Abstract
Background: The objective of this study was to compare the genomic features of TGF-β1 and MTHFR between patients with Osteoarthritis (OA) and healthy controls in Kanpur, India.
Methods: This study collected blood samples from 220 participants, including 100 OA patients and 120 healthy individuals, and analysed them use genetic polymorphism analysis techniques. The differences in the genetic makeup of TGF-β1 and MTHFR genes between the two groups were determined by using PCR techniques and statical analysis.
Results: The results showed significant variations in the expression levels of the TGF-β1 gene in OA patients compared to healthy controls, suggesting its potential role in the pathogenesis of OA. However, the MTHFR 1298A>C SNP was not significantly associated with OA disease in any genetic model, indicating that it is not associated with the pathogenesis of OA.
Conclusions: This study highlights the importance of further research to investigate the underlying mechanisms of TGF-β1 and MTHER, and its potential as a therapeutic target for the disease.
Keywords: Pathogenesis; Therapeutic targets; Genomic comparison; TGF-β1; MTHFR
Abbreviations: OA: Osteoarthritis; TGF-β1: Transforming Growth Factor-beta 1; MTHFR: Methylenetetrahydrofolate Reductase; SNP: Single Nucleotide Polymorphism; PCR: Polymerase Chain Reaction; CI: Confidence Interval; OR: Odds Ratio; BMI: Body Mass Index; HAQ: Health Assessment Questionnaire; DAS-28: Disease Activity Score 28; CRP: C-reactive Protein; ECM: Extracellular Matrix; K-L: Kellgren and Lawrence; VAS: Visual Analog Scale; ACR: American College of Rheumatology; DNA: Deoxyribonucleic Acid; PCR: Polymerase Chain Reaction; ARMS PCR: Amplification Refractory Mutation System Polymerase Chain Reaction; CT: Cytosine to Thymine; AT: Adenine to Thymine; SNPs: Single Nucleotide Polymorphisms; BP: Blood Pressure
Introduction
With more than 100 million people worldwide, Osteoarthritis (OA) is a prevalent rheumatologic disorder that usually affects the joints of hands, feet, knees, and hips. The prevalence rates of knee OA range from 7.5% (in China) to 25% (in northern Pakistan) and 22-39% in India, where it is the second most common rheumatologic problem [1,2]. The degeneration of cartilage occurs due to the direct rubbing of bones during movement and it causes pain, discomfort, swelling, and loss of motion [1,3]. Knee OA is worst affected in people aged 65 years and above [4]. The exact pathogenesis behinds OA remains unclear, but it is believed that it is a multifactorial disease influenced by both genetic and environmental factors. Several genes have been implicated in the development of OA and it accounts for more than 20% of OA's heritability. Most of them are located in noncoding regions of the genome, where they are presumed to regulate the expression of target genes [5,6]. Cartilage DNA methylation changes in cis have been shown to correlate with a large number of risk variants which indicate that epigenetics may play a role in MTHER gene expression [1]. Among the genetic factors, transforming growth factor-beta 1 (TGF-β1) and Methylenetetrahydrofolate Reductase (MTHFR) are two important genes that have been implicated in OA susceptibility and progression [7]. Studies have shown that TGF-β1 signalling is required for the formation of articular cartilage during early joint development but may also be involved in joint destruction [7]. Additionally, TGF-β has been implicated in abnormal bone remodelling and cartilage degeneration in OA [8,9]. Genetic variations of the MTHFR gene link to the incidence of spinal osteophyte formation [10]. However, studies report conflicting results on whether MTHFR gene polymorphism is associated with early primary knee osteoarthritis or not [11]. The present study is an important addition to the ongoing research on osteoarthritis, specifically by focusing on the Kanpur District of Uttar Pradesh State in India. The primary objective of the study is to investigate the association of TGF-β1 and MTHFR genes with OA disease, as well as to explain the amplification of PCR and polymorphism of these genes. The results of this study are expected to be useful for the pharmaceutical industry in developing new medications for OA.
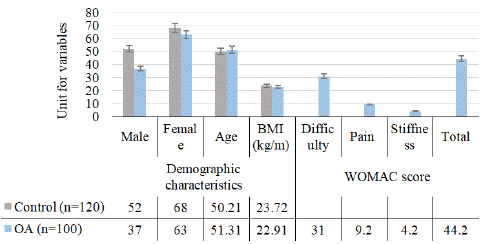
Figure 1: Graphical representation of demographic characteristics of healthy controls and patients.
