
Research Article
Ann Agric Crop Sci. 2023; 8(3): 1138.
To Analyze the Mechanism of Glycyrrhiza uralensis (Licorice) in the Treatment of COVID-19 Based on Network Pharmacology and Molecular Docking Technology
Junyi Zhang1; Xuhai Yang 1,2,3#; Lichun Zhu1; Ruiying He1; Shuangquan Xie1,4,*
1Shihezi University, China
2Xinjiang Production and Construction Corps Key Laboratory of Modern Agricultural Machinery, Shihezi University, China
3Engineering Research Center for Production Mechanization of Oasis Special Economic Crop, Ministry of Education, Shihezi University, China
4Key Laboratory of Xinjiang Phytomedicine Resource and Utilization, Ministry of Education, Shihezi University, China
*Corresponding author: Shuangquan Xie Shihezi University, Key Laboratory of Xinjiang Phytomedicine Resource and Utilization, Ministry of Education, Shihezi University, Shihezi 832002, China. Tel: 183-9983-7286 Email: xiesq0921@163.com
#These authors contributed equally to this work
Received: June 27, 2023 Accepted: July 27, 2023 Published: August 03, 2023
Abstract
The current study, the effectiveness of analyzing Glycyrrhiza uralensis for the treatment of COVID-19 was investigated using an integrated network pharmacology and molecular docking approach. Through network pharmacology, establishment of Protein-Protein Interactions (PPI), molecular docking simulations, GO analysis, and KEGG analysis, the aim was to investigate the mechanism of licorice in the treatment of COVID-19. There are 7 core genes corresponding to 3 bioactive compounds of G. uralensis. The target proteins of G. uralensis for the treatment of neocoronary pneumonia could be enriched to cancer pathway, lipid The target proteins of G. uralensis for the treatment of neocoronary pneumonia could be enriched to cancer pathway, lipid and atherosclerosis pathway, PI3K-Akt signaling pathway, and down-regulate the activity of core targets to inhibit the expression of virus SARS-CoV-2. Based on molecular docking simulations, the largest significant binding affinities were found for TNF and 7-Methoxy-2-methyl isoflavone (-6.95 kcal/mol), MAPK1 and kaempferol (-6.75 kcal/mol), and AKT1 and quercetin (-6.74 kcal/mol). Quercetin induces viral cell cycle arrest and inhibits growth and metastasis by engaging in the induction and expression of key intracellular targets. The flavonoid chemicals of kaempferol can inhibit inflammation-related signaling pathways and suppress the release of inflammation-related factors, and 7-methoxy-2-methylisoflavone may have therapeutic effects on COVID-19 by reducing hydroxyproline levels to suppress lung inflammation and fibrosis indices and modulate immune function. This suggests that G. uralensis has an interfering effect on novel coronaviruses and its main active component has a strong binding ability to the core gene of SARS-CoV-2, providing knowledge for future studies based on COVID-19.
Keywords: Glycyrrhiza uralensis; Novel coronavirus pneumonia; Network pharmacology; Molecular docking; Quercetin; Kaempferol
Introduction
Coronavirus Disease 2019 (COVID-19) is a Severe Acute Respiratory Syndrome Coronavirus that spreads through contact, aerosols, and direct contact. Acute respiratory infection brought on by the human body catching the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [1,2]. Since the outbreak of New Crown, 651918402 people worldwide have now been infected, with a morbidity and mortality rate of approximately 2%. To date, the mechanism of treatment for New Crown pneumonia is unknown, and according to the available clinical cases [3,5]. In the context of universal vaccination, the rate of severe illness and mortality of the Omicron variant has fallen to levels similar to those of seasonal influenza (a more optimistic estimate is 0.3%, 0.1%). However, its ability to spread is much greater than that of seasonal influenza, and the base of the infected population will be much larger than that of seasonal influenza. Therefore, the level of threat to the health of the entire population and the pressure on the health system from the new coronavirus is much greater than that of seasonal influenza.
To date, the pharmaceutical community has made significant progress in mitigating the SARS-CoV-2 threat through the development of small-molecule drugs [6,7]. Gilead’s Veklury was conditionally approved to combat the outbreak [8] and Merck’s Lagevrio [9] raise hope for a COVID-19 cure. However, promising bullets still do not exist. As an indispensable resource for promising compounds, traditional medicine [10] and natural products [11,12] have attracted significant attention in countering SARS-CoV-2 infection.
Traditional Chinese medicine has been fighting against disease for thousands of years [13]. The benefit of Traditional Chinese Medicine (TCM) that "Prevention precedes disease" has been adequately mirrored in the prevention and treatment of epidemics, and TCM has had successful outcomes treating COVID-19 in various parts of China [14]. The main therapeutic mechanisms of herbal medicine for neoconjunctivitis include direct antiviral, anti-inflammatory effects, protection of target organs and immunomodulation [15], especially in terms of the cure rate of clinical symptoms such as TCM symptoms and pulmonary lesions, as well as prognosis [16]; however, there are no specific therapeutic drugs for neoconjunctivitis. Clinical evidence suggests that herbal medicines are effective against viral infections such as influenza, SARS and SARS-CoV-2 by targeting viral cell entry, viral replication and host authorities for the treatment of COVID-19. According to the frequency of herbal medicine use since the epidemic [17,18], Glycyrrhiza uralensis, almond, and raw gypsum were the top 3 drugs with the highest frequency of occurrence in the treatment of COVID-19. Recent reports also suggest that Glycyrrhiza uralensis extracts may play a potential role in the fight against COVID-19 and related diseases [19]. According to the Chinese Pharmacopoeia, Glycyrrhiza uralensis is able to strengthen the spleen, clear heat, detoxify and resolve phlegm, as well as stop cough, spasm and pain, thus harmonizing the effects of other drugs [20]. However, the specific mechanism of action of licorice with COVID-19 is still unclear, so this paper uses network pharmacology and molecular docking methods to analyze it.
Network pharmacology is a contemporary science that leverages data networks to explore biological mechanisms and pharmacological molecules, in accordance with the theoretical framework of systems biology [21-23]. A detailed inspection of the therapeutic mechanisms of molecules, cells, tissues, and other drugs in complex diseases is also made possible by it, in addition to new ideas and procedures for interdisciplinary research involving Chinese and Western medicine, artificial intelligence, and large-scale biomedical data analysis [24].
