
Special Article: Plant Biology
Ann Agric Crop Sci. 2023; 8(4): 1140.
Multiple Introduction Events of Cultivated Diospyros Kaki L. of and Native Diospyros Species in Taiwan Inferred by Low Copy Nuclear Markers
Kuan-ting Hsin1#; Ya-Ling Chang5,3#; Chun-Neng Wang1; Jui-Sheng Lai2; Ching-Chang Shiesh3; Huey-Ling Lin3,4*
1Department of Life Science, College of Life Science, National Taiwan University, Taiwan
2Miaoli District Agricultural Research and Extension Station, Council of Agricultural, Taiwan
3Department of Horticulture, National Chung Hsing University, Taiwan
4Department of Horticulture and Innovation and Development Center of Sustainable Agriculture (IDCSA), National Chung Hsing University, Taiwan
5Miaoli District Agricultural Research and Extension Station, Ministry of Agriculture, Taiwan
*Corresponding author: Huey-Ling Lin Department of Horticulture, National Chung Hsing University, 145, Xingda Road, Taichung 40227, Taiwan. Tel: +886-4-22840340; Fax: +886-4-22860574 Email: hllin@dragon.nchu.edu.tw
#These authors have contributed equally to this article.
Received: July 31, 2023 Accepted: August 22, 2023 Published: August 29, 2023
Abstract
Diospyros kaki L. is one of the most important fruits in Taiwan. Cultivated D. kaki cultivars are classified into four types, according to their loss of astringency in ripening fruit and their change in flesh color, namely the Pollination-Constant Non-Astringent (PCNA), the Pollination Variant Non-Astringent (PVNA), the Pollination Variant Astringent (PVA) and the Pollination Constant Astringent (PCA). Recently, persimmon breeders are paying more attention to rootstock selection for successful grafting and agricultural production. To identify suitable rootstock and to trace the origins of extant native Diospyros species in Taiwan, the 10 known native Diospyros species together with four D. kaki cultivars were analyzed by using two low-copy, nuclear-encoded DNA markers, ncpGS and PHYA. There was moderate to strong support for the major nodes within the phylogenetic tree, obtained from both the Maximum-likelihood or the Bayesian inference methods. The constructed D. kaki phylogeny using Maximum Likelihood (ML) and Bayesian Inference (BI) algorithms formed three clades. The most basal clade (clade A) comprised species distributed in the South East Asia , then species distributed in East Asia (clade B), and then D. kaki cultivars (clade C). The extant native Taiwan Diospyros species nested into clade A and B, suggesting a northward migration pattern of native Taiwan Diospyros species. In addition, the species D. oldhamii formed a sister clade with the D. kaki cultivars, suggesting that D. oldhamii may be a compatible rootstock. Lastly, D. kaki cultivars sampled from growers that were acquaintances formed corresponding subclades within clade C (C-1, C-2 and C-3), suggesting a high discrimination ability of the ncpGS and PHYA molecular markers used here.
Keywords: Diospyros kaki, ncpGS, PhyA, phylogeny, persimmon
Introduction
The Diospyros genus contains over 500 species, making it the largest genus in the Ebenaceae family [1]. The members of genus Diospyros are distributed broadly from tropical to temperate regions. Most Diospyros species are distributed in Asia and the Pacific, making Asia and Oceania its center of diversity [2]. Some species are well known for their edible fruits, such as D. kaki, D. lotus and D. virginiana [3,4], while other species are famous for their timber, like D. ebenum, D. mollis and D. dendo. Moreover, leaves and fruits of some Diospyros species have been used as medicine, e.g. D. kaki and D. lotus [5,6].
D. kaki is the most economically important and widely cultivated species in the world. Cultivars of D. kaki are grown in Australia, Brazil, China, Iran, Israel, Italy, Japan, New Zealand, and United States of America [7,8]. The geographical origin of the D. kaki species is not yet known. However, wild D. kaki plants have been observed in forests in China, so D. kaki is believed to have originated in China [9]. According to features of the mature fruit (astringency due to tannins, presence of seeds), cultivars are classified into four types, namely 1) Pollination-Constant Non-Astringent (PCNA), 2) Pollination-Variant Non-Astringent (PVNA), 3) pollination-variant astringent (PVA) and 4) Pollination-Constant Astringent (PCA) [9] (Figure 1). Among these four types, the PCNA and PVNA types lose astringency naturally during fruit growth and become edible at maturity, while PVA and PCA types retain astringency at maturity. Further, in the PVNA cultivars, sufficient pollination can enhance the loss of astringency, but insufficient pollination will make the fruits retain astringency at maturity but may be edible when softened. In contrast, PCNA fruit consistently loses its astringency during fruit development, regardless of pollination efficiency. PCNA-type cultivars are important for commercial production of persimmon worldwide since the fruits lose their astringency naturally and do not require post-harvest astringency processing.
