
Review Article
Ann Agric Crop Sci. 2021; 6(1): 1070.
Role of ABA in Overcoming Environmental Stress: Sensing, Signaling and Crosstalk
Benderradji L¹, Saibi W² and Brini F²*
¹University of M’Sila-Algeria, Biodiversity and Biotechnology Techniques for Plant Resources Valorization Laboratory, Algeria
²University of Sfax, Biotechnology and Plant Improvement Laboratory, Tunisia
*Corresponding author: Faical Brini, University of Sfax, Biotechnology and Plant Improvement Laboratory, Centre of Biotechnology of Sfax, PO-1177, 3018, Sfax-Tunisia
Received: January 18, 2021; Accepted: February 20, 2021; Published: February 27, 2021
Abstract
The Abscisic Acid (ABA) is an isoprenoid phytohormone, regulating various physiological processes ranging from stomatal opening to protein storage. Moreover, it provides adaptation to drought, salt and cold stresses acts also as a signaling mediator during the plant’s adaptive response to environmental conditions. In addition, numbers of transcription factors are involved in regulating the expression of ABA responsive genes by interacting with their respective cis-acting elements. ABA signal transduction initiates signal perception by ABA receptors and transfer via downstream proteins, including protein kinases and phosphatases. Hence, for improvement in plants-stress-tolerance capacity, it is necessary to understand the mechanism behind it. On this ground, this article lightens the importance and also the role of ABA signaling with regard to various stresses as well as regulation of ABA biosynthetic pathway along with the transcription factors for stress tolerance.
Keywords: Abiotic stress; Abscisic acid; ABA signaling; Gene expression; Phytohormone
Abbreviations
ABA: Abscisic Acid; LRs: Lateral Roots; MAPK: Mitogen- Activated Protein Kinases; NCED: 9-cis-Epoxy Carotenoid Dioxygenase; PP2C: Protein Phosphatase 2C; ROS: Reactive Oxygen Species; Snrk2: The Sucrose Non-Fermenting 1 (SNF1)-Related Protein Kinase 2; ZEP: Zeaxanthin Oxidase
Introduction
According to various studies, abiotic stresses trigger many physiological, biochemical, and molecular responses that influence various cellular processes in plants [1]. To combat various environmental stresses novel and dynamic approaches should be devised and phytohormone engineering could be a method of choice to improve the productivity. Phytohormones are the key regulators of plant growth and development as well as mediators of environmental stress responses [2]. Among various phytohormones, Abscisic Acid (ABA), which is the central regulator of abiotic stress resistance in plants and coordinates an array of functions [3,4], enabling plants to cope with different stresses? In the plant, when environmental conditions are harsh, the level of ABA increases via ABA biosynthesis. The increased ABA binds to its receptor to initiate signal transduction leading to cellular responses to stresses [5]; therefore, ABA is also called a stress hormone [6]. ABA is significantly increased under drought or salinity stress conditions, stimulating stomatal closure, change in gene expression, and adaptive physiological responses [7-10]. ABA also plays an important role in many cellular processes including seed development, dormancy, germination, vegetative growth [11,12] and modulation of root architecture [13]. Since the discovery of ABA, several efforts have been devoted to understanding how ABA is synthesized under stress conditions. For ABA perception and signaling, two breaks through were achieved in 2009 by the discoveries of the soluble ABA receptor proteins and the core signaling complexes that perceive ABA and transmit cues to subsequent molecular events [14-16], which adds the essences of more research with new ideas in ABA signaling. Recently, Park et al., [17] engineered ABA receptor using agrochemicals, which provide new possibilities for the better development of the crop. Thus, a better understanding of ABA regulatory mechanisms will contribute to engineered stress tolerant crop plants, which is one of the primary goals of plant molecular biologists. This review focuses on the recent development of the role of ABA in understanding cellular networks of biotechnological relevance in abiotic stress responses in plants.
Abscisic Acid Biosynthesis
Abscisic acid is a type of metabolite known as isoprenoids, or terpenoids. Isopentenyl (IDP) is a five-Carbon (C5) precursor molecule from which it is derived. (Table 1) illustrates some of the abscisic acid systematic and also their chemical properties. Various enzymes are involved which utilizes β-carotene to synthesize ABA. Conversion of β-carotene to ABA is mediated via number of enzyme-catalyzed steps (Figure 1). The first step of ABA biosynthesis pathway is the conversion of zeaxanthin and antheraxanthin to all trans-violaxanthin, which will be catalyzed by Zeaxanthin Epoxidase (ZEP) in the plastid. The ZEP was identified by Marin et al., [18]. In this reaction, antheraxanthin is the intermediate formed. After that, all-trans-violaxanthin converted to 9-cis-violaxanthin or 9-cisneoxanthin. The enzyme involved in this reaction is unknown [19]. After that, oxidative cleavage of 9-cis-violaxanthin and 9-cisneoxanthin, catalyzed by the enzyme called 9-cis-Epoxy Carotenoid Dioxygenase (NCED), which yields a C15 intermediate product called xanthoxin and C25 metabolite [20]. ZmNCED gene was first isolated using the maize viviparous 14 mutant, and NCED is the key enzyme in ABA biosynthesis [21]. Then, the product xanthoxin is exported to the cytosol [22] where xanthoxin is converted to ABA. In this step, xanthoxin is converted in to ABA by two enzymatic reactions. First of all, xanthoxin is converted to an ABA aldehyde by an enzyme called Short-Chain Alcohol Dehydrogenase/Reductase (SDR) encoded by the AtABA2 gene in Arabidopsis thaliana [23]. The next and final step of ABA biosynthesis is oxidation of the abscisic aldehyde to ABA, catalyzed by the Abscisic Aldehyde Oxidase (AAO) (Figure 1). Till date, the genes for those enzymes have been classified in Arabidopsis alone.
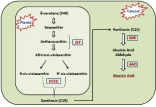
Figure 1: Schematic representation of biosynthesis of ABA in plants. ABA
is derived from β-Carotene (C40) through an oxidative cleavage reaction
in plastids. The first step of ABA biosynthesis pathway is the conversion
of zeaxanthin and antheraxanthin to all trans-violaxanthin, which will
be catalyzed by Zeaxanthin Epoxidase (ZEP). Antheraxanthin is the
intermediate product. All-trans-violaxanthin is converted to 9-cis-violaxanthin
or 9’-cis-Neoxanthin by the 9-cis-Epoxy Carotenoid Dioxygenase (NCED),
which yields a C15 intermediate product called xanthoxin. Then, the product
xanthoxin is exported to the cytosol where xanthoxin is converted to ABA.