In the present investigation, we utilized a bioinformatics approach to screen the potentially useful small molecules of Glycyrrhiza uralensis. Neo-orange and Glycyrrhiza uralensis' primary cross-targets were examined. In order to investigate the mechanisms of action and potential pathways, Protein-Protein Interactions (PPI), KEGG, and Gene Ontology (GO) were employed to assess any potential linkages between the primary crossover targets. For the purpose of to investigate the mechanism of action of Glycyrrhiza uralensis for the treatment of new coronavirus pneumonia, seven major genes were screened to acquire their corresponding proteins to which the active molecules of Glycyrrhiza uralensis were docked.
Therefore, this study investigated the targets and molecular processes of Glycyrrhiza uralensis for the treatment of novel coronavirus pneumonia, and predicted the anti-novel coronavirus pneumonia effect of Glycyrrhiza uralensis using network pharmacology and molecular docking techniques, which could provide new ideas and necessary theoretical basis for clinical treatment. And the workflow of this study is presented in Figure 1.
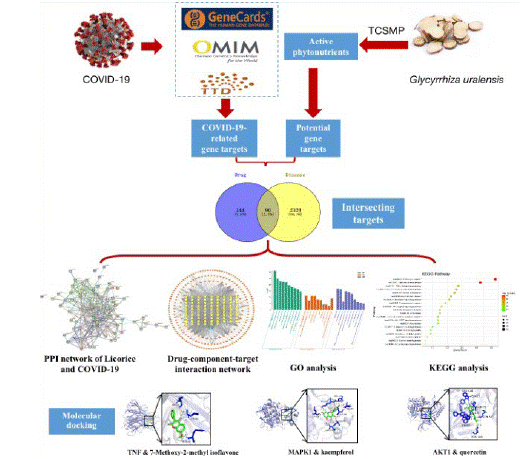
Figure 1:
Materials and Methods
Screening of Active Ingredients and Action Targets of G. Uralensis
The drug composition of G. uralensis was obtained by searching the TCMSP (http://tcmspw.com/tcmsp.php) database [25], and the extraction of G. uralensis under the conditions of Oral Bioavailability (OB) ≥30% and Drug-Likeness (DL) =0.18. The active ingredients were extracted under the conditions of oral bioavailability (OB) ≥30% and Drug Similarity (DL) ≥0.18. Then, the targets matching with the active ingredients of G. uralensis were detected by comparison and the results were imported into the UniProt platform (http://www.Unitprot.org/) to correct the gene names of the chemical composition targets and standardize the protein target information [26].
Collection of Disease Targets
The GeneCards database (https://www.genecards.org/), the Online Mendelian Inheritance in Man database (OMIM, https://omim.org/) and Therapeutic Target Database (TTD, http://bidd.nus.edu.sg/group/cjttd) [27] was searched by the keyword “Novel Coronavirus” and “COVID-19” to obtain the relevant disease target genes, and the target genes obtained from the gene screening of the three databases were intersected and de-duplicated to finally obtain the potential targets. The active ingredient targets of G. uralensis were matched to get cross-targets (common targets), which may be potential targets for G. uralensis treatment of COVID-19. Cross-gene Venn diagrams of drugs and diseases were mapped using the online website Venny 2.1.0.
Construction of Protein Interaction Network and Active Ingredient-Disease-Target Network
The intersection of potential drug targets and disease-related targets was screened using the Cytoscape 3.9.1 software with drug, ingredient and target represented by different colors and shapes respectively. The importance of the nodes in the network is reflected by the Degree value, and the core active ingredients in the network are obtained.
Construction of Protein-to-Protein Interaction (PPI) network and acquisition of core genes
With the support of STRING (https://string-db.org/) database, the intersection targets of "Glycyrrhiza-COVID-19" were imported, "Homo sapiens" was selected as the species with confidence score = 0.9, PPI network analysis was run, and the results were saved the results were saved as a "TSV" file. The file was imported into Cytoscape 3.9.1 program and a PPI network map was created. Topological analysis of the PPI network was performed using the CytoNCA plug-in of Cytoscape 3.9.1 software. The "Local Average Connectivity" (LAC), "Network Centrality" (NC), "Betweenness Centrality" (BC), "Closeness Centrality" (CC), "Degree centrality" (DC), "Eigenvector Centrality" (EC), and other characteristic parameters. All six parameters of the target node are higher than the median value in the corresponding retained PPI network to construct a new PPI network for further study. The number of edges connected to each node in the PPI network was analyzed and calculated using Cytoscape 3.9.1 software, and the stronger the interaction between the target protein and other proteins, the more dense the number of edges.
Gene Ontology Function Enrichment Analysis and KEGG Pathway Enrichment Analysis
GO functional enrichment analysis was performed using the Metascape database (http://metascape.org/), and the statistical significance threshold was set at P<0.01 in terms of Biological Processes (BP), Molecular Functions (MF), and Cellular Components (CC) of the core targets, and then KEGG enrichment analysis was performed to obtain the key pathways. The top 20 eligible results obtained from GO and KEGG enrichment analysis were selected, and the data results were visualized using the microbiology letter platform (http://www.bioinformatics. com.cn/) to analyze the pathways of glycyrrhizal tuning for the treatment of neocrown pneumonia.
Molecular Docking
Download 3D structures of the first seven potential anti-COVID-19 core targets RELA (PDBID:3QXY), TP53 (PDBID:1AIE), AKT1 (PDBID:3O96), MAPK1 (PDBID:6G9K), MAPK3 (PDBID:6GES), STAT3(PDBID:5AX3) and TNF(PDBID:1A8M) from the RCSB PDB (https://www.rcsb.org/) database, applying PyMOL software for dehydration and solvent molecule manipulation with full hydrogen addition. Set as receptor (exported to PDB file format) for backup. Apply PyMOL software to add all-hydrogen to the active small molecule of Glycyrrhiza glabra, set as ligand (automatic charge assignment), detect and set the torsion bond, and set aside. Import the above active compounds and protein structure files into Auto Dock Tools 1.5.7 software for optimization operations, set up protein activity pockets, and finally visualize the top 4 docking complexes with the lowest binding energy using PyMOL software [28].
Results
Screening of Active Ingredients and Action Targets of G. Uralensis
A total of 265 G. uralensis chemical components were queried in the TCSMP database, and the screening parameters (OB=30%, DL=0.18) were set, resulting in a total of 93 active components, as shown in Table 1. Meanwhile, the target genes corresponding to the G. uralensis components were screened using the TCSMP database and a total of 238 targets were obtained.