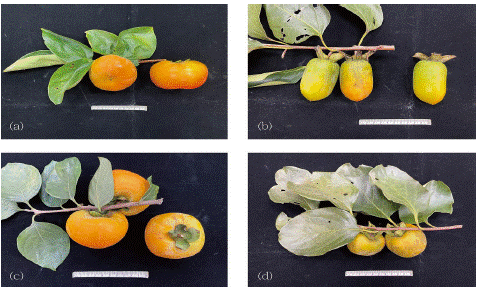
Figure 1: Four types of Diospyros kaki cultivars based on fruit classification. (a). Diospyros kaki cv. Fuyu, PCNA type. Fruit will be sweet whether hard or soft (b). Diospyros kaki cv. Fudegaki, PVNA. Unseeded fruit must soften after harvest to be sweet (c). Diospyros kaki cv. Hirataneanshi, PVA. Fruit remains astringent, except near seeds. (d). Diospyros kaki cv. Bull Heart, PCA. Fruit must become soft to be edible.
Previous phylogenetic studies revealed that two species distributed in Asia, Diospyros glandulosa (2n=2x=30) and Diospyros oleifera (2n=2x=30), are most likely ancestors of D. kaki (2n= 6x= 90) [2,7,8]. Their phylogenies also suggested that D. kaki originated in Asia through polyploidization. Although the cultivar development history is not yet clear, it is proposed that the PCA-type cultivars are thought to have appeared first in China, and then split into PVA-PVNA and PCNA lineages in Japan [10]. According to this hypothesis, PCNA-type accessions exhibit a single origin from a PCA-type progenitor. However, molecular evidence showed multiple origins of both PCNA and non-PCNA types [11]. For example, PCNA-type cultivars in Japan and in China formed clades respectively [11]. Non-PCNA type cultivars in Japan also exhibit paraphyly in their phylogeny [11]. To conclude, the multiple origin hypothesis of PCNA and non-PCNA cultivars in Asia is more likely than a single origin hypothesis as proposed by [10].
Due to lack of sterility of seeds, D. kaki is propagated via grafting of propagated scions onto wild rootstocks. The commonly used rootstocks include D. kaki, D. lotus and D. virginiana [12,13]. In Taiwan, the most common rootstock for D. kaki cultivars is D. japonica. It has been proposed that a closer genetic relationship between scion and rootstock is associated with higher grafting success [14].
There are 10 Diospyros species identified in Taiwan, including Diospyros ferrea, Diospyros japonica, Diospyros kotoensis, Diospyros morrisiana, Diospyros philippensis, Diospyros eriantha, Diospyros vaccinioides, Diospyros maritima, Diospyros oldhamii and Diospyros rhombifolia [15]. By analyzing Random Amplified polymorphic DNA (RAPD) markers, D. kaki clustered with D. oldhamii and D. japonica, suggesting that D. oldhamii and D. japonica may be potentially compatible rootstocks for grafting. However, the genetic relationship among D. kaki cultivars and other native Diospyros species remains unclear.
To reconstruct a phylogeny of closely related taxa or within the same species, low copy nuclear markers are preferable than commonly used non-coding cpDNA markers that have a high genetic variability [16]. For example, eight chloroplast markers, comprising over 8000 bases, were sequenced to resolve the relationship of Diospyros species from New Caledonian, Hawaii and islands in the Indian Ocean [17]. However, low variability and little resolution were shown among those target Diospyros species in their phylogeny. In contrast, two nuclear markers, chloroplast-expressed glutamine synthetase (ncpGS) and phytochrome A (PHYA), comprising about 1900 bases, were be able to resolve the relationship among those New Caledonian, Hawaii and Indian ocean Diospyros species [2]. Thus, ncpGS and PHYA are more satisfactory genetic markers in drawing the relationships among native Diospyros species and cultivars in Taiwan.
In this study, we aimed to define 1) the relationship among native Diospyros species in Taiwan and 2) the relationship among D. kaki cultivars. Two low-copy nuclear markers, ncpGS and PHYA, were isolated from the target species to draw the phylogenetic relationship among the native Diospyros species and the D. kaki cultivars in Taiwan.