Xanthoxin is then converted into ABA by two enzymatic reactions. Finally,
xanthoxinis converted to an ABA aldehyde by the enzyme, Short-Chain
Alcohol Dehydrogenase/Reductase (SDR), and then oxidation of the abscisic
aldehyde to ABA is catalyzed by the Abscisic Aldehyde Oxidase (AAO).
Abscisic acid
Names
Systematic IUPAC name
(2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid
Other names
(2Z,4E)-(S)-5-(1-Hydroxy-2,6,6-trimethyl-4-oxo-2-cyclohexen-1-yl)-3-methyl-2,4-pentanedienoic acid; Dormic acid
Identifiers
CAS Number
21293-29-8
3D model
B00898
Abbreviation
ABA
Beilstein Reference
2698956
EC Number
244-319-5
Properties
Chemical formula
C15H20O4
Molar mass
264.321 g·mol−1
Melting point
163ºC (325ºF; 436K)
Acidity (pKa)
4.868
Basicity (pKb)
9.129
Table 1: Illustration of the ABA Identity data.
Role of ABA in Plants
ABA is one of the most important hormonal mediators of abiotic and biotic signals, and as such, ABA acts as the interpreter of the environment, controlling key physiological processes such as modulation of root architecture, germination, stomatal movements, dormancy and plant-microbe interactions. Consequently, there is a growing awareness of the role of ABA in environmentally-regulated plant developmental processes.
Seed dormancy and germination
Seed is a crucial organ in higher plants and the transition to seed dormancy and germination signifies a key stage in the plant life cycle. The ABA plays a central role in the induction and maintenance of seed dormancy. It also inhibits the transition from embryonic to germination growth [7]. There are number of studies showing the number of genes involved in aforementioned processes. For example, the genes ABI1 and ABI2 hinder number of ABA responses which involves inhibition of germination process of seed and growth of seedling and support stomatal closure and other genes such as ABI3, ABI4, and ABI5 only display ABA insensitivity during seed germination and premature seedling development [24].
Root architecture
In roots, three main factors control the architecture of roots, i.e., lateral root (positions of branch roots), the angle form with parent root, and root length. Root structure is determined through interactions between the roots and its environment during their lifetime [13]. One of the primary functions of ABA is altering root architecture, and thus changing the pattern of growth and quiescence in plant roots. For the root system, the major abiotic stress occurs when there is a scarcity of water, or the availability of water is inconsistent. Under this scenario, the level of ABA is altered in response to the intensity of water stress. Subsequently, changes in the root environment will have both local and systemic effects on ABA-mediated responses [25].
ABA regulates root meristem function: ABA has been shown to modulate the major control points of root growth: cell division and cell elongation. Cell division in the root is restricted to meristems, regions that contain an organizing center (the Quiescent Center) surrounded by stem cells (initials) that divide to produce cells that differentiate into the various tissues of the root, as well as producing stem cells, that continue dividing and allow for indeterminate root growth [26,27]. Although high levels of ABA have long been known to inhibit root growth in well-watered conditions, analysis of the Medicago truncatula latd mutant indicated a positive role for ABA in the establishment or maintenance of root meristem function [28]. Subsequently, ABA was shown to regulate quiescence of the organizing center and inhibit differentiation in the Arabidopsis thaliana root meristem, thus maintaining the stem cell population [29]. Since the stem cells are the population of dividing cells, ABA regulates root growth by directly regulating the population of dividing cells in the root tip.
Modulation of root cell length by ABA: ABA has been shown to also control root elongation by regulating cell length, which has a profound effect on root growth. Although continued cell division is essential for continued root growth, most of the very rapid root growth is driven by cell elongation of newly formed, differentiating root cells. In M. truncatula, ABA plays a similar role, but in the absence of water stress, regulating cell length, and thus root elongation, by modulating levels of ROS via controlling expression of RESPIRATORY BURST OXIDASE HOMOLOG (RBOH) genes, encoding superoxide-generating NADPH oxidase enzymes [30]. This ability of ABA to modulate root length either up or down depending on the environmental conditions or genotypic background provides a nuanced way to fine-tune growth of the root system to provide the necessary plasticity to survive changing situations.
Control of root length by ABA: Although ABA signaling occurs in all root cell layers, analysis from José Dinneny’s lab demonstrates that ABA signaling is required within the endodermis to regulate root growth [31,32]. By expressing the dominant aba insensitive 1-1 (abi1-1) allele under different promoters to disrupt ABA signaling in specific root cell layers, they were able to determine that ABA signaling is required specifically within the endodermis for control of root elongation in response to salt both in primary roots and in lateral roots [31,32]. This finding is consistent with the observation by Ubeda-Thomas and colleagues that elongation of the endodermis is sufficient to control root elongation [33], and elevates the previously overlooked endodermis to a master role in the control of root growth. Within the context of salt stress, ABA can both stimulate growth of the primary root and inhibit growth of lateral roots by signaling within the endodermal layer [31,32]. This careful modulation of root growth by ABA in response to an environmental stimulus reveals a significant role for this hormone in shaping the overall structure of the root system. The control of the transition from proliferation to differentiation is a convenient way to coordinate cell division and cell length and is a likely target of ABA signaling. This transition is controlled by ROS molecules as well as by hormonal and genetic factors (discussed below). The regulation of ROS levels is a common signaling target for ABA in several plant species and in multiple tissues [30,34], and ROS molecules can function as a secondary messenger for both division and elongation, regulate gene expression and act directly on the cell wall, thus making it an attractive candidate for a mediator of cell length [35,36]. In Arabidopsis, ROS species control the transition between cell division and differentiation in the root apical meristem, thus modulating root length and providing an explicit link between cell division and cell length [37].
Lateral root development: While root elongation is an important component of root architecture, the huge variety in the architecture of the roots of different plants is due largely to the formation of lateral roots both on the primary root as well as on lateral roots. By controlling the number and position of these lateral roots, the architecture of an individual plant can be sculpted to accommodate existing conditions and neighbors. The role of ABA signaling in LR development is intricately connected to environmental response, and, surprisingly, appears to regulate different aspects of Lateral Roots (LRs) development and have different effects in different plant species [13]. ABA appears to be largely limited to regulating the activation of the meristem once the LR has emerged from the parent root. In Arabidopsis, high levels of nitrate block the activation of the newly formed LR meristems, resulting in arrested, but emerged, LRs [38]. ABA signaling mutants have increased stimulation of LR elongation by nitrate, indicating that ABA signaling plays an inhibitory role in LR elongation in Arabidopsis [39]. When the root system encounters localized nitrate (i.e., a patch of nitrate), LR elongation is stimulated, in a process that is inhibited by ABA signaling. In both cases, ABA is involved in mediating an environmental signal (either high nitrate or localized nitrate) and affects later stages of LR development, meristem activation and LR elongation [13].