Mol ID
Molecule Name
OB (%)
DL
MOL001484
Inermine
75.18
0.54
MOL001792
DFV
32.76
0.18
MOL000211
Mairin
55.38
0.78
MOL002311
Glycyrol
90.78
0.67
MOL000239
Jaranol
50.83
0.29
MOL002565
Medicarpin
49.22
0.34
MOL000354
Isorhamnetin
49.6
0.31
MOL000359
Sitosterol
36.91
0.75
MOL003656
Lupiwighteone
51.64
0.37
MOL003896
7-Methoxy-2-methyl isoflavone
42.56
0.2
MOL000392
Formononetin
69.67
0.21
MOL000417
Calycosin
47.75
0.24
MOL000422
Kaempferol
41.88
0.24
MOL004328
Naringenin
59.29
0.21
MOL004805
(2S)-2-[4-hydroxy-3-(3-methylbut-2-enyl)phenyl]-8,
8-dimethyl-2,3-dihydropyrano[2,3-f]chromen-4-one31.79
0.72
MOL004806
Euchrenone
30.29
0.57
MOL004808
Glyasperin B
65.22
0.44
MOL004810
Glyasperin F
75.84
0.54
MOL004811
Glyasperin C
45.56
0.4
MOL004814
Isotrifoliol
31.94
0.42
MOL004815
(E)-1-(2,4-dihydroxyphenyl)-3-(2,2-dimethylchromen-6-yl)prop-2-en-1-one
39.62
0.35
MOL004820
Kanzonols W
50.48
0.52
MOL004824
(2S)-6-(2,4-dihydroxyphenyl)-2-(2-hydroxypropan-2-y)
-4-methoxy-2,3-dihydrofuro[3,2-g]chromen-7-one60.25
0.63
MOL004827
Semilicoisoflavone B
48.78
0.55
MOL004828
Glepidotin A
44.72
0.35
MOL004829
Glepidotin B
64.46
0.34
MOL004833
Phaseolinisoflavan
32.01
0.45
MOL004835
Glypallichalcone
61.6
0.19
MOL004838
(6-hydroxy-2-benzofuranyl)-2,
2-dimethyl-5-chromenol58.44
0.38
MOL004841
Licochalcone B
76.76
0.19
MOL004848
Licochalcone G
49.25
0.32
MOL004849
3-(2,4-dihydroxyphenyl)-8-(1,1-dimethylprop-2-enyl)-7-hydroxy-5-methoxy-coumarin
59.62
0.4
MOL004855
Licoricone
63.58
0.47
MOL004856
Gancaonin A
51.08
0.4
MOL004857
Gancaonin B
48.79
0.45
MOL004860
Glycyrrhiza uralensis glycoside E
32.89
0.27
MOL004863
(3,4-dihydroxyphenyl)-5,7-dihydroxy-8-(3-methylbut-2-enyl)chromone
66.37
0.41
MOL004864
5,7-dihydroxy-3-(4-methoxyphenyl)-8-(3-methylbut-2-enyl)chromone
30.49
0.41
MOL004866
2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-6-(3-methylbut-2-enyl)chromone
44.15
0.41
MOL004879
Glycyrin
52.61
0.47
MOL004882
Licocoumarone
33.21
0.36
MOL004883
Licoisoflavone
41.61
0.42
MOL004884
Licoisoflavone B
38.93
0.55
MOL004885
Licoisoflavanone
52.47
0.54
MOL004891
Shinpterocarpin
80.3
0.73
MOL004898
(E)-3-[3,4-dihydroxy-5-(3-methylbut-2-enyl)phenyl]-1-(2,4-dihydroxyphenyl)prop-2-en-1-one
46.27
0.31
MOL004903
Liquiritin
65.69
0.74
MOL004904
Licopyranocoumarin
80.36
0.65
MOL004905
3,22-Dihydroxy-11-oxo-delta(12)-oleanene-27-alpha-methoxycarbonyl-29-oic acid
34.32
0.55
MOL004907
Glyzaglabrin
61.07
0.35
MOL004908
Glabridin
53.25
0.47
MOL004910
Glabranin
52.9
0.31
MOL004911
Glabrene
46.27
0.44
MOL004912
Glabrone
52.51
0.5
MOL004913
1,3-dihydroxy-9-methoxy-6-benzofurano
[3,2-c]chromenone48.14
0.43
MOL004914
1,3-dihydroxy-8,9-dimethoxy-6-benzofurano
[3,2-c]chromenone62.9
0.53
MOL004915
Eurycarpin A
43.28
0.37
MOL004917
Glycyroside
37.25
0.79
MOL004924
(-)-Medicocarpin
40.99
0.95
MOL004935
Sigmoidin-B
34.88
0.41
MOL004941
(2R)-7-hydroxy-2-(4-hydroxyphenyl)chroman-4-one
71.12
0.18
MOL004945
(2S)-7-hydroxy-2-(4-hydroxyphenyl)-8-(3-methylbut-2-enyl)chroman-4-one
36.57
0.32
MOL004948
Isoglycyrol
44.7
0.84
MOL004949
Isolicoflavonol
45.17
0.42
MOL004957
HMO
38.37
0.21
MOL004959
1-Methoxyphaseollidin
69.98
0.64
MOL004961
Quercetin der.
46.45
0.33
MOL004966
3'-Hydroxy-4'-O-Methylglabridin
43.71
0.57
MOL000497
Licochalcone a
40.79
0.29
MOL004974
3'-Methoxyglabridin
46.16
0.57
MOL004978
[(3R)-8,8-dimethyl-3,4-dihydro-2H-pyrano [6,5-f]chromen-3-yl]-5-methoxyphenol
36.21
0.52
MOL004980
Inflacoumarin A
39.71
0.33
MOL004985
Icos-5-enoic acid
30.7
0.2
MOL004988
Kanzonol F
32.47
0.89
MOL004989
6-prenylated eriodictyol
39.22
0.41
MOL004990
7,2',4'-trihydroxy-5-methoxy-3-arylcoumarin
83.71
0.27
MOL004991
7-Acetoxy-2-methylisoflavone
38.92
0.26
MOL004993
8-prenylated eriodictyol
53.79
0.4
MOL004996
Gadelaidic acid
30.7
0.2
MOL000500
Vestitol
74.66
0.21
MOL005000
Gancaonin G
60.44
0.39
MOL005001
Gancaonin H
50.1
0.78
MOL005003
Licoagrocarpin
58.81
0.58
MOL005007
Glyasperins M
72.67
0.59
MOL005008
Glycyrrhiza flavonol A
41.28
0.6
MOL005012
Licoagroisoflavone
57.28
0.49
MOL005013
18a-hydroxyglycyrrhetic acid
41.16
0.71
MOL005016
Odoratin
49.95
0.3
MOL005017
Phaseol
78.77
0.58
MOL005018
Xambioona
54.85
0.87
MOL005020
Dehydroglyasperins C
53.82
0.37
MOL000098
Quercetin
46.43
0.28
Table 1: Active ingredients of G. uralensis.