Materials and Methods
DNA Extraction, Molecular Marker Amplification and Sequencing
Genomic DNA from 56 samples Diospyros species and cultivars (Table 1) was extracted from leaf samples dried in silica gel following a CTAB method (Doyle & Doyle, 1987). Two nuclear-encoded genes (PHYA and the chloroplast-expressed glutamine synthetase,e ncpGS) were sequenced for phylogeny reconstruction [2]. Initial PCR products of PHYA and ncpGS were obtained using primer pairs from [19,20], respectively. As these primers were not specifi c enough, we sliced the major PCR products from agarose gels and sequenced the major PCR products to be able to design Diospyros-specifi c PHYA PCR primers (phyA_F: GAGCTTGGAAAGGCTTTGTG; phyA_R: CAGCTGCCATCCCACATACT) and ncpGS PCR primers (ncpGS_F: TTCTTGGTCTGGTAGTGGGAAT; ncpGS_R: TGCTTTTTCTAGTCTCGGTATGC).
Taxon
collection site
Collection codes
Accession no. for ncpGS
Accession no. for PHYA
Diospyros eriantha Champ. ex Benth.
Wufeng, TaichungΔ
ON351647
ON351614
Diospyros ferrea (Lour.) A.Chev.
Wufeng, TaichungΔ
ON351643
ON351612
Diospyros japonica Sieb.& Zucc.
Heping, TaichungΔ
ON351670
ON351634
Diospyros kotoensis T. Yamaz.
Wufeng, TaichungΔ
ON351644
none
Diospyros maritima Blume
Wufeng, TaichungΔ
ON351645
ON351613
Diospyros morrisiana
Zhongzheng, TaipeiΔ
ON351678
ON351642
Diospyros oldhamii
Anle, KeelungΔ
ON351677
ON351641
Diospyros philippensis (Desr.) Gürke
Wufeng, TaichungΔ
ON351646
none
Diospyros rhombifolia Hemsl.
Wufeng, TaichungΔ
ON351648
ON351615
Diospyros vaccinioides Lindl.
Wufeng, TaichungΔ
ON351649
ON351616
Diospyros andamanica
Thailand*
KF291447
KF291624
Diospyros apiculata
Thailand*
KF291449
KF291626
Diospyros borbonica
Reunion*
KF291453
KF291630
Diospyros discolor
Thailand*
KF291473
KF291650
Diospyros dictyoneura
Thailand*
KF291471
KF291648
Diospyros diepenhorstii
Thailand*
KF291472
KF291649
Diospyros glandulosa
Thailand*
KF291492
KF291669
Diospyros kupensis
Cameroon*
KF291501
KF291678
Diospyros lotus
Living coll.
Kew 1882-3501*KF291507
KF291684
Diospyros macrocarpa
New Caledonia*
KF291509
KF291686
Diospyros mollis
Thailand*
KF291522
KF291699
Diospyros oubatchensis
New Caledonia*
KF291529
KF291706
Diospyros rhodocalyx Kurz
Thailand*
KF291567
KF291744
Diospyros samoensis
Cult. Hawaii Bot Garden*
KF291569
KF291746
Diospyros sp.(Madagascar)
Madagascar*
KF291571
KF291748
Diospyros texana Scheele
Middle America*
KF291575
KF291752
Diospyros venosa
Thailand*
KF291596
KF291773
Diospyros virginiana L.