Leaf senescence
Leaf senescence is an essential part of the final stage of plant development, which is regulated by a complex range of endogenous and environmental factors [40]. Among these factors, ABA has been put forward to affect leaf senescence significantly [41]. Foliar spraying with ABA has been revealed to promote leaf senescence in rice [42] and similar results were shown in maize by He and Jin [43]. An increase in endogenous ABA appears to coincide with senescence of leaves [44], which supports the notion that ABA functions in the leaf senescence. Likewise, ABA can promote senescence and induce expression of several specific senescence-associated genes [3,41]. Many studies also reported that, in Arabidopsis mutants with deficiencies in ABA biosynthesis or signaling, exhibit altered or delayed senescence [45]. Lee et al., [46] identified a Receptor Kinase (RPK1) that mediates age-and ABA-induced senescence in old leaves. Recently, Takasaki et al., [47] reported that SNAC-A, a subfamily of stress responsive NAC transcription factors, plays critical roles in ABA-induced leaf senescence signaling in Arabidopsis [47]. Similarly, a cotton NAPlike transcription factor (NAC like, activated by apetala3/pistillata, GhNAP) has been shown to regulate leaf senescence through ABAmediated pathways [48]. All these studies point to the significance of NAC-type transcription factors in ABA-mediated leaf senescence.
Stomata regulation
Stomata are small pores on the leaf surfaces formed by guard cells, which control plant gas exchange processes. Light usally stimulates stomatal opening whereas ABA and elevated CO2 levels promote partial or complete closure of stomata [10,49,50]. In drought conditions, ABA alteration of guard cell ion transport, which promotes stomatal closure and prevents stomatal opening, reducing water loss [10,51]. The ABA-Activated Protein Kinase (AAPK) is a guard cell-specific protein kinase whose catalytic activity is activated by ABA [52]. Further, Li et al., [53] showed that AAPK is a positive regulator of ABA- induced stomatal closure through activating plasma membrane anion channels. Constant efflux of both anions and K+ from guard cells drives water efflux and contributes to ward to the loss of guard cell turgor, leading to stomatal closure [54]. For depolarization of the guard cell membrane, ABA also inhibits the activity of guard cell plasma membrane H+-ATPase [55,56]. Rapid stomatal responses to environmental stimuli might mainly rely on guard cell-synthesized ABA whereas ABA synthesized in the vasculature might contribute more to stomatal regulation during long-term soil water deficit [57]. In this way, ABA has a very useful role in regulating stomatal aperture, which helps plants to adapt and survive stress conditions.
Regulation of ABA Biosynthesis through Abiotic Stress
The increase in de novo biosynthesis of ABA is due to the rise in abiotic stress, which plays a role to inhibit its degradation and is thought to be stimulated by stress relief. Gene identified for ABA biosynthesis is zeaxanthin oxidase (ZEP) and has been cloned and expressed in numerous plant species. This gene is found to be present in every plant part but is highly associated for basal expression in leaves [58]. Moreover, level of ABA biosynthesis through ZEP gene is regulated not only in different plant portions and development phases but also in different plant species. The ZEP gene in Arabidopsis has a same basal transcript level in non-stressful conditions as in tobacco and tomato. These variations in the expression of ZEP genes are partly associated to basal transcript levels, which also cover stress inducibility of genes as identified in different experiments. However, another ABA biosynthetic gene expression (NCED, AtAAO3 and AtSDR1) are less debatable. The ABA biosynthesis is notably achieved after cleavage in the rate-limiting step, and thus expression of NCED gene(s) has received a significant importance. NCED gene is found to be overexpressed in drought stress condition in maize (Zea mays; [21]), tomato (Lycopersicon esculentum; [59]), bean (Phaseolus vulgaris; [60]), Arabidopsis [61] and cowpea (Vigna unguiculata; [62]). A remarkable rise in NCED transcript levels has been reported following 15-30 min of leaf extrication or induced dehydration [11], providing an evidence for the instant activation of NCED genes. Since ABA biosynthesis mechanism up-regulates drastically in response to stress, it can be deduced that protein levels of the related genes increase with the transcript levels, which were similarly noticed in NCED gene [11]. Since the product of NCED gene regulates the rate-limiting step in the ABA biosynthesis pathway, the information regarding the control of this gene product by ABA is very limited in terms of auto-regulation of ABA biosynthesis. In tomato plants and cowpea, this NCED gene was found to be not affected by exogenous ABA [62]. Hence, it can be concluded that ABA cannot stimulate its production but have the potential for its degradation. The transcript measures 9-cis-Epoxycarotenoid Dioxygenase (NCED1) (an enzyme catalyzing the first step of ABA biosynthesis) and grows significantly in grape berries [63]. The resultant will be accumulation of ABA in different tissue pertaining to hormonal response ultimately causing a berry development and ripening [64]. However, Xiong and Zhu [11] have shown that there is an up-regulation of ZEP and AAO3 in Arabidopsis by ABA, even though being stimulated by stress. Exogenous ABA significantly regulates expression of such genes. Though, ABA biosynthesis production and degradation both are of greater significance in regulating ABA expression and adjusting plant stress responses and development strategies. As DRE and ABRElike cis elements are promoters of stress-inducible ABA genes [65], consequently, these genes are in the same way controlled as DRE/CRT class of stress-responsive genes [66]. Biosynthesis of ABA is augmented by cold stress which helps plants to withstand the adverse conditions [67, 68]. In agreement with this observation, exogenous application of ABA stimulates freezing tolerance, and ABA biosynthesis mutants demonstrate cold sensitivity [69,70]. Increased biosynthesis of phytohormone reflects the upregulation of biosynthetic genes or downregulation of the genes in catabolic pathways. Under cold stress, ABA content has been shown to be increased in cauline leaves and inflorescence meristem of Arabidopsis thaliana and shoot area of Oryza sativa L. [70,71]. In Arabidopsis, ABA biosynthetic genes, catabolic genes are selectively overexpressed at 0ºC in an organspecific manner [71]. For instance, ABA1, ABA2, AAO3, NCED2, NCED5, NCED6, CYP707A1, CYP707A2, CYP707A3, and CYP707A4 in cauline leaves; ABA1, ABA2, ABA4, AAO3, NCED3, CYO707A1, CYP707A2, and CYP707A4 in inflorescence meristem; and ABA1, NCED2, NCED3, NCED5, NCED6, CYP707A2, CYP707A3, and CYP707A4 in developing silique are overexpressed during cold stress [71]. With the help of ABA in stress response condition, screening can be done in vegetative tissues and would probably help to identify new loci, which can be essential for regulating ABA metabolism [72]. ABA does specific types of acts, which contain complicated regulatory mechanisms, production, degradation, signal perception, and transduction. Figuring out the crucial position of ABA in response to plant stress and their regulatory systems will help to formulate real-time techniques to procreate or genetically regulate crops with expanded tolerance to adverse environment stipulations.