Collection of Disease Targets
The three databases of GeneCards, OMIM and TTD were searched to collect the relevant target genes of COVID-19 disease, and the disease correlation was set =1. The screening data of the three databases were de-weighted separately, and the intersection of the three results was taken, and a total of 2410 disease targets were obtained. The active ingredients and disease targets of G. uralensis were imported into the Venny 2.1.0 platform, and the cross-gene Venn diagram of G. uralensis and COVID-19 was plotted (Figure 2), and 90 intersecting targets of drugs and diseases were obtained.
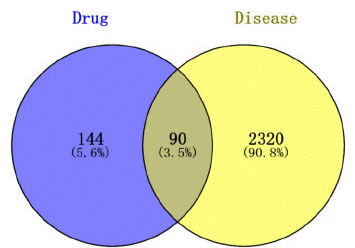
Figure 2:
Construction of Protein Interaction Network and Active Ingredient-Disease-Target Network
In order to generate and illustrate the network diagram, the drug G. uralensis and its active components, as well as the intersecting target genes, were loaded into Cytoscape 3.9.1 software and represented as triangles, hexagons, and circles, respectively (Figure 3). The network consists of 177 nodes (87 components and 90 intersecting genes), with 1395 connecting lines. To determine the target score ranking, all of the targets in this network were assessed and categorized by Degree. While there are 27 intersecting genes with greater than average degree values, a higher degree value implies that the target is addressed by more active components, demonstrating the crucial significance of targeting targets to the protein interaction network.
Gene names
BC
CC
DC
EC
LAC
NC
RELA
784.0339
0.5448
29
0.2912
6.6897
17.3825
AKT1
869.0675
0.5064
24
0.1943
4.0833
12.7715
MAPK3
330.3859
0.52667
23
0.2696
7.9130
14.5476
STAT3
635.6587
0.5032
23
0.2486
6.4348
13.5643
TP53
886.2071
0.5130
27
0.2406
5.9260
15.8523
MAPK1
303.3641
0.5097
22
0.2563
7.1818
12.8873
TNF
412.3410
0.4817
22
0.2158
6.5455
14.1546
Table 2: Topological parameters of core target proteins of protein interaction network.
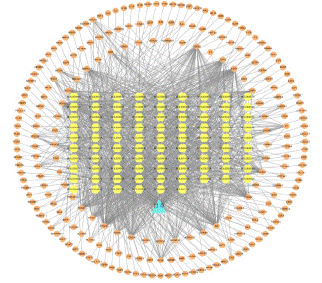
Figure 3:
Construction of Protein-to-Protein Interaction (PPI) Network and Acquisition of Core Genes
In order to generate and illustrate the network diagram, the drug G. uralensis and its active components, as well as the intersecting target genes, were loaded into Cytoscape 3.9.1 software and represented as triangles, hexagons, and circles, respectively (Figure 3). The network consists of 177 nodes (87 components and 90 intersecting genes), with 1395 connecting lines. To determine the target score ranking, all of the targets in this network were assessed and categorized by Degree. While there are 27 intersecting genes with greater than average degree values, a higher degree value implies that the target is addressed by more active components, demonstrating the crucial significance of targeting targets to the protein interaction network.
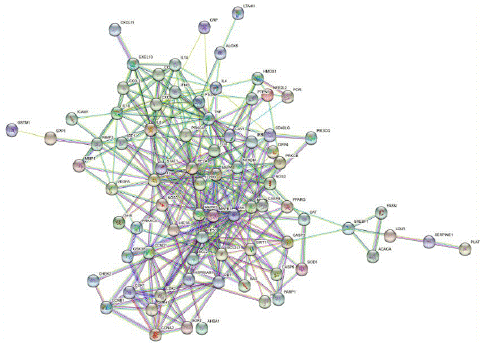
Figure 4:
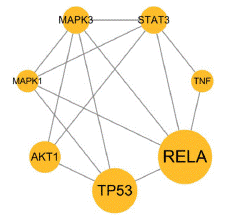
Figure 5:
Gene Ontology Function Enrichment Analysis and KEGG Pathway Enrichment Analysis
The enrichment analysis was performed in the Metascape database, and a total of 1148 Biological Processes (BP), 41 Cellular Components (CC) and 95 Molecular Functions (MF) were obtained, taking the relevant entries of each of the three branches. The information of the top 20 in number in each of the three branches was taken (Figure 6A). The top 10 most significantly enriched GO analyses of each component were screened for BP, CC, and MF triple visualization graph construction (Figure 6B). GO enrichment analysis showed that BP-related gene targets are involved in inorganic substances, reactive oxygen species, cellular response to chemical stress, apoptotic process, negative regulation of gene expression, etc.; gene targets in CC are mainly involved in transcriptional regulation, RNA polymerase II transcriptional regulation, membrane raft, membrane microdomain, etc.; gene targets in MF are mainly involved in Kinase binding, protein kinase binding, histone deacetylase binding, etc. The main focus of BP, CC and MF is on cell membrane activity, control of apoptotic signaling pathway, lipid Apoptosis signaling pathway control, lipopolysaccharide response and receptor binding. 95 signaling pathways were generated from KEGG Pathway analysis, and the top 20 most enriched signaling pathways were selected to create bubble plots (Figure 7). x-axis indicates the number of generations, i.e. the number of background genes concentrated in all genes of the pathway. Pathways are represented by the y-axis, and the size of the node indicates the number of targets converging to the pathway; the larger the node, the greater the number of enriched targets. The color of the nodes ranges from green to red to indicate the ranking of p-values from high to low, so the more important the role of the signaling pathway is, the larger the red nodes are. As shown in Figure 7, the top three most significant enriched pathways were cancer pathway, lipid and atherosclerosis pathway, PI3K-Akt signaling pathway, and the other pathways involved can be broadly classified as cancer inhibition related pathway, inflammation related pathway, inhibition of viral infection pathway and immune response related pathway. Among them, the cancer pathway involves 17 genes such as STAT3, TP53, PTEN, AKT1, MAPK, etc. The lipid and atherosclerosis pathway is an important pathway containing 27 target genes, covering many target genes including Src, STAT3, etc. The PI3K-Akt signaling pathway involves 15 genes such as MAPK 1, EGFR, IL6, VEGFA, AKT1, etc. It is predicted that licorice may inhibit the progression of COVID-19 by interfering with these signaling pathways.
