
Research Article
Ann Agric Crop Sci. 2022; 7(4): 1121.
A Major Recessive Gene Associated with Anthracnose (Colletotrichum capsici) Resistance in Chilli Pepper
Mohammad Siddiqur Rahman1* and Akanda AM2
¹Mohammad Siddiqur Rahman, Senior Scientific Officer, Plant Pathology Division, Bangladesh Agricultural Research Institute, Gazipur-1701
²Abdul Mannan Akanda, Professor, Department of Plant Pathology, Bangabandhu Sheikh Mujibur Rahman Agricultural University, Gazipur-1706
*Corresponding author: Mohammad Siddiqur Rahman, Senior Scientific Officer, Plant Pathology Division, Bangladesh Agricultural Research Institute, Gazipur-1701, Bangladesh
Received: September 23, 2022; Accepted: October 21, 2022; Published: October 28, 2022
Abstract
Chilli anthracnose caused by Colletotrichum sp., is an important disease in Bangladesh and many other Asian countries. Recently the local Bangladeshi genotype ‘Comilla-2’ was found resistant to C. capsici. Inheritance of resistant to C. capsici was analyzed in segregating populations derived from a cross of ‘BARI chilli-1’ x ‘Comilla-2’. BARI chilli-1 as susceptible and Comilla-2 as resistant parent was used in the study. Detached matured green fruits were inoculated using the microinjection method. Disease response was evaluated using disease incidence and over all lesion diameter at 8 days after inoculation. The disease reaction of F1 plants in case of disease incidence and overall lesion diameter were clearly skewed to the susceptible parent, with average values of 46.70% and 13.2 mm, respectively. The distribution of disease incidence in the F2 population was skewed toward the susceptible parent and the distribution of overall lesion diameter in the F2 population showed a similar trend. Based on the scale of resistance and susceptibility, less than 25.0% disease incidence or less than 9.0 mm overall lesion diameter were evaluated as resistance. In the cross ‘BARI chilli-1’ x ‘Comilla-2’, the segregation ratios of resistance and susceptibility scored by disease incidence and overall lesion diameter in the F2, BCr and BCs populations and chi-squared test significantly fitted one recessive gene model i.e. 1:3 Mendelian model. The result indicates that the resistance of ‘Comilla-2’ to C. capsici is controlled by a single recessive gene.
Keywords: Chilli (Capsicum annum); anthracnose (Colletotrichum capsici); Inheritance.
Introduction
Chilli (Capsicum annuum L.) is the most popular species grown worldwide. Anthracnose disease of chilli caused by several Colletotrichum spp. is responsible for serious yield loss and affects crop quality in tropical and subtropical regions. The crop is severely infected by anthracnose which causes yield losses up to 50% [1]. Colletotrichum species reduces marketable yield of chilli fruits up to 80% [2]. The damage caused by the pathogen is very serious in many Asian counties including Bangladesh. Colletotrichum capsici is mainly responsible for chilli fruit anthracnose in Bangladesh. In other countries like Thailand and Indonesia, the primary causal agents of anthracnose are C. gloeosporioides and C. capsici [3]. Colletotrichum gloeosporioides attacks chilli fruits at both the green and red stages, while C. capsici mainly attacks the fruits at the red stage [4]. Many fungicides are used to control the disease in developing countries like Bangladesh, but these are not only increase production cost but also cause consumer antipathy. The most economic and environmentally friendly method to control the disease is the use of resistant varieties. Currently, no resistant chilli cultivar is available in Bangladesh against anthracnose. Anthracnose resistant varieties in this species are rarely available [5]. Only one variety of C. annuum, which was a local Korean variety ‘Daepoong-cho’ was recently reported to be resistant to Colletotrichum capsici [6]. However, study of inheritance of resistant for the development of anthracnose resistant chilli variety is essential.
