Abstract
Objective: C-reactive protein is a sensitive and dynamic systemic marker of inflammation. Pro- and anti-inflammatory cytokines have been well documented with initiation, control and susceptibility to periodontal diseases. This study applied high-sensitivity CRP (Hs-CRP), pro- and anti-inflammatory cytokines to predict risk of periodontal diseases.
Methods: 2200 subjects (1200 healthy subjects and 1000 chronic periodontal patients) were recruited from community of Hong Kong, Department of Periodontology and Oral Medicine, Xiangya Hospital, Central South University, Hunan, China, respectively. Blood was drawn from subjects and DNA was extracted. The polymorphic sites of C-Reactive Protein (CRP), proinflammatory (interleukin-1a (IL-1a), IL-1β, Tumor Necrosis Factor-a (TNF-a), IL-6 and IFN-γ) and anti-inflammatory cytokines (IL-4, IL-10) were amplified and measured via polymerase chain reaction for further analysis. Chi-square test and logistic regression analysis were applied to analyze genotype distribution differences, allele frequencies and carriages rates between healthy and disease groups. Soluble protein levels of cytokines were evaluated by the t-test.
Results: Genotype frequencies of CRP, pro- and anti-inflammatory cytokines was statistically higher in periodontal-diseased patients than healthy controls (p<0.05). Prevalence of periodontal diseases was statistically correlated with carriage of single nucleotide polymorphisms of the cytokine parameters.
Conclusions: Hs-CRP and cytokine gene polymorphisms may be applied as biomarkers to predict periodontitis susceptibility, clinical behavior and severity.
Keywords: C-Reactive Protein; Pro-Inflammatory Cytokines; Anti- Inflammatory Cytokines; Periodontitis; Gene Polymorphisms
Introduction
The presence of periodontitis is mainly caused by the interaction of various factors, including the susceptibility of the host, the presence of pathogenic organisms, and the absence of beneficial species [1,2]. The main cause of plaque-induced inflammatory periodontal diseases is the result of bacteria, but progression and clinical characteristics of these diseases are influenced by both acquired and genetic factors that can modify susceptibility to infection [3]. Genetic factors have been demonstrated to play a significant role in the risk of periodontal diseases [4,5].
C-Reactive Protein (CRP) is an acute phase protein and its levels increase rapidly during infection and inflammation [6]. Thus, CRP is recognized as a sensitive and dynamic systemic marker of inflammation. High sensitive CRP (hs-CRP) is reported to be a strong predictor of cardiovascular risk, in spite of conventional cardiovascular risk factors [7]. The A/G polymorphism at position -717 of the human CRP gene was found to be associated with coronary heart disease [8,9]. The infection-induced elevation of systemic CRP might account for the relationship between inflammatory and cardiovascular diseases.
Cytokines are soluble proteins that are secreted by cells to act as a messenger for transmitting signals to other cells. They initiate, mediate and control immune and inflammatory responses, as well as regulate growth and differentiation of cells [10]. Gingival epithelial cells produce a broad range of cytokines, among which, Interleukin-1a (IL-1a), Interleukin-1β (IL-1β), Tumor Necrosis Factor-a (TNF-a), Interleukin-6 (IL-6) and Interferon-γ (IFN-γ) are classified as proinflammatory cytokines, and Interleukin-4 (IL-4) and Interleukin-10 (IL-10) are categorized as anti-inflammatory cytokines [11-13].
IL-1 consists of at least two separate gene products, IL-1a and IL- 1β, which have common biological activities but limited homology at nucleotide and peptide levels [14]. Concentrations of IL-1a and IL-1β are significantly greater at diseased sites (p<0.05) [15]. In comparison to other pro-inflammatory cytokines, IL-1β was most prevalent in the presence of active inflammation [16]. The gene encoding IL-1 is assigned to chromosome 2q13–21 [17,18,14]. The carriage of certain alleles of IL-1a and IL-1β is associated with the incidence and the severity of periodontal diseases, in particular Chronic Periodontitis (CP), because these carriers produce more IL-1 in response to plaque than genotype negative individuals by different studies [3,19-22].
TNF-a is located in 6p21.3 of chromosome 6 within the major histocompatibility complex [23,24]. Eight Single Nucleotide Polymorphisms (SNP) in the promoter region of this gene have been studied at positions -1031T/C, -863C/A, -857C/T, -575G/A, -376G/ A, -308G/A, -244G/A, and -238G/A [25-28]. Many researchers investigated the possible link between the -308 polymorphism in the TNF-a gene and periodontitis because a G to A polymorphism at the -308 position of the TNF-a promoter region was suggested to influence TNF-a production and monocytes of patients with periodontitis [29-32].
The IL-6 gene is assigned to chromosome 7p21. Various SNPs in the promoter region of this gene have been studied at positions -174G/C, -190C/T, -572C/G, -597G/A, -1363G/T, -1480C/G and -6106A/T [33,34,35]. Periodontitis patients carrying one or two copies of the rare allele in the IL-6 (-174) polymorphism displayed significantly higher serum IL-6 and C-reactive protein concentrations [36]. Carriers of the rare allele at this position was associated with less reduction in probing depths among chronic periodontitis patients after delivery of standard non-surgical periodontal therapy [37].
The mRNA expression and/or concentration of IFN-γ in gingival crevicular fluid, gingival tissues, and serum were able to affect gingivitis, probing depths and alveolar bone loss [38,39]. Polymorphism in gene IFN-γ was found to be functionally relevant and causes differences in the immunoregulatory activity of its cytokine molecules. The T allele of the IFN-γ 874 T/A is found in high producers of IFN-γ [40,41].