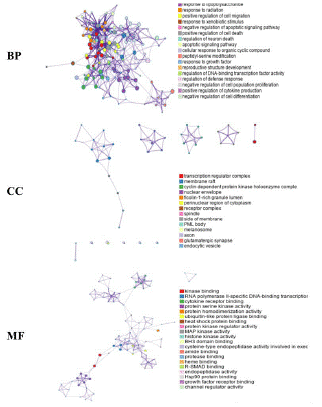
Figure 6A:
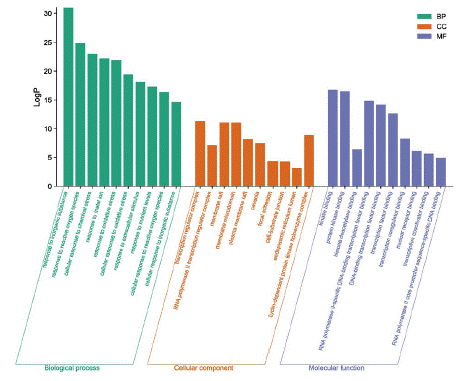
Figure 6B:
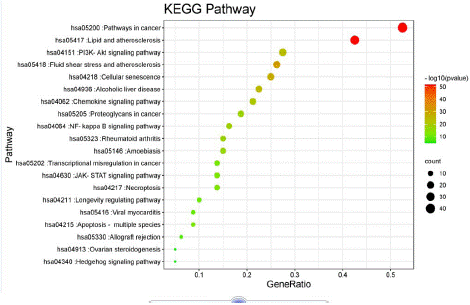
Figure 7:
Molecular Docking
In order to validate the intrinsic mechanism of Glycyrrhiza uralensis treatment of COVID-19, a final molecular docking study was performed. The current study addresses the function of the active phytonutrients of Glycyrrhiza uralensis, as well as the potential anti-COVID-19 core targets. The binding affinity of the first three active phytonutrients of Glycyrrhiza uralensis to the first seven potential anti-COVID-19 core targets (RELA, TP53, AKT1, MAPK1, MAPK3, STAT3 and TNF) was assessed. The crystal structures of all core targets (PDB ID: 3QXY, 1AIE, 3O96, 6G9K, 6GES, 5AX3 and 1A8M) were retrieved from the Protein Data Bank in PDB format, respectively [28]. The chemical structures of three active phytonutrients of Glycyrrhiza uralensis, quercetin (MOL000098), kaempferol (MOL000422), and 7-methoxy-2-methylisoflavone (MOL003896), were verified in TCMPS.
Molecular docking technology is used to simulate the main active ingredient and the key target protein of the disease through computer-aided drug design, so as to simply verify the results of network pharmacology analysis, the smaller the free energy of binding, the easier it is for the ligand and the receptor to bind better, thus supporting the interaction between the active ingredient and the key target. Generally, a binding free energy less than -5.0 kcal-mol-1 is considered a successful simulation, and less than -7.0 kcal-mol-1 is considered a good binding ability [29]. The first three active components obtained in the network pharmacological analysis were individually docked to the core genes to calculate the binding free energy [30], using the minimum binding conformation, and the results are shown in Table 3. The findings showed that all three Glycyrrhiza uralensis's skey active phytonutrients had good to excellent binding affinity scores (<-4.0 to <-7.0 kcal/mol), respectively, with all anti-COVID-19 core targets [31]. The affinities The top four best performers were TNF and 7-Methoxy-2-methyl isoflavone, MAPK1 and kaempferol, AKT1 and quercetin, RELA and kaempferol, 7-methoxy-2-methyl isoflavone and ASGR1, with affinities of -6.95, -6.75, - 6.74, -6.62 molecular docking plots for all active compounds are shown in Figure 8. Hence, the findings of molecular docking analysis corroborated that the Glycyrrhiza uralensis may ameliorate COVID19 disease by modulating the functions/expressions of these targets.
Gene
Compoundquercetin
C15H10O7kaempferol
C15H10O67-Methoxy-2-methyl isoflavone
C17H14O3RELA
-5.61
-6.62
-6.59
TP53
-5.88
-6.13
-5.85
AKT1
-6.74
-5.49
-5.32
MAPK1
-5.38
-6.75
-6.33
MAPK3
-5.09
-5.53
-6.36
STAT3
-4.82
-5.64
-5.48
TNF
-6.03
-5.67
-6.95
Table 3: Molecular docking efficiency of core active ingredients with SARS-CoV-2 binding site(kcal•mol-1).
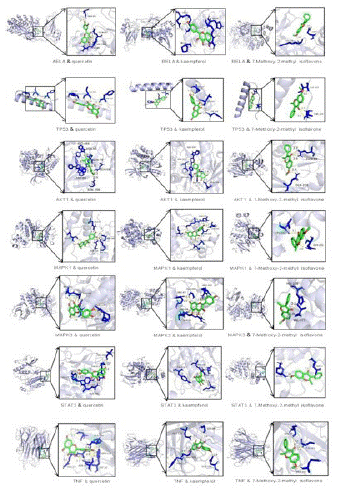
Figure 8:
It is shown that drug small molecules and protein complexes can maintain a very stable binding state in the treatment of COVID-19 and thus exert pharmacological effects.
Discussion
By considerably diminishing the percentage of patients who developed severe disease and died [32], Chinese medicine therapeutic approaches played a vital role in the management of this COVID-19 outbreak [33]. To expel cold and disperse the lung, clear heat and detoxify the lung, and dispel dampness and cold, patients were treated with moistening and moistening medicines, heat-clearing medicines, expectorant and cough-suppressing medicines, and qi-regulating medicines during medical observation thereby achieving the purpose of regulating qi and preventing further spread of the disease [34]. G. uralensis was chosen as the subject of this paper even though, according to the herbal examination of G. uralensis [35], it has the efficacy of clearing heat and detoxifying, expectorant, relieving discomfort, and coordinating other drugs [36]. Given the wide range of active ingredients and targets of herbal medicines, the therapeutic mechanism of G. uralensis for COVID-19 needs to be clarified.