Materials and Methods
Inclusion and Exclusion Criteria for Control and Subject
This study involved the selection of control subjects according to strict criteria of inclusion and exclusion. The controls were free of any clinical manifestations and radiological evidence of joint pain, crepitus, or reduction of joint space on X-ray. Normal healthy individuals were chosen from the medical college, departmental staff, aged between 30 and 60 (50.21±2.10) years with a male (89) to female (131) ratio of ≈ 1:1.47. The selection criteria for the study population were defined to include patients with OA in the knee joint. The radiological grading of the Kellgren and Lawrence (K-L) score was used to screen OA patients. The evaluation of OA patients was conducted based on three different criteria: the WOMAC (total=44.2±9.8) score, the VAS (4.7±1.4) score, and the American College of Rheumatology (ACR) classification. Only patients who met six or more criteria were included in the study and these characteristics are knee pain (asymmetrical) lasting more than six months, stiffness (less than 30 minutes), swelling, cripitus, tenderness on the medical side of the joint, X-ray with more than one-third decrease (Grade II-78 and Grade III-22) in joint space and/or presence of osteophytes, and decreased range of motion in the knee joint. Patients with normal ligament stability (anterior cruciate, posterior cruciate) were included. The inclusion criteria for the diagnosis of OA also included the duration of symptoms (3.3±1.49 years), range of movements (0-140/42±20.2), VAS pain on movement (4.7±1.4 cm) and WOMAC score of patients (difficulty 31.0±8.1; pain 9.2±2.2; stiffness 4.2±1.3 and total 44.2±9.8), which are depicted in Fig. 1. In the study, only normal weight, BMI 23.72±0.78 for control and BMI 22.91±0.62 for the subject, was selected. Within a range of 80-120, BP is considered normal and was considered in this study [12]. To gather data regarding the patient’s medical history, demographic characteristics, awareness of the disease, and treatment adherence, a structural Health Assessment Questionnaire (HAQ) was administered to all patients via a personal interview. The questionnaire was modelled on DAS-28 with a C-Reactive Protein (CRP) score, and it was appropriately adjusted to include queries about disease awareness. The exclusion criteria for both groups were defined to exclude patients with infectious diseases, such as diabetes mellitus, hypertension, thyroid dysfunction, neurological disorders, cancer, and any other forms of arthritis. Finally, the study was conducted in accordance with ethical standards, as it received approval from the Institutional Ethics Committee (CSJMU/BSBT/BT/EC-20), and all patients, including both controls and OA subjects, provided written informed consent.
DNA Isolation
DNA isolation from blood tissue was performed by using a standard phenol-chloroform protocol [13]. Blood (600 μl) was washed in TE buffer and centrifuged (20°C; 8500 rpm; 10 min) [14]. The pellet was homogenized with SET buffer (pH 8) twice and centrifuged (20°C; 8500 rpm; 10 min). This step was repeated twice; 1 ml TE buffer (pH 8), 600 μl 10% SDS and 5 μl of 20 mg/ml proteinase K were added to the pellet and incubated overnight at 37°C [15]. Saturated phenol:chloroform:isoamyl alcohol (25:24:1) was added to the tube, and the tube was centrifuged at 20°C and 8500 rpm for 10 min. The supernatant was removed, and the pellet was resuspended in sodium acetate and chilled absolute alcohol and incubated for 2 hours. Centrifugation at 4°C; 8500 rpm; 10 min was carried out [16]. The DNA pellet was then washed twice with 70% alcohol and resuspended in TE buffer (pH 8) [17]. Finally, a 0.8% TAE agarose gel was used to check the integrity of DNA [17]. The quantity and purity of DNA were checked by measuring the OD at 260 nm [18,19]. The concentration of DNA was obtained by the following formula: The concentration of DNA = The quality and purity were confirmed by 0.8% agarose gel electrophoresis in 1X TBE buffer (Green & Sambrook, 2019; How Do I Determine the Concentration, Yield and Purity of a DNA Sample?, n.d.).