The gene for IL-4 is localized in chromosome 5q31.1 [42]. The presence of IL-4-producing cells and the percentage of IL-4- expressing cells were significantly higher in established and advanced periodontitis lesions than in gingivitis tissues. IL-4 levels in the serum of patients were higher in chronic periodontitis but these levels did not correlate with the degree of bone loss or pocket formation [43]. Mout et al. identified promoter SNP at position (-590) and a 70-bp Variable Numbers of Tandem Repeat (VNTR) polymorphism at intron 2 [44]. However, the reports about the connection of IL-4 polymorphism and periodontitis seem controversial [45-47].
The gene encoding IL-10 was mapped to chromosome 1q31- 32 [48]. The (-1082) G/A locus was not associated with chronic periodontitis susceptibility in most Caucasian populations except in one Swedish study but was linked to chronic periodontitis severity [49]. The (-1082) single nucleotide polymorphism was associated with high in vitro interleukin-10 production [50,51]. There was a complete absence of the N-allele carriage at position -1082 among the Japanese in contrast to Caucasians where the -1082 N-allele is the most occurring variant [50,51,6].
Since Hs-CRP, pro- and anti-inflammatory cytokines play a significant role in inflammatory diseases, the aim of this study is to investigate the association between polymorphisms in gene of Hs- CRP, pro- and anti-inflammatory cytokines and chronic periodontitis subjects in Chinese populations.
Materials and Methods
Selection of subjects and inclusion criteria
This study took place from 2010 to 2017, recruiting a total of 2200 subjects: 1200 healthy subjects and 1000 chronic periodontal patients. The healthy control group, comprising 1200 subjects, was randomly recruited from community of Hong Kong, and Changsha while patients of the study group were recruited from and Department of Periodontology and Oral Medicine, Xiangya Hospital, Central South University, Hunan, China.
All subjects were screened against the inclusion criteria of the study prior to enrollment in the study. For eligibility to be included as healthy subjects, participants could be either male or female, between the age of 23-66, non-smoking, have no past history of smoking, BMI (Body Mass Index) of 25, no systemic diseases, sites % with gingival recession < 5, no severe caries and no periodontal diseases. For the study group, prescreening was conducted to confirm the following criteria: between the age of 30-70, probing depth >5mm, and clinical attachment loss>4, gingival recession, and tooth mobility (for confirmation of chronic periodontal diseases).
Among the 1200 control samples, 432 were female and 768 were male, with an average age of 45.6 years old. The study group comprising 1000 Chinese periodontitis patients (410 females and 590 males) was on average 49.2 years of age. Approval was obtained from the Ethics Committee, Xiangya Hospital, Central South University, and informed consent was obtained from subjects prior to study start-up.
Oral clinical examination
Prior to enrollment in the study, potential healthy and study subjects were invited to take a full mouth examination at Keenlink Dental Clinic, Hong Kong, Department of Periodontology and Oral Medicine, Xiangya Hospital, Central South University, Hunan, respectively (Table 1) for prescreening against the inclusion in the study. In both prescreening sites, oral examinations were carried out by one periodontist with a minimum of 10 years of experience. An intra-oral examination was performed to determine periodontal conditions, including supragingival/subgingival calculus, gingival recession, Bleeding On Probing (BOP), Probing Depth (PD), Clinical Attachment Loss (CAL), gingival recession and tooth mobility.
Parameters
Control subjects (N=1200)
Periodontitis patients (N=1000)
Age (years)
45.6± 8.6
49.2±11.2
Age range (years)
24 - 65
30 - 68
Male/female
768/432
590/410
PD (mm)
2.2±0.9
5.98±2.5*
Sites% with BOP
35.3±7.2
78.2±19.8*
Sites% with gingival recession
0.9±0.5
38.9±25.9*
Sites% with calculus
30.2±8.6
65.0±12.5
Clinical Attachment Loss (mm)
1.1±0.76
5.8 ±1.8
Significant difference from the control subjects, *p<0.05.
Table 1: The clinical data (Mean±SD) of 1200 control subjects and 1000 periodontitis patients.
In the prescreening process for inclusion of subjects in the healthy group, potential subjects were recruited to the study if all inclusion criteria for healthy subjects were met. Study group subjects were recruited after the oral examination to determine if chronic periodontitis was present. The basis of diagnosing chronic periodontitis was made following the criteria defined by the American Academy of Periodontology in 2017 [52].
Study subject sample size calculation
The sample size calculation for this study was decided based on the reports of [53]. In the report, the sample size was calculated based on a 0.05 level of significance for two arms to achieve 90% power [53]. The required number of periodontal subjects for the study was determined according to a data survey in Hong Kong that measured the periodontal Pocket Depth (PD) and Bleeding On Probing (BOP).
Blood sample preparation
Two tubes of blood were collected in Essence Medical Laboratory, Hong Kong from all subjects by direct venipuncture from each subject: 20ml in lithium heparin tubes and 10ml in clot Blood Tubes (BD Vacutainer, NJ USA), respectively. The blood samples were centrifuged for 10 min at 1,500 rpm. Serum and plasma was removed for Enzyme Linked Immunosorbent Assay (ELISA) analysis. The remaining cellular components were transferred to a 50 ml centrifuge tube with an addition of red blood cell lysis buffer, up to a final volume of 45 ml. The mixture in the tube was inverted several times, and then centrifuged for 10 min at 1,500 rpm. The supernatants were discarded and the remaining components were washed with 0.9% PBS used for DNA extraction.