In the present investigation, databases based on the notion of network pharmacology were used to search for and test 93 active components of G. uralensis. A considerable fraction of the 2410 possible targets that were discovered were linked to the targets of neoconjunctivitis, indicating both psychological effects and a molecular foundation for neoconjunctivitis caused by G. uralensis. The main active ingredients of the main pharmacodynamic material basis for the treatment of COVID-19 may include quercetin, kaempferol, and 7-Methoxy-2-methyl isoflavone, according to this study's analysis of the core components of the "herbal-component-target gene" network with values greater than the average degree. The most obvious finding to emerge from the analysis is that, quercetincan induce viral cell cycle arrest, inhibit growth and metastasis, and reduce apoptosis and inflammation of bronchial epithelial cells by participating in the induction and expression of phosphorylation of key intracellular targets [37,38], which proves the mechanism of quercetin against COVID-19; flavonoid chemicals of kaempferol play anti-inflammatory, antioxidant, ischemic injury and other roles in the nervous system [39], which can inhibit inflammation-related signaling pathways and the release of inflammation-related factors. Examples include Mitogen-Activated Protein Kinase (MAPK), Protein Kinase C (PKC), Phosphatidylinositol-3-Kinase (PI3K) and Janus Kinase (JAK)/Signal Transducer and Activator of Transcription (STAT), which exert anti-inflammatory effects; 7-methoxy-2-methylisoflavone may have therapeutic effects on COVID-19 by reducing hydroxyproline levels to suppress lung inflammation and fibrosis indices and modulating immune function [40].
The PPI network analysis ruled out RELA, TP53, AKT1, MAPK1, MAPK3, STAT3, and TNF, the seven main targets for the treatment of neocrown pneumonia, which may have a repercussions on immune control, transcription factor activity, cytokine receptor binding, phosphatase binding, and cellular stress, leading to the development of COVID-19. One of the targets is RELA. High on the list of targets is RELA, a crucial member of the NF-κB family whose post-translational modifications dynamically adjust the transcriptional activity of NF-κB and are extremely vital for the control of crucial bodily functions the same as inflammation, tumor growth, metabolism, and immune response in addition to emergence of associated diseases [41] and can regulate the accessibility of promoters to transcription factors, thereby indirectly regulating gene expression. MAPK1, MAPK3 MAPK3 are members of the MAPK family and can be involved in the response to potentially harmful abiotic stress stimuli. TNF (tumor necrosis factor) is considered a primary inflammatory cytokine that drives cytokine production, cell survival or cell death [42], and overproduction of the pro-inflammatory factor TNF induces viral progression and thus aggravates the disease.
GO enrichment analysis revealed that the anti-COVID-19 effects of key active phytonutrients of G. uralensis may be attributed to multiple gene targets associated with multiple BPs, such as protein phosphorylation, peptidyl serine phosphorylation, inflammatory response, apoptotic processes, positive regulation of gene expression, negative regulation of gene expression, and positive regulation of protein kinase B signaling. Reports suggest that SARS-CoV-2 infection stimulates host kinases, leading to high phosphorylation in both host and virus [43]. Many studies have shown that excessive activation of the innate and adaptive immune system after SARS-CoV-2 invasion induces an inflammatory response. This further stimulates several intracellular cytokines, such as NF-kB, mTOR, and TNF-a [44]. Apoptosis is a programmed cell death process that can be induced by the host to limit viral replication. Excessive apoptosis damages the bronchoalveolar network and causes lung injury and ARDS. The anti-COVID-19 effect of key active phytonutrients of G. uralensis may be attributed to multiple genetic targets found mainly in the cytoplasm, cytosol, nucleus, nucleoplasm, membrane, and mitochondria.
The results of KEGG enrichment analysis showed that the target proteins of G. uralensis a for neocrown pneumonia could be enriched to pathways in cancer, lipid and atherosclerosis, PI3K-Akt signaling pathway and so on. These pathways are closely related to cancer, pancreatic and liver inflammation and cell proliferation, apoptosis, and differentiation. These pathways are closely related to pancreatic and hepatic inflammation and cell proliferation, apoptosis, and differentiation. The results suggest that G. uralensis may inhibit viral SARS-CoV-2 expression by down-regulating the activity of core targets through these pathways. Among them, PI3K-AKT signaling pathway is widely present in cells and involved in the regulation of cell growth, proliferation and differentiation [45], and SARS-CoV-2 S protein can activate the Reactive Oxygen Species (ROS)-inhibited phosphoinositide 3 kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway, which in turn induces autophagy-triggered apoptosis and inflammation. Hayashi et al. [46] found that anticancer drugs blocked MAPK and inhibited SARS-CoV-2 proliferation. In SARS-CoV-2 infection, the (NF-κB) pathway has been found to be activated, involved in cellular responses to stimuli such as cytokines and stress, upregulates the transcriptional expression of some important apoptosis suppressors, and plays a key role in regulating the immune response to infection [47]; many cytokines involved in the pathogenesis of autoimmune and inflammatory diseases transmit intracellular signals through the JAK-STAT signaling pathway to deliver intracellular signals [48].
The molecular docking of the key active phytonutrients of the first three licorice to the first seven anti-COVID 19 core targets, including RELA, TP53, AKT1, MAPK1, MAPK3, STAT3 and TNF, was further analyzed. The results of the molecular docking analysis in terms of binding affinity (kcal/mol) scores are presented in tabular form as shown in Table 3. In addition, their docking complexes are shown in 2D form in Figure 7. The lower binding affinity represents the superior binding capacity of the phytonutrient and the target [49]. The results of the study showed that the binding affinity of the key active phytonutrients of the three licorices to all core anti-COVID-19 targets was rated as good to excellent (<-5.0 to <-7.0 kcal/mol), respectively. Among them, MOL003896 (7-Methoxy-2-methyl isoflavone) exhibited superior binding affinity scores, and MOL0038968 had affinities of -6.95, -6.59, -6.36 and -6.33 kcal/mol with TNF, RELA, MAPK3 and MAPK1, respectively. MOL000422 (kaempferol) exhibited superior binding affinity scores (-6.75, -6.62, -6.13 and -5.67kcal/mol) for MAPK1, RELA, TP53 and TNF, respectively. MOL000098 (quercetin) exhibited superior binding affinity scores (-6.75, -6.62, -6.13 and -5.67kcal/mol) for AKT1, TNF, TP53, respectively, RELA had affinities of -6.74, -6.03, -5.88, and -5.61 kcal/mol, respectively. In addition, MOL000422 (kaempferol) and MOL000098 (quercetin) showed similar binding affinity scores for MAPK1 and AKT1, respectively (-6.75 and -6.74). Although MOL000098 (quercetin) presented poor binding affinity scores compared to MOL003896 (7-Methoxy-2-methyl isoflavone) and MOL000422 (kaempferol), it has shown excellent binding affinity scores against AKT 1 and TP53. In addition, MOL003896 (7-Methoxy-2-methyl isoflavone) and MOL000422 (kaempferol) showed good binding affinity scores <-5.0 kcal/mol for RELA, TP53, AKT1, MAPK1, MAPK3, STAT3 and TNF. Thus, the findings of molecular docking analysis confirm that licorice can improve COVID 19 disease by modulating the function/expression of these targets.