ARMS PCR
ARMS PCR is a relatively economical method for SNP genotyping involving a single PCR followed by agarose gel electrophoresis [21,22]. All reagents used for PCR amplification [23] (dNTPs, Taq polymerase, PCR buffer and primers) were obtained from Bangalore Genei (India), Genetix Biotech Asia Pvt Ltd. and Fermentas (Lithuania). Cytosine to Thymine (CT) in the transition dinucleotide repeat of TGF-β1 at positions 869 and 1298 (T869C and A1298C, respectively) positioned in the signal sequence region of TGF-β1 is related to an elevated prevalence of spinal osteophytosis and ossification of the posterior longitudinal ligament [24]. The following primers were used: forward primer 5'-CAAGCAGAGTACACACAGCA-3' and reverse primer 5'-GATGCTGGGCCCTCTCAAGC-3' [25]. Furthermore, MTHER genotypes for dinucleotide Adenine-Thymine (AT) repeats in humans are responsible for the production of functional methylenetetrahydrofolate enzyme production [21]. The polymorphism was genotyped with forward primer 5'-TGAACAGGTGGAGGCCAGCCTCT-3' and reverse primer 5'-AGGACGGTGCGGTGAGAGTH-3' [26]. PCR conditions were used as standardized in the lab. Polymorphic alleles were detected for the MTHFR and TGF-β1 gene polymorphisms according to the length of the PCR product, and three genotypes were assigned to the individuals according to the positions of the dominant and recessive as follows: homozygous recessive genotype (CC), heterozygous recessive (CA), and homozygous dominant (AA) [27].
Results
The association between the TGF-β polymorphism and OA susceptibility was assessed by the Odds Ratio (OR) with a 95% Confidence Interval (CI). The OR at 95% CI for TGF-β C869T was calculated for five genetic models: CC versus CT + TT in a dominant model, TT versus CC + CT in a recessive model, CC versus TT in a codominant I model, CC versus CT in codominant II models and C versus T in the allelic model (Table 1). The ORs for the C869T polymorphism were 17.6 (95% CI: 8.01-38.18, p<0.0001) in the dominant model, 1.85 (95% CI: 0.92-3.72, P=0.1008) in the recessive model, 39.11 (95% CI: 14.74-103.69, p<0.0001) in the codominant I model, 8.38 (95% CI: 3.42-20.54, p<0.0001) in the codominant II model and 0.21 (95% CI: 0.13-0.33, p<0.0001) in the allelic model. These results indicate that the SNP TGF-β 869 C>T was significantly associated with OA disease in the dominant model. The association between the MTHFR polymorphism and OA susceptibility was also assessed by the OR with a 95% CI. The OR at 95% CI for A1298C was calculated for five genetic models: AA versus AC + CC in a dominant model, CC versus AA + AC in a recessive model, AA versus CC in a codominant I model, AA versus AC in a codominant II model (Table 2) and A versus C in the allelic model (Table 1). The ORs for the A1298C polymorphism were 1.71 (95% CI: 0.93-3.15, P=0.092) in the dominant model, 2.26 (95% CI: 0.68-7.5, P=0.263) in the recessive model, 2.74 (95% CI: 0.8-9.39, P=0.150) in the codominant I model, 1.96 (95% CI: 1.05-3.65, P=0.042) in the codominant II model and 0.74 (95% CI: 0.46-1.21, P=0.267) in the allelic model. These results indicate that the SNP MTHFR 1298A>C was not significantly associated with OA disease in any of the genetic models.
Control N (%)
Patient N (%)
Odds ratio
P value
Pearson value
TGF β-1 gene -
Genotypic distribution
TT
88(74.6)
10(14.3)
3.192
(2.111-4.827)<0.0001
74.307
CT
9(7.6)
40(57.1)
CC
21(17.8)
20(28.6)
T
185(78.4)
60(42.9)
0.21
(0.13-0.33)<0.0001
48.870
C
51(21.6)
80(57.1)
MTHFR gene -
Genotypic distribution
AA
46(58.9)
42(45.6)
1.611
(0.989-2.626)0.154
3.743
AC
28(35.9)
40(43.5)
CC
4(5.1)
10(10.9)
Allelic distribution
A
100(64.1)
124(67.4)
0.74
(0.46-1.21)0.2674
38.099
C
36(23.1)
60(32.6)
Table 1: Genotype and allele frequency distribution of the TGF β-1 gene.