USA*
KF291612
KF291789
Diospyros vera
Central African Republic*
KF291597
KF291774
Diospyros winitii Fletcher
Thailand*
KF291615
KF291792
Styrax officinalis
Indonesia*
KF291623
KF291800
Euclea undulata
Living coll. HBV*
KF291620
KF291797
Diospyros kaki cv. Amahyakume
Heping, TaichungΔ
G21
ON351663
ON351627
Diospyros kaki cv. Aoso
Heping, TaichungΔ
G18
ON351660
none
Diospyros kaki cv. Bull Heart
Kungkuan, MiaoliΔ
G08
ON351650
ON351617
Diospyros kaki cv. Diamond Bull Heart
Heping, TaichungΔ
G11
ON351653
ON351619
Diospyros kaki cv. Fudegaki
Dongshi, TaichungΔ
G09
ON351651
none
Diospyros kaki cv. Fuyu
Heping, TaichungΔ
G25
ON351667
ON351631
Diospyros kaki cv. Fuyu
Heping, TaichungΔ
G19
ON351661
ON351625
Diospyros kaki cv. Fuyu
Heping, TaichungΔ
G17
ON351659
ON351624
Diospyros kaki cv. Fuyu
Sinyi, NantouΔ
G23
ON351665
ON351629
Diospyros kaki cv. Fuyu
Heping, TaichungΔ
G22
ON351664
ON351628
Diospyros kaki cv. Fuyu
Heping, TaichungΔ
G29
ON351671
ON351635
Diospyros kaki cv. Fuyu
Dongshi, TaichungΔ
G10
ON351652
ON351618
Diospyros kaki cv. Hana-gosho
Heping, TaichungΔ
G16
ON351658
none
Diospyros kaki cv. Hirataneanshi
Wufeng, TaichungΔ
G13
ON351655
ON351621
Diospyros kaki cv. Maekawa-Jirow
Sinyi, NantouΔ
G24
ON351657
ON351623
Diospyros kaki cv. Maekawa-Jirow
Heping, TaichungΔ
G15
ON351666
ON351630
Diospyros kaki cv. Soshu
Heping, TaichungΔ
G14
ON351656
ON351622
Diospyros kaki cv. Syh Jou
Dongshi, TaichungΔ
G12
ON351654
ON351620
Diospyros kaki cv.Taishuu
Sinyi, NantouΔ
G26
ON351662
ON351626
Diospyros kaki cv.Taishuu
Heping, TaichungΔ
G20
ON351668
ON351632
Diospyros kaki cv. Fuyu
Wufeng, HsinchuΔ
C017
ON351671
ON351635
Diospyros kaki cv. Fuyu
Wufeng, HsinchuΔ
C019
ON351672
ON351636
Diospyros kaki cv. Fuyu
Wufeng, HsinchuΔ
J26
ON351673
ON351637
Diospyros kaki cv. Fuyu
Wufeng, HsinchuΔ
G44
ON351675
ON351639
Diospyros kaki cv.Tonewase
Heping, TaichungΔ
G27
ON351669
ON351633
Δ: sequences obtained in this study. *: sequences obtained from Turner et al. (2013).
Table 1: Sequences of Diospyros species and D. kaki cultivars used in this study.
The PCR reactions were made up to a total volume of 25 μl, containing 13 μl of 2x Ampliqon master mix Red (Ampliqon, Denmark), 0.25 μl each primer at 2 μM, 10.5 μl ddH2O and 1 μl template DNA at 20 ng/μl. The PCR profile for amplification of both the PHYA and ncpGS markers was an initial denaturing for 3 min at 94°C followed by 37 cycles of 30 s at 94°C, 30 s at 56°C, and 1 min at 72°C with a final extension for 10 min at 72°C, . All PCR products were examined by 1.0% (w/v) agarose gel electrophoresis, and then the band of the expected size was sent for sequencing by ABI 3700 automatic sequencer (manufacturer). All sequences were examined by Sequence Scanner v1.0 first and then aligned for phylogenetic reconstruction.
Phylogeny Reconstruction
The sequences of PHYA and ncpGS of the native Diospyros species in Taiwan were used as an outgroup, considering the close phylogenetic relationship of native species to cultivars. In addition, representatives of genus Diospyros were chosen based on [2] (Details see Table 1). The sequences were automatically aligned using MUSCLE [21] in MEGA v.6 [22]. After alignment, the PHYA and ncpGS datasets were concatenated to obtain all genetic information of these two nuclear markers. For both Maximum Likelihood (ML) and Bayesian Inference (BI) phylogeny, the web interface PhyML 3.0 was applied [23]. The best-fit nucleotide substitution model of each dataset was evaluated by Smart Model Selection (SMS), which was implementing in PhyML 3.0 [24]. The GTR + G model was also the best-fit model for concatenated PHYA and ncpGS dataset. The phylogeny is visualized in FigTree v1.4.3. [25].