Isolation of purified DNA
Genomic DNA was extracted from collected blood samples using a commercially available Genomic DNA Blood Mini Kit (QIAGEN, MD, USA), following the manufacturer’s protocols. The concentration of DNA was estimated by measurements of OD260 by a spectrophotometer (U1800, Hitachi, Japan). The extracted DNA was labeled and stored at -80°C.
Polymerase Chain Reaction (PCR) for amplification of polymorphic sites
The extracted genomic DNA from samples was amplified with a PCR kit (Promega Corporation, U.S.A.)-consisting of nuclease-free water and PCR Master Mix-according to provided protocols. All procedures were carried out in a sterile and stable environment to prevent external contamination.
PCR was undertaken in a thermal cycler (MJ, U.S.A.) with a mixture containing 20 μl of nuclease-free water, 25 μl of PCR Master Mix, 0.5 μl of each designed cytokine genes primer (Invitrogen, USA) [45,46,54-60]. All primers were designed using the Roche UPL Primer Design Program, and 4 μl of the extracted DNA sample were mixed to undergo thermal cycling. All products from the thermal cycling were labeled accordingly and stored at -80°C until use. In this study, GAPDH were used (Table 2) with the forward primer 5’-AGAAGGCTGGGGCTCATTTG-3’ and reverse primer 5’-AGGGGCCATCCACAGTCTTC-3’. GAPDH is a constitutive housekeeping gene for PCR of DNA, utilized for comparison of changes in specific gene expressions [45,46,60].
Gene
Product size (bp)
Denaturation
Annealing
Extension
Cycles
IL-1a
229
94°C,1min
55°C, 30s
72°C, 60s
35
IL-1β
305
94°C, 5mins
56°C, 45s
72°C, 60s
35
IL-6
296
95°C, 60s
60°C, 60s
72°C, 60s
35
TNF-a
133
94°C, 1min
61°C, 1min
72°C, 60s
35
IFN-γ
366
95°C, 5mins
56°C, 30s
72°C, 5mins
30
IL-4
195
95°C, 5mins
51°C, 60s
72°C, 60s
35
IL-10
139
94°C, 30s
60°C, 45s
72°C, 60s
35
CRP
376
95°C, 60s
61°C, 60s
72°C, 5mins
35
β-actin
211
94°C, 30s
55°C, 30s
72°C, 60s
30
Table 2: PCR conditions for various genes.
Restriction Digest using Fnu4H1, Aval, Nla III, Alw, Mnl I, AvaII and Bsh1236 for IL-1a, IL-1β, IL-6, IFN-γ, IL-10, TNF-a, IL-4 and CRP respectively.
The amplified fragments generated from PCR were digested: 1) the 229bp fragment on IL-1a was recognized by Fnu4H1 (Fermentas Life Sciences, U.S.A.) [61,62]; 2) the 305bp fragment on IL-1β was recognized by Aval (Fermentas Life Sciences, U.S.A.) [59,62]; 3) the 296bp fragment on IL-6 was recognized by NlaIII (Fermentas Life Sciences, U.S.A.) [56,35]; 4) the 296bp fragment on IFN-γ was recognized by Alw (Fermentas Life Sciences, U.S.A.) [63,57]; 5) the 139bp fragment on IL-10 was recognized by Mnl I (Fermentas Life Sciences, U.S.A.) [64,58]; 6) the 296bp fragment on TNF-a and 195bp fragment on IL-4 were recognized by Ava II (Fermentas Life Sciences, U.S.A.) [52,63,56]; and 7) the 376 bp fragment on CRP was recognized by Bsh1236 (Fermentas Life Sciences, U.S.A.) [9].
For each digest, 10μl of amplified PCR product was mixed with 2.5-5 μl of the corresponding restriction enzyme, 10 μl of nuclease free water and 0.5-1 μl of restriction enzyme buffer and incubated for over 4 hours at 37°C (Table 3). All digestion reagents were kept on ice before incubation to prevent denaturation. To ensure amplicons were consistent throughout the procedure, all samples were digested twice.
Ctokines
Primers
Sequence
Position
Restriction Enzyme
Digestion Time (hours)
References
IL-1a
Forward
5'-ATGGTTTTAGAAATCATCAAGCCTAGGCA-3'
-889
Fnu4H1
>12
[61]
Reverse
5'-AATGAAAGGAGGGGAGGATGACAGAAATGA -3'
IL-1β
Forward
5'-TGGCATTGATCTGGTTCATC-3'
-511
Aval
[62]
Reverse
5'-GTTTAGGAATCTTCCCACTT-3'
IL-6
Forward
5'-TTGTCAAGACATGCCAAGTGCT-3'
-174
Nla III
4
[67]
Reverse
5'-GCCTCAGAGACATCTCCAGTCC-3'
TNF-a
Forward
5'-GAAGCCCCTCCCAGTTCTAGT TC-3'
-238
Ava II
4
[68]
Reverse
5'-CACTCCCCATCCTCCCTGGTC-3'
IFN-γ
Forward
5'-GCTGTCATAATAATATTCAGAC-3'
-874
Alw
4
[21]
Reverse
5'-CGAGCTTTAAAAGATAGTTCC-3'
IL-4
Forward
5'-TAAACTTGGGAGAACATGGT-3'
-590
Ava II
>12
[22]
Reverse
5'-TGGGGAAAGATAGAGTAATA-3'
IL-10
Forward
5'-CTCGCTGCAACCCAACTGGC-3'
-1082
Mnl I
4
[64]
Reverse
5'-TCTTACCTATCCCTACTTCC-3'
CRP
Forward
5’-GACTCCTGCCT-GAAGCTTTACATA-3’
-717
Bsh1236
4
[9]
Reverse
5’-ATACATGTGCC-ATGCTGGTGTG-3’
Table 3: The primer sequences and restriction enzyme used for detection of cytokine DNA polymorphism genes.