Genetic models
Controls, N (%)
Patients, N (%)
Odds ratio
P value
Pearson value
TGF β-1 gene -
Dominant CC:CT+TT
88(74.6) : 30(25.4)
10(14.3) : 60(85.7)
17.6
(8.01-38.68)<0.0001
63.998
Recessive TT:CC+CT
97(82.2) : 21(17.8)
50(71.4) : 20(28.6)
1.85
(0.92-3.72)0.1008
2.991
Codominant I CC:TT
88(74.6) : 09(7.6)
10(14.3) : 40(57.1)
39.11
(14.75103.69)<0.0001
74.258
Codominant II CC:CT
88(74.6) : 21(17.8)
10(14.3) : 20(28.6)
8.38
(3.42-20.54)<0.0001
25.417
MTHFR gene -
Dominant AA : AA + CC
46(59.0) : 32(41.0)
42(45.7) : 50(54.3)
1.71(0.93-3.15)
0.092
3.000
Recessive AA + AC : CC
74(94.9) : 04(5.1)
82(89.1) : 10(10.9)
2.26(0.68-7.5)
0.2631
27.530
Codominant I AA : CC
46(59.0) : 04(5.1)
42(45.7) : 10(10.9)
2.74(0.8-9.39)
0.150
3.000
Codominant II AA : AC
46(59.0) : 28(35.9)
42(45.7) : 50(54.3)
1.96(1.05-3.65)
0.0422
4.488
Table 2: Logistic regression analysis of the TGF β-1 gene and MTHFR gene.
Discussion
TGF-β1 T869C SNP displayed a significant association for the risk of development of OA in dominant and codominant models. The risk was highest in the dominant model (OR, 17.6), followed by the codominant model-I (OR, 39.11). Together, these results indicate that the heterozygous TC genotype has a greater risk of developing OA. This fact is well reflected in the genotypic distribution of TC in control and OA subjects. Thus, the frequency of TC heterozygotes was higher in OA subjects (40%) than in controls (0.75%). Similar results have been reported in different populations [28]. The upregulation of the D-14 allele in OA cartilage has been observed to hinder the synthesis of cartilage-specific Extracellular Matrix (ECM) components such as collagen and proteoglycans in chondrocytes by inhibiting TGF-β signalling [29]. Changes in the MH2 protein interface domain ultimately impact the three-dimensional structure of the protein, which is crucial for Smad3 interactions necessary for TGF-β signalling. This information was reported in studies conducted by Tzavlaki et al. [29] and Che et al. [30]. Additionally, our study suggests that TC heterozygotes are predisposed to OA. For instance, a heterozygous carrier with OA in the first degree of blood relatives should take the necessary steps to safeguard him/her from potential OA complications. Furthermore, unlike TGF-β, the MTHFR A1298C SNP displayed no association with the risk of OA development. For the A1298C polymorphism in the MTHFR gene, the ORs were 1.71 (95% CI: 0.93-3.15. P=0.092) in the dominant model and 2.26 (95% CI: 0.68-7.5 P=0.263) in the recessive model. It is evident from the results that the SNP in MTHFR was not significantly associated with OA disease in each of the models examined. A similar trend has been reported in other populations [31]. However, it is important to note that we have not tested other SNPs for MTHFR in our subjects. Genotyping with other SNPs together with this could provide further insight into the risk of the development of OA.
Conclusion
In conclusion, this study compared the genomic features of the TGF-β1 and MTHFR genes between OA patients and healthy individuals in the Kanpur District of India. The findings showed significant variations in the expression levels of the dominant (CC:CT+TT) and codominant (I CC:TT and II CC:CT) TGF-β1 genes and allelic distribution in OA patients compared to the healthy controls, indicating their potential role in the pathogenesis of OA. SNP MTHFR 1298A>C was not significantly associated with OA disease in dominant recessive or other genetic models, indicating that it is not associated with the pathogenesis of OA. These results highlight the importance of further research to investigate the underlying mechanisms of TGF-β1 and MTHFR in OA and their potential as therapeutic targets for the disease. The study adds valuable insights to ongoing research on OA, which is a significant public health concern worldwide, particularly in the Indian population. The study findings may help identify potential biomarkers for the early detection and diagnosis of OA and develop personalized treatment strategies.