Several sources of resistance to C. capsici have been reported [7,8,3,9,1]and using this genetic resources, researchers have studied the inheritance of anthracnose resistance. The inheritance patterns vary depending on the source of resistance and the Colletotrichum isolate. For instance, resistance to C. dematium was inherited partially dominantly as reported by [8]. The authors also found that resistance to C. gloeosporioides was inherited as over-dominant or partially dominant in F1 plants [4]. In contrast, some reports have demonstrated that resistance to anthracnose is inherited recessively. For example, [7] found that resistance to C. capsici was inherited recessively with epistatic effects and the resistance of C. chinense Jacq. PBC932 to C. capsici was observed to be inherited through a single recessive gene [1]. The AVRDC has evaluated the resistance to anthracnose of many pepper accessions and has detected several resistance resources [10].
Reported that field resistance against anthracnose is present in Comilla-2 local Capsicum annuum genotype in Bangladesh [11]. On the other hand, this genotype was also found resistant in the earlier screening experiment. Therefore, the present study was carried out to determine the inheritance of resistance to anthracnose (C. capsici) in Capsicum annuum (Comilla-2).
Materials and Methods
Plant materials
Two parents ‘Comilla-2’ (selected as resistant chilli germplasm) and ‘BARI chilli-1-1’(susceptible) were used for inheritance study of anthracnose resistance to C. capsici. F1 plants were obtained by crossing ‘BARI chilli-1-1’ as female and ‘Comilla-2’ as male parents. F2 population was obtained by self-pollination of F1 plants. The backcross populations BCR and BCS were produced by backcrossing F1 plants to ‘Comilla-2’ and ‘BARI chilli-1’ respectively.
In the cross ‘BARI -1’ and ‘Comilla-2’ for the inheritance study, populations consisting of resistant parent (five plants), susceptible parents (five plants), F1 (five plants), BCR (36 plants), BCS (10 plants) and F2 (84 plants) were grown in pot Plant Pathology Department of BSMRAU.
Fungal isolate
Colletotricum capsici was isolated from infected chilli fruits of BSMRAU experimental farm following standard procedures [12,13]. The isolate was purified following single spore isolation method and identified following a standard key [14,15]. The isolate was maintained on Potato Dextrose Agar (PDA) medium at 250C in an incubator. After 8 days of incubation, the plates were flooded with distilled water. Conidia were collected by scraping the
culture surface with sterilized glass slides. Density of spore suspension was adjusted to 5x105 conidia/ml using a hemacytometer.
Inoculation
Healthy fruits of chilli were harvested from individual plants at the matured green and ripen stage. The fruits were washed with distilled water to remove associated microbes from the fruit surface. The fruits punctured with multi pointed needles and inoculated with 10μml of the spore suspension with a micropipette. Each fruit was swabbed with 10μml prepared conidial suspension and raped with poly bags to maintain humidity. Inoculation was conducted in glass petridish with three to five replications. The inoculated fruits were incubated at room temperature for 8 days.
Disease evaluation
Severity of disease was evaluated and expressed in percentage of infected sites and overall lesion diameter at 8 days after inoculation (DAI), as described previously [9]. Chi-square goodness of-fit test were used for statistical analysis.
Results and Discussion
Inheritance of resistance to Colletotrichum capsici in ‘Comilla-2’
The values of disease incidence and overall lesion diameter of parents and F1 plants are presented in Table 1. The symptoms of anthracnose started to develop at 2 DAI in susceptible parent ‘BARI chilli-1’ and there was no further change in disease incidence after 8 DAI. Therefore, the time of disease evaluation for the inheritance study was set at 8 DAI.
Population
Disease incidence
Overall lesion diameter (mm)
Comilla-2
15.20 ± a1.96
3.42 ± -0.19
BARI chilli-1
67.10 1.67
18.74 ± 0.94
F1 (BARI-1 x Comilla-2)
46.70 ± 6.87
13.2 ± 3.12
aMean ± standard deviation
Table 1: Disease incidence and overall lesion diameter of ‘Comilla-2’, ‘BARI chilli-1’ and their F1 progenies 8 days after inoculation with C. capsici.