ABA Regulation of Plant Responses to Drought and Salt Stresses
Exogenous application of ABA induces many genes that normally respond to drought and high salinity [73,74]. ABA controls stress adaptation responses and regulates water balance and is the most studied stress-responsive hormone [75]. The ABA-responsive genes increased by drought and high salinity can be classified into two general groups. The first group includes genes that produce important metabolic proteins to protect cells and tissues from stressful conditions. These genes encode enzymes for the biosynthesis of osmolyte, acid metabolism, and cell detoxification, as well as those for water channel proteins, membrane transporters, proteins for protection of macromolecules such as Late Embryogenesis Abundant (LEA) proteins and osmotins, etc [76]. The second group includes genes primarily encoding regulatory proteins for modulating downstream genes expression in response to stressful conditions, such as protein kinases, protein phosphatases, transcription factors, and so on [76]. The transcription factors interact with cis-acting elements to regulate the expression of stress-inducible genes and finally confer tolerance to stress on the plants [8].
In the first group of genes, overexpressing Rab16A (LEA gene), which originated from a salt-tolerant rice variety called pokkali, in tobacco resulted in enhanced tolerance to salt stress as well as increased gene expression under ABA, drought, and high salinity [77]. Similarly, overexpressing the Rab16A gene in indica rice also conferred enhanced salt tolerance [78]. The expression of ABA responsive genes is important for plant adaptation and survival under drought and salt stresses. Fine regulatory mechanisms are necessary for a correct expression of these stress-inducible genes and the transcriptional regulation of gene expression responsive to drought and high salinity depends on the interaction of transcription factors with regulatory elements, which usually results in the expression of multiple stress-inducible genes, thus conferring improved plant stress tolerance. Many ABA-inducible genes contain a conserved ABA-responsive element, ABRE, an 8-bp cis-acting sequence (PyACGTGGC), in the promoter regions. ABRE, often together with a Coupling Element (CE), forms an ABA-Responsive Complex (ABRC) [79]. This complex is the most important cis-acting DNA element involved in ABA-responsive gene expression. The ABRE element interacting with transcription factors, also known as ABREBinding Factors (ABFs) or ABRE-binding proteins (AREBs), belong to the basic leucine Zipper (bZIP) family [80]. In Arabidopsis, approximately 75 distinct bZIP-type transcription factors have been identified [81]. Among them, AREB1/ABF2, AREB2/ABF4, and ABF3 were induced by drought, high salinity, and ABA in vegetative tissues [80]. Overexpression of ABF3 and ABF4 in Arabidopsis led to ABAhypersensitive and enhanced drought-tolerant phenotypes, as well as an up-regulated expression of some ABA-responsive genes [82]. Overexpression of the active form of ABF2/AREB1 also resulted in significantly improved drought tolerance in transgenic Arabidopsis plants [83]. In addition to ABRE, MYC and MYB elements [84] and the NAC Recognition Sites (NACRS) [85] have also been identified as functioning in ABA-inducible gene expression under drought and salt stress conditions. MYC and MYB recognition sites, present in the promoter region of the RD22 gene, play key roles in ABA-mediated dehydration-inducible expression of RD22. Transgenic Arabidopsis overexpressing AtMYC2, AtMYB2, or the two genes simultaneously were hypersensitive to ABA and showed enhanced tolerance to osmotic stress [84]. The cis-acting elements, which usually contain CATGT and harbor CACG core sequences, that are bound by NAC transcription factors are called NACRS. The Arabidopsis RD26/ ANAC072 gene encodes a NAC protein, which can recognize NACRSs in the promoters of some stress-inducible genes and activate their expression. Moreover, the promoter itself also contains a NACRS that is induced by drought, high salinity, and ABA [85]. Additionally, an ABA-independent pathway exists in plants, which is mediated by the interaction of other transcription factors and cis-acting elements in response to drought and salt stresses. It has been widely reported that the dehydration-responsive element/C-repeat (DRE/CRT), with a 9-bp core sequence (TACCGACAT), acts in response to drought and high salinity through an ABA-independent signal transduction pathway [86]. DREB2, an APETALA2 (AP2)-type transcription factor induced by drought and high salinity, transactivates the promoter containing the DRE element of stress-responsive genes. Transgenic Arabidopsis plants overexpressing the constitutively active DREB2A (DREB2A-CA) showed enhanced drought tolerance [87]. Another AP2-type transcription factor, DREB1/CBF, has downstream genes that always contain DRE/CRT elements [88]. Overexpressing DREB1A in transgenic rice increased stress tolerance to drought and high salinity [89].
ABA Regulation of the Cold Stress Response in Plants
ABA is an important stress hormone in plants that has been demonstrated to be involved in the cold stress response through regulation of a set of specific stress-responsive genes such as C-repeat binding factor/DRE-binding factor (CBF/DREB) transcriptional regulatory cascade [70]. However, there are sets of genes, which are altered by cold stress but not linked to CBF [90-92]. Hence, cold stress-mediated gene expression can be divided in two groups: CBFdependent and CBF-independent [93]. As ABA does not alter CBF expression, it was hypothesized that ABA-mediated cold response follows CBF-independent pathway [94]. However, some recent results support the idea that ABA-mediated cold response may be mediated through CBF-dependent pathway. The transcription factor MYB96 is induced by both ABA and cold [95]. Studies from loss of function and overexpression lines suggest that MYB96 contributes for cold tolerance [95]. Mechanistically, MYB96 interacts with Heptahelical Protein (HHP). Interestingly, HHP1, HHP2, and HHP3 interact with ICE1, CAMTA, and ICE2, respectively [95,96], supporting the notion that ABA-mediated cold stress resistance response is also regulated through CBF-dependent pathway [93].