DNA gel electrophoresis and isualization
The 10 μl of digestion product and 1 μl of Ready-Load 1 Kb DNA Ladder (Invitrogen, Spain) were loaded into 2-4% agarose gel (Invitrogen, Spain) containing 0.5 μg/ml of ethidium bromide. The gel underwent electrophoresis at 100 volts, 100 milliAmperes for 30 minutes. Afterwards, the gel was visualized using a Dolphin-DOC ultraviolet illuminator (Wealtec, South Africa).
Sera measured by ELISA
100 μl of the standard group solutions and serum of each subject were pipetted into a 96-well plate included in the cytokines ELISA kit (Diaclone, France) [65]. The plate was incubated for 2-3 hours at 350 rpm and washed with washing buffer three times. Then the wells were dried and 200 μl of substrate tetramethylbenizidine was added into each well for 20 min in the dark at room temperature. The plates were read at 450 nm wavelength using Universal Microplate Reader (Sunrise, TECAN, Austria). The levels of cytokines in the samples were obtained by comparison with the standard curve generated from standards supplied by the manufacturer [65]. ELISA was performed according to the manufacturer’s protocols from cytokines ELISA Kit (Diaclone, France) and the serum samples. The normal detection ranges of biomarker were showed in Table 4.
Parameters
Control
Periodontitis
Normal Range
Unit
White blood cell
6.0(±0.98)
9.2(±2.1)
4.00-11.00
109/L
Red blood cell
4.8(±0.25)
5.0(±0.10)
3.8-6.0
1012/L
Hemoglobin
14.5(±0.80)
13.5(±0.43)
11.5-16.5
g/dL
Platelet
295(±30.2)
265(±23.31)
150-400
109/L
Neutrophils
70.0(±11.32)
*33.6(±8.3)
4.0-75.0
%
Lymphocytes
26.0(±7.18)
*49.2(±10.69)
20-45
%
Monocyte
3.0(±1.2)
*14.0(±2.89)
4.7-12.2
%
Eosinophil
0.83(±0.21)
1.5(±0.52)
0.7-7.0
%
Basophil
0.17(±0.09)
0.33(±0.24)
0.1-1.2
%
Significant difference from the control, * p<0.001
Table 4: Complete blood count (Mean±SD) of control subjects (N=1200) and periodontitis patients (N=1000).
Statistical analysis
SPSS 22.0 for windows (IBM, SPSS, U.S.A.) was used for calculations of the chi-square and independent t-tests. The chi-square test (Χ2) was applied to examine the differences in Hs-CRP, genotype distributions, allele frequencies and carriages rate between healthy and disease groups. The alleles were calculated as Odds Ratio (OR) with 95% Confidence Intervals (95% CI). The soluble protein levels of cytokines were evaluated with an independent t-test. A p-value of <0.05 was considered statistically significant.
Results
The chronic periodontitis study group showed a significantly greater mean of PD (5.98±2.5 mm vs. 2.2±0.9 mm), CAL (5.8±1.8 mm vs.1.1±0.76), and a higher percentage of sites with BOP (78.2±19.8% vs. 35.3±7.2%) and gingival recession (38.9±25.9% vs. 0.9±0.5%) than the healthy control group (p<0.05) (Table 1).
Confirmatory of the inclusion of systematically-healthy individuals, the blood results for both the healthy and study group were predominantly within the normal ranges. Statistical differences, however, in neutrophils (33.6% vs. 70.0%), lymphocytes (49.2% vs. 26.0%) and monocytes (14.0% vs. 3.0%) were observed (p<0.05) in the periodontitis patients compared to the healthy controls (Table 4).
All protein expressions of patients and control samples for IL- 1a (62.65±11.93 pg/ml vs. 25.28±4.51 pg/ml), IL-1β (81.73±17.94 pg/ml vs. 20.55±3.37 pg/ml), IL-6 (97.57±18.92 pg/ml vs. 28.77±3.13 pg/ml), TNF-a (70.99±13.18 pg/ml vs. 28.45±4.77 pg/ml), IFN-γ (99.44±10.52 pg/ml vs. 55.85±8.26 pg/ml), IL-4 (4.85±1.26 pg/ml vs. 1.18±0.21 pg/ml), IL-10 (49.23±4.88 pg/ml vs. 23.85±3.55 pg/ml), and Hs-CRP (136.50±12.92 pg/ml vs. 0.61±0.25 pg/ml) measured by ELISA confirmed a statistically significant difference (p<0.001). Agarose gel electrophoresis (Wealtec, South Africa) and visualization demonstrated that the pro-inflammatory cytokines IL-1a and TNF-a and the anti-inflammatory cytokines IL-4 and IL-10 were associated with chronic inflammation in patients (p<0.0001) (Figure 1).