The resistant parent ‘Comilla-2’ and the susceptible parent ‘BARI chilli-1’ showed the significant differences in disease incidence and overall lesion diameter. The disease incidence and overall lesion diameter were 15.20% and 3.42 mm in resistant parent (‘Comilla-2’) and 67.10% and 18.74 mm in susceptible parent (‘BARI chilli-1’). The disease reactions of F1 plants in case of disease incidence and overall lesion diameter were clearly skewed to the susceptible parent, with average values of 46.70% and 13.2 mm, respectively. The result The resistant parent ‘Comilla-2’ and the susceptible parent ‘BARI chilli-1’ showed the significant differences in disease incidence and overall lesion diameter. The disease incidence and overall lesion diameter were 15.20% and 3.42 mm in resistant parent (‘Comilla-2’) and 67.10% and 18.74 mm in susceptible parent (‘BARI chilli-1’). The disease reactions of F1 plants in case of disease incidence and overall lesion diameter were clearly skewed to the susceptible parent, with average values of 46.70% and 13.2 mm, respectively. The result
The distribution of disease incidence in the F2 population was skewed toward the susceptible parent (Figure 1) and the distribution of overall lesion diameter in the F2 population showed a similar trend (Figure 2). To determined the criteria of resistant and susceptibility, we scored the disease reactions of segregating populations using disease indices and overall lesion diameter. As a result, the distribution of disease incidence and overall lesion diameter in F2 and BCR populations was divided using the scale of 25.0% and 9.0 mm respectively (Figure 1,2). Based on the scale of resistance and susceptibility, less than 25.0% disease incidence or less than 9.0 mm overall lesion diameter were evaluated as resistance.
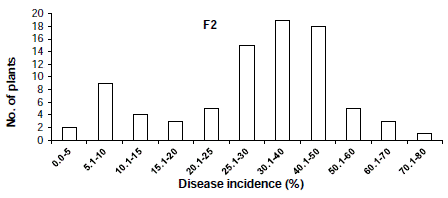
Figure 1: Distribution of disease incidence in F2 populations derived from the cross ‘BARI chilli-1’ and ‘Comilla-2’.
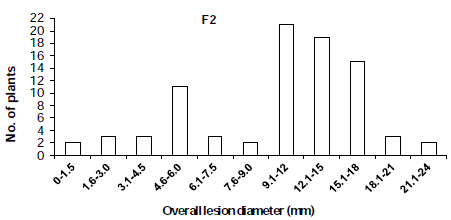
Figure 2: Distribution of overall lesion diameter in F2 populations derived from
the cross ‘BARI chilli-1’ and ‘Comilla-2’.
In the cross ‘BARI chilli-1’ x ‘Comilla-2’, the segregation of resistance and susceptibility scored by disease incidence in the F2 population was 18 to 66 (Table 2). The chi-squared and P values in the F2 population were 0.571 and 0.50-0.30, respectively, which fitted one recessive gene model i.e. 1:3 Mendelian model. The segregation ratios in the BCr and BCs populations in case of resistant and susceptibility was 16:20 and 0:10, respectively (Table 2 and Figure 3 & 4), which fitted expected segregation ratios, 1:1 and 0:1, respectively (Table 2). The segregation of resistance and susceptibility scored by overall lesion diameter in the F2 population was 23 to 61 (Table 3). The chisquared and P values in the F2 population were 0.254 and 0.70-0.50, respectively, which fitted one recessive gene model i.e. 1:3 Mendelian model. Segregation ratios in BCr and BCs populations were 21:15 and 0:10 respectively (Table 3 and Figure 5, 6). This result was also fitted to the one recessive gene model.
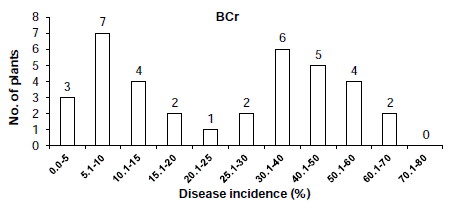
Figure 3: Distribution of disease incidence in BCr populations derived from
the cross ‘F1’ and ‘Comilla-2’.
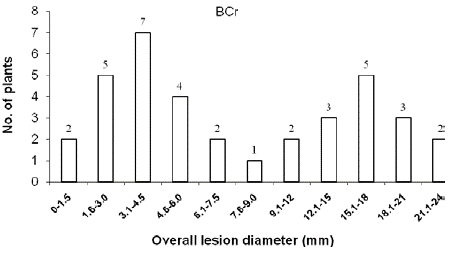
Figure 4: Distribution of overall lesion diameter in BCr populations derived
from the cross ‘F1’ and ‘Comilla-2’.