ABA and Abiotic Stress Signaling
ABA is a key endogenous messenger in plants, and it has a crucial role in various plant stresses. Therefore, understanding the signaling mechanism of ABA is critical for improving plant performance under stress environments. There are three core components of ABA signaling; Pyrabactin Resistance (PYR)/Pyrabactin Resistance-Like (PYL)/Regulatory Component of ABA Receptors (RCAR), Protein Phosphatase 2C (PP2C: acts as negative regulators) and (Sucrose Non-Fermenting) SNF1- related protein kinase 2 (SnRK2: acts as positive regulators). In the presence of ABA, PYR/PYL/RCAR-PP2C complex formation leads to inhibition of PP2C activity, which allows the activation of SnRK2. Activated SnRK2 then phosphorylates downstream substrate proteins such as transcription factors, and thus facilitating transcription of ABA-responsive genes (Figure 2).
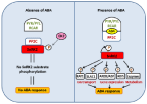
Figure 2: The schematic representation of major ABA signaling pathway
in plants within and without ABA presence. The core components in ABA
signaling include ABA receptors (PYR/PYL/RCAR), PP2C phosphatases
(negative regulators), and SnRK2 kinases (positive regulators). In the
absence of ABA, PP2Cs associate with SnRK2s and prevent the activation
of SnRK2s. The inactive SnRK2s are unable to phosphorylate down stream
substrates, and thus signal transduction is not occurring. In the presence of
ABA, PYR/PYL/RCAR receptors bind to ABA and interact with PP2Cs, which
release SnRK2s. The SnRK2s are then activated by autophosphorylation
of the activation loop. The active SnRK2s can phosphorylate down stream
substrate proteins, including transcription factors, ion channels, and enzymes
such as NADPH oxidases, there by inducing ABA responses. ABF, ABAresponsive
element binding factor; ABI5, ABA insensitive 5; AREB, ABAresponsive
element binding protein; KAT1, potassium channel in Arabidopsis
thaliana 1; PP2C, Protein phosphatase 2C; PYR, pyrabactin resistance; PYL,
PYR-related; RCAR, regulatory component of ABA receptor; SLAC1, slow
anion channel 1; SnRK2, sucrose nonfermenting-1-related protein kinase2.
The PYR/PYL/RCAR ABA receptors
Many of PYR/PYL/RCARs including PYR1, PYL1 and PYL2 have been shown to directly bind ABA [97, 98]. Once bound to ABA this receptor can bind to group A Protein Phosphatases (PP2C), including ABI1 (ABA-insensitive 1) and ABI2, the negative regulator of ABA signaling [15,16,99]. These ABA receptors played a critical role in ABA perception, as quadruple mutants’ pyr1, pyl1, pyl2 and pyl4 are insensitive toward ABA [16,100]. The PYR/PYL/RCAR family in Arabidopsis (except PYL13) is capable of activating ABA signaling response, suggesting that nearly all members can function as ABA receptors [14]. Antoni et al., [101] reported that ABA-dependent inhibition of PP2Cs by PYR/PYLs is essential for the perception of moisture gradient. The structural and molecular studies conducted by many researchers have convincingly shown that PYR/PYL/RCAR receptors play a central role in ABA perception [15,102].
PP2C: The negative regulators
Reversible protein phosphorylation catalyzed by protein kinases and phosphatases play a significant role in cellular signal transduction, which helps in the transmission of signals from external to the internal environment of the cell. There are 112 phosphatases encoded in the Arabidopsis genome, among them 76 genes encode for PP2Cs [103]. At least 6 of the nine members of the group A PP2Cs have been shown to be involved in ABA signaling, among them, ABI1 and ABI2 are the well characterized. The PP2C group A genes first characterized are ABI1 and ABI2, which was isolated from a genetic screen in Arabidopsis. Both abi1 and abi2 mutants show insensitivity in various tissues, developmental stages, suggesting that they act as a negative regulator of ABA signaling. Both of these mutants display reduced seedling growth, seed dormancy, drought tolerance and stomatal regulations [104]. It has been shown that all of the loss- of-function mutants of group APP2Cs exhibit significant ABA hypersensitivity, which establishes them as the major negative regulators of ABA signaling [101,105]. ABI1 and ABI2 showed a dominant role in ABA signaling pathways in both seeds and vegetative tissues; the evidence is supported by gene expression data and genetic analysis. These results are consistent with the fact that PP2Cs function as negative regulators of ABA signaling [6].
SnRK2 protein kinases: the positive regulators
Protein kinases are known to be involved in ABA signaling pathway. The Sucrose Non-Fermenting 1 (SNF1)-Related Protein Kinase 2 (SnRK2) family of protein kinases are plant specific serine/ threonine kinases participating in plant response and has a central role in cellular responses to drought and dehydration [106]. ABAactivated protein kinases have been identified in Arabidopsis [107] and rice [108]; they belong to the SnRK2 family. These ABA-activated protein kinases mediate ABA signaling by phosphorylating their substrate proteins such as transcription factors, ion channels, and metabolic enzymes [105,109]. SnRK2 family is the key regulator of plant response to abiotic stress and is divided into three sub-groups depending upon the affinity toward ABA. Subgroup-I kinases do not respond to ABA whereas subgroup-III actively respond to ABA and believed to be the key regulator of ABA-dependent pathway gene expression. Subgroup-II weakly responds or did not provide a response to ABA [107,110]. There are altogether 10 SnRK2 members found in Arabidopsis thaliana, i.e., SnRK2.1- SnRK2.10 or SnRK2ASnRK2J. Among them, 5 SnRK2 members (SnRK2.2, SnRK2.3, SnRK2.6, SnRK2.7, and SnRK2.8) can be activated by ABA. The all of the SnRK2s except SnRK2.9 can be activated by osmotic stress, which indicated that most SnRK2 members are involved in ABA and stress signaling [104,111]. Many studies reported that phosphorylation at the activation loop is critical for the activation of SnRK2s. Several phosphorylation sites have been identified in SnRK2.6; ABA stimulates phosphorylation of Ser175 in the activation loop, whereas PP2C can dephosphorylate the Ser175 of SnRK2.6 in the absence of ABA, resulting in the deactivation of SnRK2.6 [14-16,112].
Understanding how SnRK2 kinases exert their effects in ABA signaling network requires knowledge of SnRK2 targets. Several phosphorylation substrates of SnRK2 kinases have been identified using biochemical and molecular approaches. The ABFs (ABAresponsive- element Binding Factor)/AREBs (ABA-Responsive Element Binding factors) transcription factors involving in regulating ABA-responsive genes are SnRK2 substrates as ABA-activated SnRK2 kinases have been shown to phosphorylate ABF/AREB proteins in vitro and in vivo [24,111,113]. Another transcription factor involved in ABA signaling, ABI5, can be phosphorylated in vitro by ABAactivated SnRK2 kinases [24,114]. In addition to nuclear transcription factors, SnRK2 can also phosphorylate plasma membrane proteins. The guard cell OST1/SnRK2.6 has been shown to phosphorylate the anion channel SLAC1, the potassium channel KAT1, and the NADPH oxidase AtrbohF, all of which are plasma-membrane proteins important in controlling stomatal aperture [115,116].