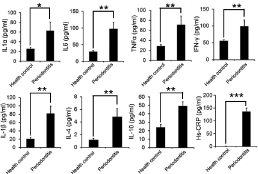
Figure 1: 1: Protein expression (Mean±SD) of various cytokines in patient and control groups as measured by ELISA. *p<0.05, **p<0.01, ***p<0.001.
The homozygous A alleles of CRP-717 were represented by a DNA band of 376 bp, and homozygous G alleles presented 2 DNA bands of 376 and 319 bp. The homozygous G alleles of CRP-717 were found to be significantly higher in periodontitis patients compared to healthy subjects, corresponding to 72% vs. 54%, respectively (p=0.008; OR=2.19; 95% CI=1.22-3.94). Heterozygotes for CRP at position -717 site of the CRP gene displayed a combination of both G and A alleles. A comparison of A/A genotype in periodontitis patients (67%) with respect to healthy controls (55%) demonstrated a higher frequency (p=0.0004). The odds ratio for carriage of CRP-717 allele (A/A and G/G genotypes combined in comparison to A/A genotype) was computed to be 2.80 (95% CI=1.58-4.99) for the patient group (Table 5).
Genotypes
CP Patients n=1000 (%)
Healthy subjects n=1200 (%)
CP versus Controls
Alleles
CP patients n=2000 (%)
Healthy subjects n=2400 (%)
CP versus Controls
OR (95% CI)
Χ2 p-values
OR (95% CI)
Χ2 p-values
CRP-717
*A/A
670(67)
660(55)
2.8
0.0004
*A
1440(72)
1764(54)
2.19
0.008
G/G
150(15)
132(11)
1.58-4.99
G
560(28)
636(46)
1.22-3.94
A/G
180(18)
408(34)
IL-1a
*C/C
540(54)
216(18)
5.35
<0.0001
*C
1280(64)
1248(42)
2.46
0.002
T/T
270(27)
132(11)
2.81-10.18
T
720(36)
1152(58)
1.39-4.34
C/T
190(19)
852(71)
IL-1β
*T/T
300(55)
456(35)
2.27
0.005
*T
1120(66)
1416(49)
2.02
0.02
C/C
120(12)
252(21)
1.28-4.01
C
880(34)
984(51)
1.14-3.57
C/T
580(33)
492(44)
IL-6
*G/G
240(24)
600(50)
0.32
0.0002
*G
1100(65)
1512(43)
2.46
0.002
C/C/
70(7)
276(23)
0.17-0.58
C
900(35)
888(57)
1.39-4.36
G/C
690(69)
324(27)
TNF-a
*G/G
450(45)
192(16)
4.3
<0.0001
*G
1100(55)
768(32)
2.59
0.0012
A/A
310(31)
564(47)
2.21-8.35
A
900(45)
1632(68)
1.46-4.62
A/G
240(24)
444(37)
IFN-γ
*T/T
550(55)
660(35)
2.27
0.005
*T
1280(65)
1584(32)
3.95
<0.0001
A/A
220(22)
276(43)
1.28-4.01
A
720(35)
816(68)
2.19-7.10
T/A
230(23)
264(22)
IL-4
*C/C
30(3)
312(26)
0.09
0.0001
*C
440(22)
1248(52)
0.26
<0.0001
T/T
530(53)
240(20)
0.03-0.30
T
1560(78)
1152(48)
0.14-0.48
T/C
440(44)
648(54)
IL-10
*A/A
650(65)
1080(90)
0.21
0.0001
*A
1300(65)
2280(95)
0.1
<0.0001
G/G
290(29)
84(7)
0.09-0.45
G
700(35)
120(5)
0.04-0.26
A/G
60(6)
36(3)
1440(72)
1764(54)
2.19
0.008
1.22-3.94
CRP: G/G+A/G versus A/A; IL-1a: T/T+C/T versus C/C;
IL-1β: C/C+ C/T versus T/T; IL-6: C/C+G/C versus G/G;
TNF-a: A/A+A/G versus G/G; IFN-γ: A/A+T/A versus T/T;
IL-4: T/T +T/C versus C/C; IL-10: G/G+A/G versus A/A
OR: Odds Ratio; CI: Confidence Interval;
Significant difference from healthy controls, *p<0.05
Table 5: Genotype and allele frequency of cytokines in periodontitis patients (CP) and control subjects.
Digesting Fnu4H1 for IL-1a formed the DNA products of 153- and 76-bp for homozygous C allele, and 124-, 76- and 29-bp for homozygous T alleles. The results of detection frequency of the homozygous C allele of IL-1a were similar in the chronic periodontitis study group (64%) as the healthy control group (42%) (Χ2, p=0.002). Varying results were demonstrated for the C/C genotype, where frequency was 54% in periodontitis patients in comparison to 18% in the control group. The odds ratio for carriage of IL-1a allele (T/T and C/T genotypes combined to compare with the C/C genotype) was 5.35 (95% CI: 2.81-10.18, Χ2: p<0.0001) in periodontitis subjects (Table 5).
The frequency of homozygous T alleles of IL-1β (305 bp) digested with Aval were discovered to be higher in the study group (66%) compared to healthy controls (49%) with significance (p=0.02; OR=2.02; 95% CI=1.14-3.57). The T/T genotype was detected to be higher also the periodontal study group compared to healthy subjects, despite an absence of significance (55% vs. 35%, p=0.05). The odds ratio for IL-1β allele (C/C and C/T genotypes combined compared with the T/T genotype) was 2.27 (95% CI: 1.28-4.01) in periodontitis patients (Table 5).