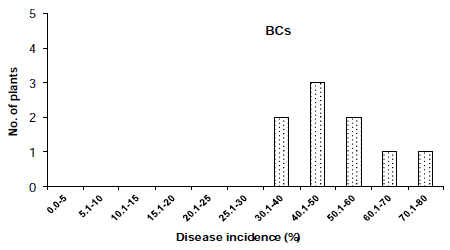
Figure 5: Distribution of disease incidence in BCs (Back cross with
susceptible parent) populations derived from the cross ‘F1’ and ‘BARI chilli-1’.
Distribution was skewed to susceptible parent.
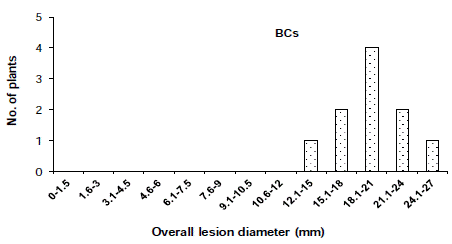
Figure 6: Distribution of overall lesion diameter in BCs (Back cross with
susceptible parent) populations derived from the cross ‘F1’ and ‘BARI chilli-1’.
Distribution was skewed to susceptible parent.
Population
Expected ratio
Observed frequency
Pprobability
(R : S)
R
S
Disease incidencea
Comilla-2
5
0
BARI chilli-1
0
5
F1 (Bari-1 x comilla-2)
1 : 3
0
5
0.571
0.50 - 0.30
F2
1 : 1
18
66
0.444
0.70 - 0.50
BCr
0 : 1
16
20
BCs
0
10
aLess than 25 % disease incidence was evaluated as resistance (R).
bLess than 9.0 mm overall lesion diameter was evaluated as resistance
BCR-F1 back crossing with resistant parent (‘Comilla-2’)
BCS-F1 back crossing with susceptible (S) parent (‘BARI chilli-1’)
Table 2: Segregation ratio of resistance and susceptibility scored by disease incidence in segregating populations derived from the cross ‘Comilla-2’, and ‘BARI chilli-1’.
Population
Expected ratio
Observed frequency
Pprobability
(R:S)
R
S
Overall lesion diameterb
Comilla-2
5
0
BARI chilli-1
0
5
F1 (Bari-1 x comilla-2)
0
5
F2
01:03
23
61
0.254
0.70-0.50
BCr
01:01
21
15
1
0.50-0.30
BCs
00:01
0
10
aLess than 25 % disease incidence was evaluated as resistance.
bLess than 9.0 mm overall lesion diameter was evaluated as resistance.
Table 3: Segregation ratio of resistance and susceptibility scored by overall lesion diameter in segregating populations derived from the cross ‘Comilla-2’ and ‘BARI chilli-1’.
The distribution of segregating population and results of chisquired tests in the cross ‘BARI chilli-1’ x ‘Comilla-2’ indicate that the resistance of ‘Comilla-2’ to C. capsici is controlled by a single recessive gene. The continuous distribution of disease incidence was displayed in all of the segregating populations, which indicates that minor genes may have affected the resistance. Also, in the crosses, F1 plants showed somewhat different responses depending on the susceptible parent.
In several reports on the inheritance of resistance to C. acutatum or C. gloeosporioides in chili peppers resistance was inherited dominantly [4,5,9]. [1] showed that the resistance of C. chinense Jacq. ‘PBC 932’ to C. capsici inherited through a single recessive gene. [16] reported that the resistance to C. acutatum was inherited through a single recessive gene. [6] suggested that the resistance of chilli germplasm ‘Daepoong-cho’ to C. capsici is controlled by a single recessive gene. Therefore, the results of the present investigation are in conformity with the findings of previous reports.
It was reported that for pepper breeding perspective, dominant resistance is more useful than recessive resistance because it will be manifested in F1 hybrids even if only one parent has the allele additionally. In case of dominant resistance, hybrid production is easier as compared to resistance conferring with recessive gene. Producing F1 varieties using recessive resistance sources requires much time and effort. However, recessive resistance is more durable than dominant resistance. This information can benefit chilli pepper breeding programs in the production of anthracnose-resistant varieties.
Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be interpreted as a potential conflict of interest.
Author’s Contribution
Md. Siddiqur Rahman was involved in the execution of the research work; collection, analysis and interpretation of the data; manuscript writing, etc. Prof. Abdul Mannan Akanda was assist during work and editing of the manuscript.
Acknowledgments
The authors greatly acknowledge the financial grant from Bangladesh Agricultural Research Institute, Bangladesh for accomplishing this PhD research.
References
- Pakdeevaraporn P, Wasee S, Taylor PWJ, Mongkolporn O. Inheritance of resistance to anthracnose caused by Colletotrichum capsici in Capsicum. Plant Breeding. 2005; 124: 206-208.
- Poonpolgul S, Kumphai S. Chili Pepper Anthracnose in Thailand. The First International Symposium on Chili Anthracnose, 23, Convention Center, Seoul National University, Korea 2007.
- Lin Q, Kanchana UC, Jaunet T. Mongkolporn O. Genetic analysis of resistance to pepper anthracnose caused by Colletotrichum capsici. Thai J Agric Sci. 2002; 35: 259-264.
- Park HK, Kim BS, Lee WS. Inheritance of resistance to anthracnose (Colletotrichum spp.) in pepper (Capsicum annuum L.). I. Genetic analysis of anthracnose resistance by diallel crosses. J Korean Soc Hort Sci. 1990a; 31: 91-105.
- Yoon JB. Identification of genetic resources, interspecific hybridization, and inheritance analysis for breeding pepper (Capsicum annuum) resistant to anthracnose. PhD Thesis, Seoul Natl. Univ. Seoul, Korea. 2003.
- Kim SH, Yoon JB, Wahng JD, Park HG. A major recessive gene associated with anthracnose resistance Colletotrichum capsici in Chilli Pepper (Capsicum annuum L.). Breeding Sci. 2008; 58: 137-141.
- Cheema DS, Singh DP, Rawal RD, Deshpande AA. Inheritance of resistance to anthracnose disease in chillies. Capsicum Eggplant Newsletter. 1984; 3: 44.
- Park HK, Kim BS, Lee WS. Inheritance of resistance to anthracnose (Colletotrichum spp.) in pepper (Capsicum annuum L.) II. Genetic analysis of resistance to Colletotrichum dematium. J Korean Soc Hort Sci. 1990b; 31: 207-212.
- Voorrips RE, Finkers R, Sanjaya L, Groenwold R. QTL mapping of anthracnose (Colletotrichum spp.) resistance in a cross between Capsicum annuum and C. chinense. Theor Appl Genet. 2004; 109: 1275-1282.
- AVRDC. Off-season tomato, pepper and eggplant. In: AVRDC Report 1998. Tainan, Taiwan, pp. 20-30.
- Rahman MS, Akhter MS, Maya MA, Rahman AHMA, Akanda AM. Field Resistance of Chilli Cultivars against Anthracnose Disease Caused by Colletotrichum capsici. Thai J Agril Sci. 2011; 44: 243-250.
- Dasgupta B. Sporulation and relative virulence among isolates of Colletotrichum capsici causing anthracnose of betel vine. Indian Phytopath. 1981; 32: 196-199.
- Agostini JP, Timmer LW. Selective isolation procedure for differentiation of two strains of Colletotrichum gloeosporioides from citrus. Plant Disease. 1992; 76: 1176-1178.
- Sutton BC. The Coelomycetes, Fungi Imperfecti with Pycnidia, Acervuli and Stromata. Commonwealth Mycological Institute, Kew, Surrey, Uk. 1980; 523- 538.
- Sutton BC. The genus Glomerella and its anamorph Colletotrichum. In: J A Biology Pathology and Control. CABI International. Oxon Dx10 8DE, UK. 1992; 1-25.
- Kim SH, Yoon JB, Wahng JD, Park HG. Resistance to Anthracnose Caused by Colletotrichum acutatum in Chilli Pepper (Capsicum annuum L.). J Crop Sci Biotech. 2007; 10: 277-280.