Reactive Oxygen Species (ROS) and ABA signaling
Various abiotic and biotic stress conditions result in the accumulation of both ABA and Reactive Oxygen Species (ROS) in plants. The adaptation of plants to these stress conditions is a very complex process, which including stress signal perception, transduction and change of genes expression. The signaling pathways between ABA and ROS are independent of each other or crosstalk at various levels [117]. Substantial effort has been expended on attempts to discover possible crosstalk between ABA and ROS in plant stress damage and signaling upon exposure to different stress conditions. For example, the time course of ABA accumulation in the leaves of maize plants exposed to water stress was monitored at the same time as the increased generation of ROS (such as O2− and H2O2) and the induction of several antioxidant enzymes (such as SOD, CAT, APX and GR) [118].
Guard cells, which form stomata, have been investigated as a single-cell system that is useful for the elucidation of individual signaling mechanisms that function in the cellular signaling network. Given that ABA plays a key role in controlling stomatal closure during drought stress, substantial work has focused on ABA signaling in guard cells [9,10]. It has also been reported that H2O2 acts as a key regulator that mediates stomatal movement [119]. Considering that ROS signaling has been shown to be intertwined with other hormonal signals, it is likely that ROS are the vital hub that connects the external growth conditions with ABA signals. The relationships between ROS and ABA signals mainly involve regulation of guard cell movement. ROS also regulate the ABA response of guard cells through MAPK cascades. Time-course analysis showed that the accumulation of H2O2 activates MAPK activity, which promotes stomatal closure [120]. Double-mutant mpk9/12 showed increased transpirational water loss and decreased sensitivity in terms of the stomatal response to ABA and H2O2, when compared with wild-type plants. Both ABA and H2O2 can enhance the protein kinase activity of MPK12. These results suggested that these two MPK act upstream of anion channels and downstream of ROS to promote ABA signaling in guard cells [121]. The biological roles of ROS in the ABA-regulated root growth signaling pathway remain largely unknown. However, secondary messenger calcium has been proved to be important in Pro-Rich Extensin-Like Receptor Kinase 4 (PERK4)- regulated root growth in ABA signaling [117]. It is possible that changes in Ca2+ might be an element common to the signaling pathways downstream of both ROS and ABA.
Conclusion and Future Prospects
ABA is a vital hormone and acts as a central regulator of different plant stresses like drought, low temperature, and salinity. Due to the advancement of molecular genetics and genome- wide technologies, a deeper understanding of the underlying mechanisms of the involvement of ABA in stress tolerance is achieved, although more is yet to be revealed. The different physiological reactions that ABA regulates have been described at molecular level like stomatal closure, change in gene expression and accumulation of osmoprotectants. Understanding of ABA signal transduction pathways and epigenetic modifications can be further exploited to produce better transgenic plants with improved stress tolerance without a yield penalty that can withstand adverse climatic conditions. The mechanisms by which the abiotic stress upregulate ABA biosynthesis genes are still needed to know. Furthermore, the molecular mechanisms of cross talk between ABA and other phytohormone signaling pathways remain to be elucidated. Although many questions are still open, the current advances in ABA signaling in Arabidopsis pave the way to address the molecular events underlying stress responses in other plant species, with the prospect to improve the abiotic stress performance of crop plants.
Acknowledgements
This work was supported by the Ministry of Higher Education and Scientific Research-Tunisia and Ministry of Higher Education and Scientific Research-Algeria.
Authors’ Contributions
LB, WS and FB wrote the manuscript and FB edited the final version of the manuscript.
References
- Hasanuzzaman M, Nahar K, Alam MM, Roychowdhury R, Fujita M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int J Mol Sci. 2013; 14: 9643-9684.
- Sreenivasulu N, Harshavardhan VT, Govind G, Seiler C, Kohli A. Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene. 2012; 506: 265-273.
- Finkelstein R. Abscisic acid synthesis and response. Arabidopsis Book. 2013; 11: e0166.
- Wani SH, Kumar V. Plant stress tolerance: engineering ABA: a Potent phytohormone. Transcriptomics: An Open Access. 2015; 3: 1000113.
- Ng LM, Melcher K, Teh BT, Xu HE. Abscisic acid perception and signaling: structural mechanisms and applications. Acta Pharmacol Sin. 2014; 35: 567- 584.
- Mehrotra R, Bhalothia P, Bansal P, Basantani MK, Bharti V, Mehrotra S. Abscisic acid and abiotic stress tolerance- different tiers of regulation. J Plant Physiol. 2014; 171: 486-496.
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006; 57: 781-803.
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007; 58: 221-227.
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010; 61: 651- 679.
- Kim TH, Bohmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2C Signaling. Annu Rev Plant Biol. 2010; 61: 561-591.
- Xiong L, Zhu JK. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003; 133: 29-36.
- Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008; 59: 387-415.
- Harris J. Abscisic acid: hidden architect of root system structure. Plants. 2015; 4: 548-572.
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, et al. In vitro reconstitution of an abscisic acid signaling pathway. Nature. 2009; 462: 660-664.
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Sci. 2009; 324: 1064-1068.
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, et al. Abscisic acid inhibits PP2Cs via the PYR/PYL family of ABA-binding START proteins. Sci. 2009; 324: 1068-1071.
- Park SY, Peterson FC, Mosquna A, Yao J, Volkman BF, Cutler SR. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature. 2015; 520: 545-548.
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, et al. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996; 15: 2331-2342.
- Seiler C, Harshavardhan VT, Rajesh K, Reddy PS, Strickert M, Rolletschek H, et al. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J Exp Bot. 2011; 62: 2615-2632.
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. Specific oxidative cleavage of carotenoids by VP14 of maize. Sci. 1997; 276: 1872- 1874.
- Tan BC, Schwartz SH, Zeevaart JAD, McCarty DR. Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA. 1997; 94: 12235-12240.
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005; 56: 165-185.
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002; 14: 2723-2743.
- Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007; 19: 485-494.
- Puertolas J, Conesa MR, Ballester C, Dodd IC. Local root Abscisic Acid (ABA) accumulation depends on the spatial distribution of soil moisture in potato: implications for ABA signaling under heterogeneous soil drying. J Exp Bot. 2015; 66: 2325-2334.