Results
Relationships of Diospyros Species and Cultivars in Taiwan
To infer the relationships among the native Taiwan Diospyros species and the currently grown Diospyros kaki cultivars, we reconstructed the phylogeny using sequences isolated from the target species (number of samples) as well as sequences obtained from the NCBI database of other Diospyros species (listed in Table 1, number of dB data). Only nodes with a Maximum Likelihood (ML) and Bayesian Inference (BI) probability value over 0.7 (BI>0.7 or aLRT > 0.7) were regarded as a reliable clade. According to our criteria, the native Taiwan Diospyros species were distributed into two clades (Figure 2). Sequences of D. philippensis and D. kotoensis were grouped with published sequences from species originating in Indo-China, Indonesia and Philippines (Figure 2, Clade A; BI: 0.98, aLRT: 1), while D. ferrea, D. rhombifolia, D. maritime, D. eriantha, D. vaccinioides, D. morrisiana, D. japonica and D. oldhamii grouped together (Figure 2, Clade B; BI: 1, aLRT: 1). All D. kaki cultivars formed a single clade, topologically the sister clade of D. oldhamii (Figure 2, clade C). D. japonica is a Taiwan native persimmon species used as the rootstock of D. kaki cv. Fuyu. The test results showed that D. kaki cv. Fuyu was closely related to cv. Aoso. On the other hand, the native Taiwan persimmon species D. oldhamii is closely related to D. kaki cv. Fuyu, while D. kaki cv. Fuyu is distantly related to D. japonica.
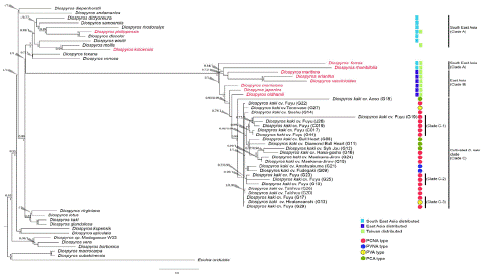
Figure 2: Phylogeny inferred from PHYA and ncpGS sequences. Numbers above a branch represent posterior probability estimated using Bayesian Inference (BI) algorithm; Numbers below a branch represent the value estimated using Maximum Likelihood (ML) algorithm. Species name colored in red represent native Diospyros species distributed in Taiwan. Light blue squares denote Diospyros species distributed in South East Asia; Blue squares denote Diospyros species distributed in East Asia; Green squares denote Diospyros species distributed in Taiwan. Red circle represents PCNA-type cultivars; green circle represents PCA-type cultivars; blue represents PVNA-cultivars; and yellow represents PVA-type cultivars.
According to the classification based on fruit characteristics, D. kaki is divided into four types: PCNA, PVNA, PVA, and PCA. The two sampled PVNA types formed a single clade, namely D. kaki cv. Amahyakume and D. kaki cv. Fudegaki. It is noteworthy that the PCNA, PVA, and PCA types of D. kaki did not form single clades based on fruit ripening characteristics. PCNA types were distributed among at least four subclades within clade C, within which both PCA and PVA types were also nested.
Fuyu is the most grown cultivar in Taiwan and forms three subclades within clade C, namely C-1, C-2, and C-3 (Figure 2). Fuyu samples G19, J26, C019, C017, and G44 are in subclade C-1 and were collected from Heping District, Taichung City (G19) and Wufeng Township, Hsinchu County (J26, C019, C017, and G44), respectively. Samples G23, G25, and G10 are subclades of Clade C-2 and were collected from Xinyi Township (G23 and G25), Nantou County and Dongshi District, Taichung City (G10), while G17 and G29 are in subclade C-3 and were both collected from Heping District, Taichung City, but cultivated by different farmers.
Discussion
By sampling all known native Diospyros species and the most widely grown D. kaki cultivars in Taiwan, we reconstructed a comprehensive phylogeny which allowed the inference of two potential dispersal routes of native Diospyros species and indicted that multiple introduction events of cultivated D. kaki cultivars had occurred. Native Taiwan Diospyros species formed two distinct clades, including the South East Asia clade and the East Asia clade (Figure 2 & Table 2). In addition, the East Asia clade nested within the South East Asia clades. Extant native Diospyros species in Taiwan within these two clades may hint that these species in Taiwan entered their current habitat through two colonization routes. The evidence showing that the East Asia clades nested within the South East Asia clade indicates that a dispersal route from Indo-China through the Asian continent to Taiwan is likely. A second dispersal route would have been through the Philippine Archipelago to Taiwan. The Quaternary, glacial and inter-glacial periods, caused by climatic oscillation, lead to repeated lowering and raising of sea levels in East Asia [26,27]. During a glacial period, submerged sea floor may have be exposed due to lower sea levels, forming land bridges connecting isolated landmass [28]. Land bridges provide