Digesting Nla III for IL-6 formed the DNA products of 296 76-bp for homozygous G allele [66]. Significant differences were detected for the frequency of the homozygous G allele of IL-6 in the chronic periodontitis study group (65%) than the healthy control group (43%) (Χ2, CI=1.39-4.36, p=0.002). Varying results were demonstrated for the G/G genotype, where a lower frequency was discovered for periodontitis patients (24%) than the control group (50%). The odds ratio for carriage of IL-6 allele (C/C and G/C genotypes combined to compare with the G/G genotype) was 0.32 (95% CI: 0.17-0.58, X2: p<0.001) in periodontitis subjects (Table 5).
Digesting AvaII for TNF-a formed the DNA products of 70- and 63-bp for homozygous G alleles, and 63-, 49- and 21-bp for homozygous A alleles. The homozygous G allele of TNF-a was shown to be higher in the study group (55%) than healthy controls (32%) (Χ2: p=0.0012, OR: 2.59, CI: 1.46-4.62). The detection frequency of the G/G genotype in periodontitis patients was higher than the controls (45% vs. 16%, p<0.0001). For the periodontal study group, the OR was 4.30 (95% CI: 2.21-8.35) for carriage of TNF-a allele (A/A and A/G genotypes combined for comparing G/G genotype) (Table 5).
The homozygous T alleles of IFN-Γ were represented by a DNA band size of 366 bp, digested by Alw. A significantly higher detection frequency (p<0.0001) was noted for the patient group (65%) compared to healthy subjects (32%) (OR=3.95, 95% CI: 2.19-7.10). Comparison of the T/T genotype of IFN-γ indicated a higher frequency also for periodontal patients (55%) than healthy subjects (35%), albeit a lack of significance. The odds ratio for carriage of IFN-γ allele (A/A and T/A genotypes combined compared with the T/T genotype) was 2.27 (95% CI: 1.28-4.01) in the study group of periodontal patients (Table 5).
The homozygous T alleles of IL-4 were represented by a DNA band size of 195bp, and homozygous C alleles were represented by DNA bands sizes 18 and 177bp [67]. The chronic periodontitis patient group had a higher detection frequency of the homozygous T alleles of IL-4 (78%) than the control group (48%), and lower frequency for the homozygous C alleles for periodontitis patients (22%) compared to healthy subjects (52%). A comparison of the study and healthy group demonstrated significant differences (p<0.0001, OR=0.26, 95% CI: 0.14-0.48). Comparing T/T genotype, the study group presented a higher frequency of 53% than 20% in healthy controls (p<0.0001). The OR for carriage of IL-4 allele (T/T and T/C genotypes combined compared with the C/C genotype) was 0.09 (95% CI: 0.03-0.30) in the study group of periodontal patients (Table 5).
The homozygous A alleles of IL-10 were represented by a DNA band size of 139bp, homozygous G alleles were represented by DNA band sizes 106 and 33bp [46]. Compared to the study group (65%), the detection frequency of the homozygous a alleles of IL-10 was higher in healthy controls (95%). Statistical significance was observed between the study and control group (p<0.0001, OR=0.10, 95% CI: 0.04-0.26). A significantly lower frequency was detected for the A/A genotype for periodontitis patients than healthy controls 65% vs. 90%, (p<0.001). The OR for carriage of IL-10 allele (G/G and A/G genotypes combined compared with the A/A genotype) was 0.21 (95% CI: 0.09-0.45) in periodontitis patients (Table 5).
Discussion
The presence of inflammation in diseased periodontal tissues or gingival crevicular fluid can be observed with increased levels of CRP and pro-inflammatory cytokines IL-1a, IL-1β, TNF-a, IFN-γ and IL-6 [68-70]. Recent study results showed that subjects with CP have increased levels of inflammation as a results of increased serum levels of Hs-CRP, IL-1, IL-6, TNF-a, and IFN-γ in comparison to healthy individuals. Hs-CRP and IL-6 are generally the most sensitive markers of the acute phase response to infections and inflammation [71,9].
Anti-inflammatory cytokines IL-4 and IL-10 work by balancing raised levels of pro-inflammatory cytokines. The progression of inflammation in periodontal diseases can be explained by an absence of response of the anti-inflammatory cytokines IL-4 and IL-10 in chronic periodontitis [68,69,72]. Genetic polymorphisms cause a change in the protein or its expressions altering the immune response.
The results of this study parallel past and current research documenting the prevalence of elevated levels of CRP inflammatory diseases as periodontitis [6]. Statistical differences were observed for A/G allele frequencies for the CRP gene between periodontal patients and orally healthy subjects (p=0.008). A higher frequency of homozygous G/G genotype and heterozygous A/G genotype was detected in orally healthy individuals than orally-diseased persons (25% vs. 15%; and 33% vs. 18%). The CRP-717 A/A genotype was more highly associated with periodontal-diseased individuals (67%) than healthy subjects (42%) (OR= 2.80; 95% CI: 1.58-4.99). Similarly, ELISA investigations for the protein expression of the Hs-CRP cytokines also demonstrated significant differences in the study and healthy subjects. A higher association of A/A genotype in periodontal patients may be an indicator of periodontal-diseased tissues in the Chinese population (Figure 1).
Significant differences in detection of C and T alleles of IL- 1a (-889) were also discovered in the study and control groups. As observed, genetic polymorphism likely differs between ethnic populations as the carriage rate of C allele was shown to be lower in our population of Chinese subjects than other Asian cohorts [73-75]. The C allele frequency in chronic periodontal patients was comparably higher than healthy controls (64% vs. 42%), while the reverse was true for the frequency of T allele, where the orally healthy control group (58%) was observed to be higher than the patient group (36%). In line with other studies, the IL-1a CC genotype (OR=5.35; 95% CI=2.81 to 10.18) was significantly associated with chronic periodontitis [56,76].