- Petricka JJ, Benfey PN. Root layers: Complex regulation of developmental patterning. Curr Opin Genet Dev. 2008; 18: 354-361.
- Heidstra R, Sabatini S. Plant and animal stem cells: Similar yet different. Nat Rev Mol Cell Biol. 2014; 15: 301-312.
- Liang Y, Mitchell DM, Harris JM. Abscisic acid rescues the root meristem defects of the Medicago truncatula latd mutant. Dev Biol. 2007; 304: 297- 307.
- Zhang H, Han W, DeSmet I, Talboys P, Loya R, Hassan A, et al. ABA promotes quiescence of the quiescent center and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 2010; 64: 764-774.
- Zhang C, Bousquet A, Harris JM. Abscisic acid and Lateral Root Organ Defective/Numerous Infections and Polyphenolics modulate root elongation via reactive oxygen species in Medicago truncatula. Plant Physiol. 2014; 166: 644-658.
- Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, et al. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell. 2013; 25: 324-341.
- Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PM, et al. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013; 25: 2132-2154.
- Ubeda-Tomas S, Swarup R, Coates J, Swarup K, Laplaze L, Beemster GT, et al. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol. 2008; 10: 625-628.
- He J, Duan Y, Hua D, Fan G, Wang L, Liu Y, et al. DEXH box RNA helicasemediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell. 2012; 24: 1815-1833.
- O’Brien JA, Daudi A, Butt VS, Bolwell GP. Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta. 2012; 236: 765-779.
- Schmidt R, Schippers JHM. ROS-mediated redox signaling during cell differentiation in plants. Biochim Biophys Acta. 2015; 1850: 1497-1508.
- Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010; 143: 606-616.
- Zhang H, Jennings A, Barlow PW, Forde BG. Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA. 1999; 96: 6529-6534.
- Signora L, de Smet I, Foyer CH, Zhang H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 2001; 28: 655-662.
- Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu Rev Plant Biol. 2007; 58: 115-136.
- Xue-Xuan X, Hong-Bo S, Yuan-Yuan M, Gang X, Jun-Na S, Dong-Gang G, et al. Biotechnological implications from Abscisic Acid (ABA) roles in cold stress and leaf senescence as an important signal for improving plant sustainable survival under abiotic-stressed conditions. Crit Rev Biotechnol. 2010; 30: 222-230.
- Ray S, Mondal WA, Choudhuri MA. Regulation of leaf senescence, grainfilling and yield of rice by kinetin and abscisic acid. Physiol Plant. 1983; 59: 343-346.
- He P, Jin JY. Relationships among hormone changes, transmembrane flux of Ca2C and lipid peroxidation during leaf senescing in spring maize. Acta Bot Sin. 1999; 41: 1221-1225.
- Yang J, Zhang J, Wang Z, Zhu Q, Liu L. Abscisic acid and cytokinins in the root exudates and leaves and their relationship to senescence and remobilization of carbon reserves in rice subjected to water stress during grain filling. Planta. 2002; 215: 645-652.
- Passioura J. The drought environment: physical, biological and agricultural perspectives. J Exp Bot. 2007; 58: 113-117.
- Lee IC, Hong SW, Whang SS, Lim PO, Nam HG, Koo JC. Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 2011; 52: 651-662.
- Takasaki H, Maruyama K, Takahashi F, Fujita M, Yoshida T, Nakashima K, et al. SNAC-As, stress-responsive NAC transcription factors, mediate ABAinducible leaf senescence. Plant J. 2015; 84: 1114-1123.
- Fan K, Bibi N, Gan S, Li F, Yuan S, Ni M, et al. A novel NAP member GhNAP is involved in leaf senescence in Gossypium hirsutum. J Exp Bot. 2015; 66: 4669-4682.
- Devireddy AR, Zandalinas SI, Gomez-Cadenas A, Blumwald E, Mittler R. Coordinating the overall stomatal response of plants: Rapid leaf-to-leaf communication during light stress. Plant Biol. 2018; 11: eaam 9514.
- Prakash V, Singh VP, Tripathi DK, Sharma Sh, Corpas FJ. Crosstalk between Nitric Oxide (NO) and Abscisic Acid (ABA) signaling molecules in higher plants. Env Exp Bot. 2019; 161: 41-49.
- Kuromori T, Seo M, Shinozaki K. ABA Transport and Plant Water Stress Responses. Trends Plant Sci. 2018; 23: 513-522.
- Li J, Assmann SM. An abscisic acid-activated and calcium- independent protein kinase from guard cells of fava bean. Plant Cell. 1996; 8: 2359-2368.
- Li J, Wang XQ, Watson MB, Assmann SM. Regulation of Abscisic acidinduced stomatal closure and anion channels by guard cell AAPK Kinase. Sci. 2000; 287: 300-303.
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol. 2015; 28: 154-162.
- Hayashi M, Inoue S, Takahashi K, Kinoshita T. Immunohistochemical detection of blue light-induced phosphorylation of the plasma membrane H+- ATPase in stomatal guard cells. Plant Cell Physiol. 2011; 52: 1238-1248.
- Jezek M, Blatt MR. The membrane transport system of the guard cell and its integration for stomatal dynamics. Plant Physiol. 2017; 174: 487-519.
- Merilo E, Jalakas P, Kollist H, Brosche M. The role of ABA recycling and transporter proteins in rapid stomatal responses to reduced air humidity elevated CO2, and exogenous ABA. Molecular Plant. 2015; 8: 657-659.
- Xiong L, Lee H, Ishitani M, Zhu JK. Regulation of osmotic stress responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem. 2002a; 277: 8588-8596.
- Burbidge A, Grieve TM, Jackson A, Thompson A, McCarty DR, Taylor IB. Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize vp14. Plant J. 1999; 17: 427-431.
- Qin X, Zeevaart JAD. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA. 1999; 96: 15354-15361.
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, et al. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001; 27: 325-333.
- Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. A stressinducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000; 123: 553-562.
- Berli FJ, Bottini R. UV-B and abscisic acid effects on grape berry maturation and quality. J Berry Res. 2013; 3: 1-14.
- Wheeler S, Loveys B, Ford C, Davies C. The relationship between the expression of abscisic acid biosynthesis genes, accumulation of abscisic acid and the promotion of Vitis vinifera L. berry ripening by abscisic acid. Aust J Grape Wine Res. 2009; 15: 195-204.
- Bray EA. Abscisic acid regulation of gene expression during water deficit stress in the era of the Arabidopsis genome. Plant Cell Environ. 2002; 25: 153-161.
- Xiong L, Shumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002b; 14: S165-S183.