opportunity allowing terrestrial plant species to migrate between the Asian mainlands and adjacent islands, like Taiwan and Japan. For example, a haplotype (H9) of Quercus variabilis was identified from both the Asia continent and some adjacent islands (including Taiwan and Japan), suggesting plant migration through land bridges [29]. The phylogeny of some herbaceous plants also support landbridging. Lysionotus pauciflorus exhibits similar distribution pattern to Q. variabilis, inhabiting the Asia continent and adjacent islands [30]. and a, few individuals from both Asia and adjacent islands formed a monophyletic clade with a calculated time of divergence during the maximal glacial period [30]. In our study, Diospyros species belonging to the East Asia clade (including D. maritima, D. eriantha, D. vaccinioides, D. morrisiana, D. japonica and D. oldhamii) were distributed in both the Asia continent and the adjacent islands of Taiwan and Japan), suggesting migration through exposed East Asia seafloor likely. The aforementioned second possible dispersal route through the Philippine archipelago to Taiwan has no historical geological event (glacial-interglacial period) that connected the Philippine archipelago and Taiwan [31], making dispersal from the Philippine archipelago through a land bridge to Taiwan unlikely. One possible scenario is through human activity. In South East Asia, Austronesian-speaking people migrate among islands using boats. To build a reliable boat, solid timber is necessary. Austronesian-speaking people in Taiwan, like the Tao people in Lanyu, use timber from D. philippensis to build a boat [32]. Austronesian-speaking people carrying necessary and useful plants with them is traceable. For example, paper mulberry (Broussonetia papyrifera) was carried by Austronesian-speaking people for its bark [33,34]. Molecular phylogeny conducted using cpDNA of B. papyrifera offers genetic evidence supporting the transport of this species by Austronesian-speaking people [35]. the migratory transport of D. philippensis by Austronesian-speaking people requires further study.
Species
Distribution range
Diospyros eriantha
China to Nansei-shoto and W. & Central Malesia, Taiwan
Diospyros ferrea
Tropical Africa to SW. Pacific.
Diospyros japonica
S. China, Central & S. Japan to Nansei-shoto, Taiwan
Diospyros kotoensis
Taiwan
Diospyros maritima
China (Yunnan) to Nansei-shoto and N. Australia, Taiwan
Diospyros morrisiana
Central & S. Japan to SE. China, Taiwan
Diospyros oldhamii
Nansei-shoto to Central & E. Taiwan.
Diospyros philippensis
E. & S. Taiwan to E. Borneo
Diospyros rhombifolia
SE. China, Taiwan
Diospyros vaccinioides
China (Guangxi, Guangdong) to Hainan, Taiwan
Table 2: Distribution range of native Diospyros species in Taiwan.
Taxonomy and genetic affinity are prerequisites for grafting affinity [15]. The greater the genetic distance between the rootstock and the scion, the lesser the chance of forming a successful graft union [36,37]. To achieve a successful grafting combination, the genetic distance between the two ranks as intraclonal > interclonal > intraspecific > interspecific > intrageneric > intergeneric > intrafamilial [38]. An autografted combination between rootstock and scion indicates close similarity in their taxonomy and a good affinity. If the rootstock and scion are different species but belong to the same genus, the graft is more or less compatible. While graft combinations are generally rarely compatible, those between families are incompatible and unsuccessful [39]. D. lotus was used as the rootstock for cultivating persimmon in California, whose grafting incompatibility resulted in dwarfism and came into bearing early of persimmon, and declined within a few years. On the other hand, D. virginiana, native to the United States, is an ideal rootstock for persimmon. Compared with D. lotus and D. virginiana, the parent species of cultivated persimmon, D. kaki is even more compatible with grafting [40].
The relationships of 19 kiwifruit species were analyzed through the chloroplast genome. The greater the pairwise genetic distance between the rootstock (Actinidia deliciosa) and the xenografted kiwifruit, the lower the survival rate of the scion [15]. In this study, the sequencing of two marker genes confirmed that D. kaki, D. japonica, cultivar Aoso, and D. oldhamii belong to the same genus. It also confirmed that D. kaki and cultivar Aoso are the same species. Considering their close kinship, D. oldhamii and D. kaki may show a better affinity after grafting, and D. oldhamii should be tested for agricultural use as a rootstock in the Taiwan persimmon industry.