Conclusions
In the current investigation, we successfully investigated the primary active phytonutrients, molecular targets, and molecular pathways of Glycyrrhiza uralensis for the therapy of COVID-19. Three important active phytonutrients were found, along with 124 possible significant anti-COVID-19 targets, 27 potential core anti-COVID-19 targets, and 144 potential major anti-COVID-19 targets. The first seven of the 27 potential core anti-COVID-19 targets (RELA, TP53, AKT1, MAPK1, MAPK3, STAT3 and TNF) were identified as key active phytonutrients nutrients, which may be associated with improved COVID-19-related molecular targets of key active phytonutrients in Glycyrrhiza uralensis. We further determined the mechanism of action of key active phytonutrients of licorice on COVID-19. Key components such as quercetin, kaempferol and 7-methoxy-2-methylisoflavone were found to be involved in cancer, lipid and atherosclerosis pathways, PI3K-Akt signaling pathway by targeting key proteins such as RELA, AKT1, MAPK1 and TNF, ultimately ameliorating inflammation, inhibiting viral SARS-CoV-2 activity and proliferation, and curing patients with neocrown pneumonia. The therapeutic approach may be to inhibit or modulate several biological processes, such as phosphorylation, inflammatory response and apoptotic processes. The results of molecular docking analysis further confirm that licorice may ameliorate COVID-19 disease by regulating the expression of these targets. The results of the network pharmacology study are consistent with the results of molecular docking, thus confirming the validity of network pharmacology.
At the same time, this study has some limitations, and some key questions, including the effects of Glycyrrhiza uralensis in clinical practice in patients with neocrown pneumonia and the exact modulation of target genes by bioactive components, need to be explored in subsequent studies to further validate the expected theoretical results and provide knowledge for future COVID-19-based studies.
Author Statements
Funding
Young Innovative Talent Program of Shihezi University (CXPY202107); the Open Project of Xinjiang Key Laboratory of Botanical Resources and Utilization, Ministry of Education (XPRU202202); and the Financial Science and Technology Program of Xinjiang Production and Construction Corps (2021CB042).
References
- Diagnosis and treatment protocol for novel coronavirus pneumonia (trial version 9). Chin J Viral Dis. 2022; 12: 161-9.
- World Health Organization. Novel coronavirus (COVID-19) [EB/OL]; 2022-07-04. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- Min H, Yong Y, Jing F, et al. Epidemic characteristics of COVID-19 patients retested positive for nucleic acid in Chengdu. Public Health China. 2022; 38: 758-61.
- Sookaromdee P, Wiwanitkit V. Persistent COVID-19 symptoms after diagnosis: correspondence. J Infect Public Health. 2022; 15: 877.
- Chen G, Lan M, Chen X, et al. A case-control study of severe risk factors for COVID-19 patients in Fujian Province. Chin J Dis Control. 2021; 25: 1327-31.
- Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020; 586: 113-9.
- Wang Zhonglei, Yang L. Broad-spectrum prodrugs with anti-SARS-CoV-2 activities: strategies, benefits, and challenges. J Med Virol(preaccepted). 2022; 94: 1373-90.
- Lamb YN. Remdesivir: first approval. Drugs. 2020; 80: 1355-63.
- Megan C. A tale of two antiviral targets - and the COVID-19 drugs that bind them. Nat Rev Drug Discov. 2021; 21.
- Wang ZL, Yang LY. Chinese herbal medicine: fighting SARS-CoV-2 infection on all fronts. J Ethnopharmacol. 2021; 270: 113869.
- Yang LY, Wang ZL. Natural products, alone or in combination with FDA-approved drugs, to treat COVID-19 and lung cancer. Biomedicines. 2021; 9: 689.
- Wang ZL, Wang N, Yang LY, Song XQ. Bioactive natural products in COVID-19 therapy. Front Pharmacol. 2022; 13: 926507.
- qiao Y, Wu Huanlin, Sun Haijiao, et al. ’Danxi Xin Method’ to treat phlegm syndrome prescription drug law. World Traditional Chinese Medicine. 2021; 16: 1909-13.
- Hui W, Jin X, Bo P, et al. Analysis of clinical research scheme of TCM intervention in novel coronavirus pneumonia. Chin J Trad Chin Med. 2020; 45: 1232-41.
- Leung EL-H, Pan H-D, Huang YF, Fan XX, Wang WY, et al. The scientific foundation of Chinese herbal medicine against COVID-19. Engineering (Beijing). 2020; 6: 1099-107.
- Jun Z, Tingting S, Weidong L, et al. Association between early treatment with Chinese herbal medicine and clinical outcomes in patients with severe coronavirus disease 2019: A retrospective study. J Trad Chin Med. 2021; 62: 1046-51.
- Yang X, Zhang H, Tang D, et al. Analysis of drug use in prevention period and medical observation period of traditional Chinese medicine for prevention and control of novel coronavirus pneumonia. Chinese modern applied pharmacy. 2021; 38: 985-90.
- Zhen Z, Zhu C, Bin Z. Study on drug use rules of traditional Chinese medicine for treating novel coronavirus pneumonia based on data mining. Chin J Trad Chin Med. 2020; 45: 1248-52.
- Yanling F, Xun M, Xinrong W. Discuss the clinical application and dosage of Glycyrrhiza uralensis. Beijing J Trad Chin Med. 2017; 36: 554-5, 564.
- Li B, Zhao Shipeng, Bo L, et al. Study on the philology and pharmacology of Shennong materia medical. Beijing Trad Chin Med. 2022; 41: 417-20.
- NiuMing SZ, Bo Z, et al. Interpretation of guidelines for network pharmacological evaluation methods. Chin Herb Med. 2021; 52: 4119-29.