- Gusta L, Trischuk R, Weiser C. Plant cold acclimation: the role of abscisic acid. J Plant Growth Regul. 2005; 24: 308-318.
- Mega R, Meguro-Maoka A, Endo A, Shimosaka E, Murayama S, Nambara E, et al. Sustained low abscisic acid levels increase seedling vigor under cold stress in rice (Oryza sativa L.). Sci Rep. 2015; 5: 13819.
- Cuevas JC, Lopez-Cobollo R, Alcazar R, Zarza X, Koncz C, Altabella T, et al. Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiol. 2008; 148: 1094-1105.
- Eremina M, Rozhon W, Poppenberger B. Hormonal control of cold stress responses in plants. Cell Mol Life Sci. 2016; 73: 797-810.
- Baron KN, Schroeder DF, Stasolla C. Transcriptional response of Abscisic Acid (ABA) metabolism and transport to cold and heat stress applied at the reproductive stage of development in Arabidopsis thaliana. Plant Sci. 2012; 188: 48-59.
- Zhang D. Abscisic Acid: Metabolism, Transport and Signaling. New York, NY: Springer. 2014.
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003; 6: 410-417.
- Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002; 53: 247-273.
- Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol. 2011; 14: 290-295.
- Ma Y, Qin F. ABA Regulation of Plant Responses to Drought and Salt Stresses. In Abscisic Acid: Metabolism, Transport and Signaling, edited by Zhang DP. Springer. 2014; 315-336.
- RoyChoudhury A, Roy C, Sengupta DN. Transgenic tobacco plants overexpressing the heterologous lea gene Rab16A from rice during high salt and water deficit display enhanced tolerance to salinity stress. Plant Cell Rep. 2007; 26: 1839-1859.
- Ganguly M, Datta K, Roychoudhury A, Gayen D, Sengupta DN, Datta SK. Overexpression of Rab16A gene in indica rice variety for generating enhanced salt tolerance. Plant Signal Behav. 2012; 7: 502-509.
- Hobo T, Asada M, Kowyama Y, Hattori T. ACGT-containing Abscisic Acid Response Element (ABRE) and Coupling Element 3 (CE3) are functionally equivalent. Plant J. 1999; 19: 679-689.
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Nat Acad Sci USA. 2000; 97: 11632-11637.
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002; 7: 106-111.
- Kang JY, Choi HI, Im MY, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002; 14: 343-357.
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, et al. AREB1 is a transcription activator of novel ABRE dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005; 17: 3470-3488.
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003; 15: 63-78.
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABAdependent stress-signaling pathway. Plant J Cell Mol Biol. 2004; 39: 863-876.
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994; 6: 251-264.
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought responsive gene expression. Plant Cell. 2006; 18: 1292-1309.
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, et al. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J Cell Mol Biol. 2004; 38: 982-993.
- Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, et al. Arabidopsis CBF3/ DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005; 138: 341-351.
- Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007; 12: 444-451.
- Li H, Ye K, Shi Y, Cheng J, Zhang X, Yang S. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol Plant. 2017; 10: 545-559.
- Xie Y, Chen P, Yan Y, Bao C, Li X, Wang L, et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBFindependent pathways in apple. New Phytol. 2018; 218: 201-218.
- Ashraf MA, Rahman A. Hormonal regulation of cold stress response. In Cold tolerance in plants, edited by SH Wani and V Herath. 2018; 65-88.
- Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci. 2014; 5: 170.
- Lee HG, Seo PJ. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF–COR pathway in Arabidopsis. Plant J. 2015; 82: 962-977.
- Chen CC, Liang CS, Kao AL, Yang CC. HHP1, a novel signaling component in the cross-talk between the cold and osmotic signaling pathways in Arabidopsis. J Exp Bot. 2010; 61: 3305-3320.
- Miyazono KI, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, et al. Structural basis of abscisic acid signaling. Nature. 2009; 462: 609-614.
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009; 16: 1230-1236.
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, et al. PYR/ PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010; 61: 290-299.
- Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012; 24: 2483-2496.
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Peirats-Llobet M, Pizzio GA, Fernandez MA, et al. PYRABACTIN RESISTANCE1-LIKE8 plays an important role in the regulation of abscisic acid signaling in root. Plant Physiol. 2013; 161: 931-941.
- Melcher K, Zhou XE, Xu HE. Thirsty plants and beyond: structural mechanisms of abscisic acid perception and signaling. Curr Opin Struct Biol. 2010; 20: 722-729.
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004; 9: 236-243.
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, et al. ABAhypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006; 140: 115-126.
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, et al. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010; 51: 1821-1839.
- Saruhashi M, Ghosh TK, Arai K, Ishizaki Y, Hagiwara K, Komatsu K, et al. Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc Natl Acad Sci USA. 2015; 112: E6388-E6396.
- Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem. 2004; 279: 41758- 41766.
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell. 2004; 16: 1163- 1177.
- Fujita Y, Yoshida T, Yamaguchi-Shinozaki K. Pivotal role of the AREB/ABFSnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant. 2013; 147: 15-27.
- Kulik A, Wawer I, Krzywinska E, Bucholc M, Dobrowolska G. SnRK2 protein kinases-key regulators of plant response to abiotic stresses. OMICS. 2011; 15: 859-872.
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA. 2006; 103: 1988-1993.
- Vlad F, Droillard MJ, Valot B, Khafif M, Rodrigues A, Brault M, et al. Phosphosite mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 2010; 63: 778-790.
- Sirichandra C, Davanture M, Turk BE, Zivy M, Valot B, Leung J, et al. The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS One. 2010; 5: e13935.
- Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J Exp Bot. 2013; 64: 675-684.
- Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, et al. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J. 2009; 424: 439-448.
- Sirichandra C, Gu D, Hu H, Davanture M, Lee S, Djaoui M, et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009; 583: 2982-2986.
- Bai L, Zhang G, Zhou Y, Zhang Z, Wang W, Du Y, et al. Plasma membrane associated proline rich extension like receptor kinase 4, a novel regulator of Ca2+ signaling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J. 2009; 60: 314-327.
- Jiang M, Zhang J. Role of abscissic acid in water stress-induced antioxidant defense in leaves of maize seedlings. Free Radic Res. 2002; 36: 1001-1015.
- Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006; 45: 113-122.
- Jiang J, Wang P, An G, Wang P, Song CP. The involvement of a P38-like MAP kinase in ABA-induced and H2O2-mediated stomatal closure in Vicia faba L. Plant Cell Rep. 2008; 27: 377-385.
- Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, et al. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci USA. 2009; 106: 20520-20525.