and PSSM into Chou’s PseAAC. J Theor Biol. 2018; 457:163-169.
- Zhang S, Yang K, Lei Y, Song K. iRSpot-DTS: Predict recombination spots by incorporating the dinucleotide-based spare-cross covariance information into Chou’s pseudo components. Genomics. 2018; 11: 457-464.
- Zhao W, Wang L, Zhang TX, Zhao ZN, Du PF. A brief review on software tools in generating Chou’s pseudo-factor representations for all types of biological sequences. Protein Pept Lett. 2018; 25: 822-829.
- Ahmad J, Hayat M. MFSC: Multi-voting based feature selection for classification of Golgi proteins by adopting the general form of Chou’s PseAAC components. J Theor Biol. 2019; 463: 99-109.
- Al Maruf MA, Shatabda S. iRSpot-SF: Prediction of recombination hotspots by incorporating sequence based features into Chou’s Pseudo components. Genomics. 2019; 111; 966-972.
- Butt AH, Rasool N, Khan YD. Prediction of antioxidant proteins by incorporating statistical moments based features into Chou’s PseAAC. Journal of Theoretical Biology. 2019; 473; 1-8.
- Javed F, Hayat M. Predicting subcellular localization of multi-label proteins by incorporating the sequence features into Chou’s PseAAC. Genomics. 2019; 111: 1325-1332.
- Pan Y, Wang S, Zhang Q, Lu Q, Su D, Zuo Y, Yang L. Analysis and prediction of animal toxins by various Chou’s pseudo components and reduced amino acid compositions. J Theor Biol. 2019; 462: 221-229.
- Tahir M, Hayat M, Khan SA. iNuc-ext-PseTNC: an efficient ensemble model for identification of nucleosome positioning by extending the concept of Chou’s PseAAC to pseudo-tri-nucleotide composition. Mol Genet Genomics. 2019; 294: 199-210.
- Tahir M, Tayara H, Chong KT. iRNA-PseKNC(2methyl): Identify RNA 2’-O-methylation sites by convolution neural network and Chou’s pseudo components. J Theor Biol. 2019; 465; 1-6.
- Tian B, Wu X, Chen C, Qiu W, Ma Q, Yu B. Predicting protein-protein interactions by fusing various Chou’s pseudo components and using wavelet denoising approach. J Theor Biol. 2019; 462: 329-346.
- Zhang L, Kong L. iRSpot-PDI: Identification of recombination spots by incorporating dinucleotide property diversity information into Chou’s pseudo components. Genomics. 2019; 111: 457-464.
- Zhang S, Yang K, Lei Y, Song K. iRSpot-DTS: Predict recombination spots by incorporating the dinucleotide-based spare-cross covariance information into Chou’s pseudo components. Genomics. 2019; 111: 1760-1770.
- Chou KC. Two kinds of metrics for computational biology. Genomics. Dihub. 2019.
- Chou KC. Proposing pseudo amino acid components is an important milestone for proteome and genome analyses. International Journal for Peptide Research and Therapeutics (IJPRT). 2019.
- Chou KC. An insightful recollection for predicting protein subcellular locations in multi-label systems. Genomics. Dihub. 2019.
- Chou KC. Progresses in predicting post-translational modification. International Journal of Peptide Research and Therapeutics (IJPRT). 2019.
- Chou KC. Recent Progresses in Predicting Protein Subcellular Localization with Artificial Intelligence (AI) Tools Developed Via the 5-Steps Rule. Japanese Journal of Gastroenterology and Hepatology. 2019.
- Chou KC. An insightful recollection since the distorted key theory was born about 23 years ago. Genomics. Dihub. 2019.
- Chou KC. Artificial intelligence (AI) tools constructed via the 5-steps rule for predicting post-translational modifications. Trends in Artificial Inttelengence (TIA). 2019; 3: 60-74.
- Chou KC. Distorted Key Theory and Its Implication for Drug Development. Current Genomics. 2020.
- Chou KC. An insightful recollection since the birth of Gordon Life Science Institute about 17 years ago. Advancement in Scientific and Engineering Research. 2019; 4: 31-36.
- Chou KC. Gordon Life Science Institute: Its philosophy, achievements, and perspective. Annals of Cancer Therapy and Pharmacology. 2019; 2: 001-26.