- Wang Z, Xin W, Zhang Daiyan, et al. Network pharmacology of Traditional Chinese Medicine:the development of the new era under the guidance of the Guide. Chin J Trad Chin Med. 2022; 47: 7-17.
- Lin Z, Fan W, Yu X, Liu J, Liu P. Research into the mechanism of intervention of SanQi in endometriosis based on network pharmacology and molecular docking technology. Medicine. 2022; 101: e30021.
- Fangyuan W, Dong W, Xianbin K, et al. Mechanism of Salviae Miltiorrhizae Radix et rhizoma-Paeoniae Radix rubra drug pair in treatment of colorectal cancer based on network pharmacology and molecular docking. Chin Herb Med. 2022; 53: 5731-41.
- Shen H, Du H, Zhou H, et al. To investigate the mechanism of Guanxintong capsule in treating heart failure based on network pharmacology and molecular docking. New Chin Med Clin Pharmacol. 2022; 33: 1093-101.
- Wu Jinpin, Na Y, Chen XD, et al. Study on network pharmacology and molecular docking of Daagaoxitang in the treatment of acute pancreatitis. J Immunol. 2022; 38: 737-44 + 752.
- Tragni V, Preziusi F, Laera L, Onofrio A, Mercurio I, et al. Modeling SARS-CoV-2 spike/ACE2 protein–protein interactions for predicting the binding affinity of new spike variants for ACE2, and novel ACE2 structurally related human protein targets, for COVID-19 handling in the 3PM context. EPMA J. 2022; 13: 149-75.
- Xin F, Duan Y, Min B, et al. Objective: to investigate the mechanism of Shenqi Yiwu prescription in treating hepatocellular carcinoma based on network pharmacology and molecular docking and in vivo experiments. Pharmacol Clin Chin Med. 2022; 38: 28-33.
- Khan SA, Lee TKW. Identifying potential pharmacological targets and molecular pathways of Meliae cortex for COVID-19 therapy. Front Immunol. 2023; 14: 1128164.
- Shaban A, Piyush B, Jitendra K, Kant PR, Dharmendra M, et al. Molecular dynamics simulation and docking analysis of NF-κB protein binding with sulindac acid. Bioinformation. 2022; 18: 170-179.
- Li HL, Zhou JP, Deng JM. Li Haili, Z Jian Peng, et al. Therapeutic mechanism of Xiaoqinglong decoction against COVID-19 based on network pharmacology and molecular docking technology. Comb Chem High Throughput Screen. 2022; 25: 2264-77.
- Leung EL-H, Pan HD, Huang YF, Fan XX, Wang WY, et al. The scientific foundation of Chinese herbal medicine against COVID-19. Engineering (Beijing). 2020; 6: 1099-107.
- Kang X, Jin D, Jiang L, Zhang Y, Zhang Y, et al. Efficacy and mechanisms of traditional Chinese medicine for COVID-19: a systematic review. Chin Med. 2022; 17: 30.
- Yang X, Zhang H, Tang DL, et al. Analysis of drug use in prevention period and medical observation period of traditional Chinese medicine for prevention and control of novel coronavirus pneumonia. Chinese modern applied pharmacy. 2021; 38: 985-90.
- Xiaojuan G, Dan Z, Jianjun Z, et al. Textual research of Glycyrrhiza uralensis herb. Chin J Exp Formulae. 2017; 23: 193-8.
- Bo Z, Zhizhou X, Yong S, et al. Investigation and analysis on the proposed new standard system under construction for pharmaceutical packaging materials in Chinese pharmacopoeia. Herald Med. 2022; 41: 1412-6.
- Juan Z, Wejing M, Qinyun B. Research progress of quercetin and its derivatives on prevention and treatment of liver injury and its mechanism. Chin Herb Med. 2021; 52: 7348-57.
- Qiuxia Z, Jiongke C, Lingli W. Experimental study of quercetin inhibiting apoptosis and inflammation of respiratory syncytial virus infected bronchial epithelial cells by upregulation of miR-140-5p. J Immunol. 2022; 38: 883-9.
- Keru L, Guang-qiang HU, WU An-guo, et al. Advances in pharmacological effects of kaempferol and its derivatives on nervous system diseases. 2021; 44: 412-6.
- Xing Y, Hua YR, Shang J, Ge W, Liao J. Traditional Chinese medicine network pharmacology study on exploring the mechanism of Xuebijing Injection in the treatment of coronavirus disease 2019. Chin J Nat Med. 2020; 18: 941-51.
- Yunfeng G, Hongbin D, Linlin L, et al. Advances in the study of post-translational modification of NF-κB family member RelA and its physiological and pathological effects. Sci Life. 2020; 32: 431-8.
- Chatterjee B, Thakur SS. SARS-CoV-2 infection triggers phosphorylation: potential target for anti-COVID-19 therapeutics. Front Immunol. 2022; 13: 829474.
- Khan SA, Lee TKW. Network pharmacology and molecular docking-based investigations of kochiae fructus’s active phytomolecules, molecular targets, and pathways in treating COVID-19. Front Microbiol. 2022; 13: 972576.
- Jaco I, Annibaldi A, Lalaoui N, Wilson R, Tenev T, et al. MK2 phosphorylates RIPK1 to prevent TNF-induced cell death. Mol Cell. 2017; 66: 698-710.e5.
- Liping W, Yan-qi L, Jie C, Wu Li. Study on mechanism of curcumin in treatment of acute pancreatitis based on regulation of PI3K-Akt signaling pathway by miR-198. Chin J Trad Chin Med. 2020; 45: 3707-12.
- Hayashi T, Konishi I. Cancer therapy with decreased SARS-CoV-2 infection rates in cancer patients. Br J Cancer. 2022; 126: 521-2.
- Long Z et al. miR-1 activates NF-kappaB to promote the differentiation of bone marrow mesenchymal stem cells in mouse models of glioma. J Biomater Tissue Eng. 2021; 11: 1327-32.
- Ezeonwumelu IJ, Garcia-Vidal E, Ballana E. Ezeonwumelu Ifeanyi Jude and G.V. Edurne and Ballana ester. JAK-STAT pathway: A novel target to tackle viral infections. Viruses. 2021; 13: 2379.
- Tang Y, Li X, Yuan Y, Zhang H, Zou Y, et al. Network pharmacology-based predictions of active components and pharmacological mechanisms of Artemisia annua l. for the treatment of the novel corona virus disease. BMC Complement Ther 2019. 2022; 22: 1-16.