Author Statements
Competing Interests
The authors report no conflicts of interest.
Author 1 declared no conflict of interest.
Author 2 declared no conflict of interest.
Author 3 declared no conflict of interest.
Author 4 declared no conflict of interest.
Funding
This research was unfunded by any public, commercial, or not-for-profit agencies.
Ethical Approval and Consent to Participate
The study was conducted in accordance with the ethical standards of India and globally, as it received approval from the institutional ethical committee (CSJMU/BSBT/BT/EC-20), and all patients, including both controls and OA subjects, provided written informed consent.
Guarantor
The article’s full responsibility lies with PY, who is the corresponding author and the third author in the list.
Authors' Contributions
Dr. VC was responsible for manuscript conceptualization, writing - original draft, ethical approvals, consent, and sample collection. VR participated in writing - review & editing. PY contributed to manuscript writing, formatting, revision and communication with all authors. Dr. TA supervised all authors and wrote, reviewed and edited the final manuscript.
Acknowledgements
The authors would like to acknowledge the Institute of Biosciences and Biotechnology, Chhatrapati Shahu Ji Maharaj University, Kanpur - 208024, India for offering the excellent laboratory to conduct this study and acknowledge the valuable contribution of the Department of the Community Health Centre, Ganesh Shankar Vidyarthi Memorial Medical College (GSVM) medical college, Kanpur – 208002, India to provide the control and subjects for this study.
Availability of Data and Materials
Data and materials are available upon request.
Consent for Publication
All authors consented to manuscript publication.
References
- Cui A, Li H, Wang D, Zhong J, Chen Y, Lu H. Global, regional prevalence, incidence and risk factors of knee osteoarthritis in population-based studies. EClinicalmedicine. 2020; 29-30: 100587.
- Venkatachalam J, Natesan M, Eswaran M, Johnson AKS, Bharath V, Singh Z. Prevalence of osteoarthritis of knee joint among adult population in a rural area of Kanchipuram District, Tamil Nadu. Indian J Public Health. 2018; 62: 117-22.
- Osteoarthritis. National Health Portal of India. Available from: https://www.nhp.gov.in/disease/musculo-skeletal-bone-joints-/osteoarthritis.
- Singh A, Das S, Chopra A, Danda D, Paul BJ, March L, et al. Burden of osteoarthritis in India and its states, 1990-2019: findings from the Global Burden of disease study 2019. Osteoarthr Cartil. 2022; 30: 1070-8.
- Aubourg G, Rice SJ, Bruce-Wootton P, Loughlin J. Genetics of osteoarthritis. Osteoarthr Cartil. 2022; 30: 636-49.
- Chandra DrV, Ashraf dr (Mohd.) T, Yadav P, Raghuvanshi MrV. Gene expression profiles of knee osteoarthritis patients and healthy controls: a microarray analysis, Study [preprint] -SSRN. 2023.
- Fang J, Xu L, Li Y, Zhao Z. Roles of TGF-beta 1 signaling in the development of osteoarthritis. Histol Histopathol. 2016; 31: 1161-7.
- Bush JR, Beier F. TGF-β and osteoarthritis—the good and the bad. Nat Med 2013 19:6. 2013; 19: 667-9.
- Shen J, Li S, Chen D. TGF-β signaling and the development of osteoarthritis. Bone Res. 2014; 2: 1-7.
- Arida A, Nezos A, Papadaki I, Sfikakis PP, Mavragani CP. Osteoprotegerin and MTHFR gene variations in rheumatoid arthritis: association with disease susceptibility and markers of subclinical atherosclerosis. Sci Rep. 2022; 12: 9534.
- Poornima S, Daram S, Devaki RK, Merugu R, Subramanyam K. Association of MTHFR gene polymorphism C677T (rs1801133) studies with early primary knee osteoarthritis in a south Indian population: a hospital-based study. Afr Health Sci. 2022; 22: 338-43.
- Pagel PS, Freed JK. Cardiac physiology. Kaplan’s essentials of cardiac anesthesia for cardiac surgery. Published online. 2018; 62-79.
- Gautam A. Phenol-chloroform DNA isolation method. Published online. 2022; 33-9.