It is hypothesized that PCA-type cultivars of D. kaki are first in China, and then split into the PVA-PVNA type and PCNA lineages in Japan [10]. However, this is not supported by recent molecular studies [7,9,11]. An extensive analysis of the genetic diversity of 146 D. kaki cultivars, obtained from China, Korea and Japan, was surveyed with 496 distinct AFLP markers [1]. By using clustering algorithm, these 146 cultivars were assigned into three clusters corresponding to geological regions. In addition, relationships among the studied D. kaki cultivars did not follow horticultural classification [11]. According to molecular data, the primary character separating them appears to be their origin instead of their astringency level. In our study, extant D. kaki cultivars in Taiwan exhibit polyphyly (Figure 2), similar to previous studies [7,9,11,41]. Unlike PCNA-type cultivars forming a monophyletic clade in previous studies, PCNA-type cultivars in Taiwan formed at least three sub-clades (Figure 2). These three sub-clades may suggest at least three origins of Taiwanese PCNA-type D. kaki cultivars. One possible reason behind multiple origins may associate with human behavior, e.g. scion sharing among farmers, since Grafting is the only way to grow cv. Fuyu. In subclade C-1, G19 was collected from Heping, Taichung, but others (including C019, C017 and G44) were collected 100 km away in Wufeng, Hsinchu. After consulting local farmers, samples collected from Wufeng originated from farmers in Heping. Scion sharing could also be found in the C-2 and C-3 sub-clades. Within the C-2 sub-clade, samples G10 (Dongshi, Taichung), G23 and G25 (Sinyi, Nantou) are close to each other. Lastly, G17 and G29 were collected from Heping, Taichung and are grown by acquaintances. To conclude, sequencing of the genes ncpGS and PhyA is able to identify the origin of D. kaki cv. Fuyu cultivars in Taiwan.
References
- Duangjai S, Wallnöfer B, Samuel R, Munzinger J, Chase MW. Generic delimitation and relationships in Ebenaceae sensu lato: evidence from six plastid DNA regions. Am J Bot. 2006; 93: 1808-27.
- Turner B, Munzinger J, Duangjai S, Temsch EM, Stockenhuber R, Barfuss MHJ, et al. Molecular phylogenetics of New Caledonian Diospyros (Ebenaceae) using plastid and nuclear markers. Mol Phylogenet Evol. 2013; 69: 740-63.
- Heywood V, Brummitt CA, Seberg O. Flowering plant families of the World 2007.
- Wallnöfer B. The biology and systematics of Ebenaceae: a review. Annalen Naturhistorischen Museums Wien S B Bot Zool. 2001; 103: 485-512.
- Rauf A, Uddin G, Siddiqui BS, Muhammad N, Khan H. Antipyretic and antinociceptive activity of Diospyros lotus L. in animals. Asian Pac J Trop Biomed. 2014; 4: S382-6.
- Xie C, Xie Z, Xu X, Yang D. Persimmon (Diospyros kaki L.) leaves: a review on traditional uses, phytochemistry and pharmacological properties. J Ethnopharmacol. 2015; 163: 229-40.
- Yonemori K, Kanzaki S, Honsho C, Akagi T, Parfitt DE. Phylogeny and cultivar development of Diospyros kaki: a survey based on molecular analyses. Adv Hortic Sci. 2008; 22: 261-8.
- Yesiloglu T, Çimen B, Incesu M, Yilmaz B. Genetic diversity and breeding of persimmon; 2018.
- Yonemori K, Sugiura A, Yamada M. Persimmon genetics and breeding. Plant Breed Rev. 2000; 19: 191-225.
- Kanzaki S. The Origin and Cultivar Development of Japanese persimmon (Diospyros kaki Thunb.). Nippon Shokuhin Kagaku Kogaku Kaishi. 2016; 63: 328-30.
- https://link.springer.com/article/10.1007/s11295-015-0848-z
- Izhaki A, Yitzhak Y, Blau T, David I, Rotbaum A, Riov J, et al. Rooting of cuttings of selected Diospyros virginiana clonal rootstocks and bud growth in rooted cuttings. Sci Hortic. 2018; 232: 13-21.
- Yakushiji H, Sugiura H, Yamasaki A, Azuma A, Koshita Y. Tree growth, productivity, and fruit quality of ’Fuyu’ persimmon trees onto different dwarfing rootstocks. Sci Hortic. 2021; 278.
- Li D, Han F, Liu X, Lv H, Li L, Tian H, et al. Localized graft incompatibility in kiwifruit: analysis of homografts and heterografts with different rootstock & scion combinations. Sci Hortic. 2021; 283: 110080.
- Lu S-Y. Yang y-p. A revision of the Ebenaceae of Taiwan. 1988; 3: 131-40.
- Krak K, Álvarez I, Caklová P, Costa A, Chrtek J, Fehrer J. Development of novel low-copy nuclear markers for Hieraciinae (Asteraceae) and their perspective for other tribes. Am J Bot. 2012; 99: e74-7.