The association of IL-1β SNPs with periodontal diseases has been commonly reported in Caucasian populations [16,77], while varying results have been published for the Chinese population [78-80]. The association of SNP IL-1β (-511) and periodontitis was examined in this study. A statistically significant correlation for the SNP IL-1β (-511) and periodontitis was established for the patients of this study, correlating with severity of periodontitis. The IL-1β SNP was shown to be elevated in the patient group, compared to healthy controls (p=0.005). The carriage rate of the T allele was found to be higher in orally diseased patients of the study, compared to orally healthy individuals (66% vs. 49%). A higher detection rate for the TT genotype was also found (55% vs. 35%). These results highlight a similar role of IL-1β in inflammation in periodontal diseases of Chinese patients as Caucasians.
The pro-inflammatory cytokine TNF-a possesses a wide range of immunoregulatory functions, including production of secondary mediators [81]. Ample research has studied the polymorphic site position 308 and concluded a lack of association with periodontitis. In contrast, the results of this present study was able to demonstrate a higher frequency of homozygous (G/G) genotype and an association of TNF-a G/G genotype with chronic periodontal diseases (OR=4.30; 95% CI=2.21-8.35). Taken together, TNF-a may be considered a risk genotype for periodontitis susceptibility.
The results of the current study showed a similar association of IL-6 in the Chinese population with periodontitis to Caucasians and Brazilians. [55,82]. The study and control groups demonstrated significantly differing carriage rates for both G and C alleles, rendering at 65% and 43% for C allele and 35% and 57% for G allele. The heterozygous genotype IL-6-174 (G/C) was observed to have a higher frequency in the study cases in contrast to healthy controls (69% and 27%). IL-6 -174 G/C genotype could be considered a risk genotype for periodontitis susceptibility.
Previous case–control studies have demonstrated varied results in the relationship between the IL-4 gene polymorphism and susceptibility to chronic periodontitis [47,83-85]. The present study showed an association of IL-4 C/C genotype (OR=0.09; 95% CI: 0.03- 0.30) with a lower frequency in the study subjects. The T/T genotype was more prevalent in the study patients (53%) than healthy controls (20%), suggesting this may be a risk genotype for periodontitis susceptibility. A comparatively lower C/C genotype in patients (3%) compared to healthy controls (26%) may be an indicative factor against the development of the disease.
In this study, the IL-10 (-1082) polymorphism demonstrated a lower frequency of A/A genotype with periodontitis patients than healthy subjects (65% vs. 90%). A higher frequency of IL-10 (-1082) G/G genotype (29%) was found in the study group compared to healthy subjects (7%), which may be considered a risk genotype for periodontitis susceptibility. The results of this study were in accordance with past studies: subjects who exhibited the G/G genotype were significantly larger in subjects with chronic periodontal diseases than in periodontal-healthy individuals [86,87].
Conclusion
The results of this study demonstrate that Hs-CRP in combination with cytokine gene polymorphisms may be associated with periodontitis susceptibility, clinical behavior and severity. Application of these biomarkers may be used as putative risk indicators for chronic periodontitis in the Chinese population.
Acknowledgement
The present study was supported by grants from the Science and Technology Project of Hunan Province, China (2018JJ2625).
The authors would like to thank Dr. Eva Loo Kwok Ying of Keenlink Dental Clinic, Hong Kong for performing pre-study screening for 1200 healthy subjects.
References
- Slots J. Subgingival microflora and periodontal disease. J Clin Periodontol. 1979; 6: 351-382.
- Genco RJ. Host responses in periodontal diseases: Current concepts. J Periodontol. 1992; 63: 338-355.
- Kornman KS, Crane A, Wang HY, di Giovine FS, Newman MG, Pirk FW, et al. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol. 1997; 24: 72-77.
- Michalowicz BS. Genetic and heritable risk factors in periodontal disease. J Periodontol. 1994; 65: 479-488.
- Hart TC, Kornman KS. Genetic factors in the pathogenesis of periodontitis. Periodontol. 1997; 2000; 14: 202-215.
- Israelsson E, Ekstrom M, Nasr A, Dolo A, Kearsley S, Arambepola G, et al. Marked differences in crp genotype frequencies between the fulani and sympatric ethnic groups in africa. Malar J. 2009; 8: 136.
- Maekawa T, Tabeta K, Kajita-Okui K, Nakajima T, Yamazaki K. Increased expression of c-reactive protein gene in inflamed gingival tissues could be derived from endothelial cells stimulated with interleukin-6. Arch Oral Biol. 2011; 56: 1312-1318.
- Chen J, Zhao J, Huang J, Su S, Qiang B, Gu D. -717a>g polymorphism of human c-reactive protein gene associated with coronary heart disease in ethnic han chinese: The beijing atherosclerosis study. J Mol Med (Berl). 2005; 83: 72-78.
- Tian Y, Li JL, Hao L, Yue Y, Wang M, Loo WT, et al. Association of cytokines, high sensitive c-reactive protein, vegf and beta-defensin-1 gene polymorphisms and their protein expressions with chronic periodontitis in the chinese population. Int J Biol Markers. 2013; 28: 100-107.