- Di Pietro F, Ortenzi F, Tilio M, Concetti F, Napolioni V. Genomic DNA extraction from whole blood stored from 15- to 30-years at -20 °C by rapid phenol-chloroform protocol: a useful tool for genetic epidemiology studies. Mol Cell Probes. 2011; 25: 44-8.
- Nasiri H, Forouzandeh M, Rasaee MJ, Rahbarizadeh F. Modified salting-out method: high-yield, high-quality genomic DNA extraction from whole blood using laundry detergent. J Clin Lab Anal. 2005; 19: 229-32.
- Guminska N, Plecha M, Walkiewicz H, Halakuc P, Zakrys B, Milanowski R. Culture purification and DNA extraction procedures suitable for next-generation sequencing of euglenids. J Appl Phycol. 2018; 30: 3541-9.
- Green MR, Sambrook J. Analysis of DNA by agarose gel electrophoresis. Cold Spring Harb Protoc. 2019; 2019: 6-15.
- Lee A, Jain A. DNA CONCENTRATION MEASUREMENT AT 260 nm USING PHOTOPETTE® BIO. Published online 2017.
- Nzilibili SMM, Ekodiyanto MKH, Hardjanto P, Yudianto A. Concentration and Purity DNA Spectrophotometer: sodium monofluorophosphate forensic impended effect. Egypt J Forensic Sci. 2018; 8: 1-7.
- How do I determine the concentration, yield and purity of a DNA sample?.
- Mesrian Tanha H, Mojtabavi Naeini M, Rahgozar S, Rasa SM, Vallian S. Modified tetra-primer ARMS PCR as a single-nucleotide polymorphism genotyping tool. Genet Test Mol Biomarkers. 2015; 19: 156-61.
- Sayed S, Galal S, Herdan OM, Mahran A. Single nucleotide polymorphism T869C of transforming growth factor-beta 1 gene and systemic lupus erythematosus: association with disease susceptibility and lupus nephritis. Egypt J Immunol. 2014.
- Xu G, Yang H, Qiu J, Reboud J, Zhen L, Ren W, et al. Sequence terminus dependent PCR for site-specific mutation and modification detection. Nat Commun. 2023; 14: 1-11.
- Han IB, Ropper AE, Jeon YJ, Park HS, Shin DA, Teng YD, et al. Association of transforming growth factor-beta 1 gene polymorphism with genetic susceptibility to ossification of the posterior longitudinal ligament in Korean patients. Genet Mol Res. 2013; 12: 4807-16.
- Martelossi Cebinelli GC, Paiva Trugilo K, Badaró Garcia S, Brajão de Oliveira K. TGF-β1 functional polymorphisms: a review. Eur Cytokine Netw. 2016; 27: 81-9.
- Poursadegh Zonouzi A, Chaparzadeh N, Asghari Estiar M, Mehrzad Sadaghiani M, Farzadi L, Ghasemzadeh A, et al. Methylenetetrahydrofolate reductase C677T and A1298C mutations in women with recurrent spontaneous abortions in the northwest of Iran. ISRN Obstet Gynecol. 2012; 2012: 945486.
- Lajin B, Alachkar A, Sakur AA. Triplex tetra-primer ARMS-PCR method for the simultaneous detection of MTHFR c.677C>T and c.1298A>C, and MTRR c.66A>G polymorphisms of the folate-homocysteine metabolic pathway. Mol Cell Probes. 2012; 26: 16-20.
- Lerer B, Segman RH, Tan EC, Basile VS, Cavallaro R, Aschauer HN, et al. Combined analysis of 635 patients confirms an age-related association of the serotonin 2A receptor gene with tardive dyskinesia and specificity for the non-orofacial subtype. Int J Neuropsychopharmacol. 2005; 8: 411-25.
- Tzavlaki K, Moustakas A. TGF-β signaling. Biomolecules. 2020; 10.
- Che X, Jin X, Park NR, Kim HJ, Kyung HS, Kim HJ, et al. Cbfβ is a novel modulator against osteoarthritis by maintaining articular cartilage homeostasis through TGF-β signaling. Cells. 2023; 12: 1064.
- Palomino-Morales R, Gonzalez-Juanatey C, Vazquez-Rodriguez TR, et al. A1298C polymorphism in the MTHFR gene predisposes to cardiovascular risk in rheumatoid arthritis. Arthritis Res Ther. 2010; 12: 1-8.