- Duangjai S, Samuel R, Munzinger J, Forest F, Wallnöfer B, Barfuss MHJ, et al. A multi-locus plastid phylogenetic analysis of the pantropical genus Diospyros (Ebenaceae), with an emphasis on the radiation and biogeographic origins of the New Caledonian endemic species. Mol Phylogenet Evol. 2009; 52: 602-20.
- Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987; 84: 9054-8.
- Mathews S, Donoghue MJ. The root of angiosperm phylogeny inferred from duplicate phytochrome genes. Science. 1999; 286: 947-50.
- Yockteng R, Nadot S. Phylogenetic relationships among Passiflora species based on the glutamine synthetase nuclear gene expressed in chloroplast (ncpGS). Mol Phylogenet Evol. 2004; 31: 379-96.
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32: 1792-7.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30: 2725-9.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010; 59: 307-21.
- Lefort V, Longueville JE, Gascuel O. SMS: smart model selection in PhyML. Mol Biol Evol. 2017; 34: 2422-4.
- Rambaut A. FigTree v1. 2012; 4.
- Ohmura A, Shi Y, editors. The Quaternary glaciations and environmental variations in China. J Glaciol. 2007. ISBN 7 5375 3247 8, hardback. 190.00 yuan. 2006: 53: 518.
- Zheng Y, Yu G, Wang S, Xue B, Liu H, Zeng X. Simulations of LGM climate of East Asia by regional climate model. Sci China Ser D-Earth Sci. 2003; 46: 753-64.
- Osozawa S, Shinjo R, Armid A, Watanabe Y, Horiguchi T, Wakabayashi J. Palaeogeographic reconstruction of the 1.55 Ma synchronous isolation of the Ryukyu Islands, Japan, and Taiwan and inflow of the Kuroshio warm current. Int Geol Rev. 2012; 54: 1369-88.
- Chen D, Zhang X, Kang H, Sun X, Yin S, Du H, et al. Phylogeography of Quercus variabilis based on chloroplast DNA sequence in East Asia: multiple glacial refugia and mainland-migrated island populations. PLOS ONE. 2012; 7: e47268.
- Kokubugata G, Hirayama Y, Peng C-I, Yokota M, Möller M. Phytogeographic aspects of Lysionotus pauciflorussensu lato (Gesneriaceae) in the China, Japan and Taiwan regions: phylogenetic and morphological relationships and taxonomic consequences. Plant Syst Evol. 2011; 292: 177-88.
- Voris HK. Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J Biogeography. 2000; 27: 1153-67.
- Hung SF, Roan SF, Chang TL, King HB, Chen IZ. Analysis of aroma compounds and nutrient contents of mabolo (Diospyros blancoi A. DC.), an ethnobotanical fruit of Austronesian Taiwan. J Food Drug Anal. 2016; 24: 83-9.
- Matisoo-Smith EA. Tracking Austronesian expansion into the Pacific via the paper mulberry plant. Proc Natl Acad Sci U S A. 2015; 112: 13432-3.
- Wu C, Rolett B. Prehistoric maritime cultures and seafaring in east Asia. 2019.
- Payacan C, Moncada X, Rojas G, Clarke A, Chung KF, Allaby R, et al. Phylogeography of herbarium specimens of asexually propagated paper mulberry [Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae)] reveals genetic diversity across the Pacific. Ann Bot. 2017; 120: 387-404.
- Dogra K, Kour K, Kumar R, Bakshi P, Kumar V. Graft-incompatibility in horticultural crops. Int J Curr Microbiol Appl Sci. 2018; 7: 1805-20.
- Rasool A, Mansoor S, Bhat KM, Hassan GI, Baba TR, Alyemeni MN, et al. Mechanisms underlying graft union formation and rootstock scion interaction in horticultural plants. Front Plant Sci. 2020; 11: 590847.
- Andrews PK, Graft SM C. Incompatibility. Hortic Rev. 1993: 183-232.
- Mudge K, Janick J, Scofield S, Goldschmidt EE. A history of grafting. Hortic Rev. 2009: 437-93.
- Hodgson RM. Rootstocks for the oriental persimmon. Proc Am Soc Hortic Sci. 1939; 37: 338-9.
- Yonemori K, Honsho C, Kanzaki S, Ino H, Ikegami A, Kitajima A, et al. Sequence analyses of the ITS regions and the matK gene for determining phylogenetic relationships of Diospyros kaki (persimmon) with other wild Diospyros (Ebenaceae) species. Tree Genet Genomes. 2008; 4: 149-58.