- Elenkov IJ, Iezzoni DG, Daly A, Harris AG, Chrousos GP. Cytokine dysregulation, inflammation and well-being. Neuroimmunomodulation. 2005; 12: 255-269.
- Yano S, Sone S, Nishioka Y, Mukaida N, Matsushima K, Ogura T. Differential effects of anti-inflammatory cytokines (il-4, il-10 and il-13) on tumoricidal and chemotactic properties of human monocytes induced by monocyte chemotactic and activating factor. J Leukoc Biol. 1995; 57: 303-309.
- Malkowska A, Kasprzak A, Stopa J. Proinflammatory cytokines in pathogenesis of periodontal disease. Pol Merkur Lekarski. 2006; 20: 93-98.
- Prasanna SJ, Gopalakrishnan D, Shankar SR, Vasandan AB. Proinflammatory cytokines, ifngamma and tnfalpha, influence immune properties of human bone marrow and wharton jelly mesenchymal stem cells differentially. PLoS One. 2010; 5: e9016.
- Gabay C, Lamacchia C, Palmer G. Il-1 pathways in inflammation and human diseases. Nat Rev Rheumatol. 2010; 6: 232-241.
- Lester SR, Bain JL, Serio FG, Johnson RB. Relationship between the gingival sulcus depth and interleukin-1 isoform concentrations within the adjacent gingival tissue. J Periodontal Res. 2009; 44: 323-329.
- McGee JM, Tucci MA, Edmundson TP, Serio CL, Johnson RB. The relationship between concentrations of proinflammatory cytokines within gingiva and the adjacent sulcular depth. J Periodontol. 1998; 69: 865-871.
- Auron PE, Webb AC, Rosenwasser LJ, Mucci SF, Rich A, Wolff SM, et al. Nucleotide sequence of human monocyte interleukin 1 precursor cdna. Proc Natl Acad Sci U S A. 1984; 81: 7907-7911.
- Cameron P, Limjuco G, Rodkey J, Bennett C, Schmidt JA. Amino acid sequence analysis of human interleukin 1 (il-1). Evidence for biochemically distinct forms of il-1. J Exp Med. 1985; 162: 790-801.
- McDevitt MJ, Wang HY, Knobelman C, Newman MG, di Giovine FS, Timms J, et al. Interleukin-1 genetic association with periodontitis in clinical practice. J Periodontol. 2000; 71: 156-163.
- Papapanou PN, Neiderud AM, Sandros J, Dahlen G. Interleukin-1 gene polymorphism and periodontal status. A case-control study. J Clin Periodontol. 2001; 28: 389-396.
- Ferreira SB, Trombone APF, Repeke CE, Cardoso CR, Martins W, Santos CF, et al. An interleukin-1β (il-1β) single-nucleotide polymorphism at position 3954 and red complex periodontopathogens independently and additively modulate the levels of il-1β in diseased periodontal tissues. Infection and Immunity. 2008; 76: 3725-3734.
- Trevilatto PC, de Souza Pardo AP, Scarel-Caminaga RM, de Brito Jr RB, Alvim-Pereira F, Alvim-Pereira CC, et al. Association of il1 gene polymorphisms with chronic periodontitis in brazilians. Arch Oral Biol. 2011; 56: 54-62.
- Ragoussis J, Bloemer K, Weiss EH, Ziegler A. Localization of the genes for tumor necrosis factor and lymphotoxin between the hla class i and iii regions by field inversion gel electrophoresis. Immunogenetics. 1988; 27: 66-69.
- Wilson AG, di Giovine FS, Duff GW. Genetics of tumour necrosis factor-alpha in autoimmune, infectious, and neoplastic diseases. J Inflamm. 1995; 45: 1-12.
- Wilson AG, di Giovine FS, Blakemore AI, Duff GW. Single base polymorphism in the human tumour necrosis factor alpha (tnf alpha) gene detectable by ncoi restriction of pcr product. Hum Mol Genet. 1992; 1: 353.
- Brinkman BM, Giphart MJ, Verhoef A, Kaijzel EL, Naipal AM, Daha MR, et al. Tumor necrosis factor alpha-308 gene variants in relation to major histocompatibility complex alleles and felty’s syndrome. Hum Immunol. 1994; 41: 259-266.
- Higuchi T, Seki N, Kamizono S, Yamada A, Kimura A, Kato H, et al. Polymorphism of the 5’-flanking region of the human tumor necrosis factor (tnf)-alpha gene in japanese. Tissue Antigens. 1998; 51: 605-612.
- Uglialoro AM, Turbay D, Pesavento PA, Delgado JC, McKenzie FE, Gribben JG, et al. Identification of three new single nucleotide polymorphisms in the human tumor necrosis factor-alpha gene promoter. Tissue Antigens. 1998; 52: 359-367.
- Louis E, Franchimont D, Piron A, Gevaert Y, Schaaf-Lafontaine N, Roland S, et al. Tumour necrosis factor (tnf) gene polymorphism influences tnf-alpha production in lipopolysaccharide (lps)-stimulated whole blood cell culture in healthy humans. Clin Exp Immunol. 1998; 113: 401-406.
- Tang GJ, Huang SL, Yien HW, Chen WS, Chi CW, Wu CW, et al. Tumor necrosis factor gene polymorphism and septic shock in surgical infection. Crit Care Med. 2000; 28: 2733-2736.
- Fassmann A, Holla LI, Buckova D, Vasku A, Znojil V, Vanek J. Polymorphisms in the +252(a/g) lymphotoxin-alpha and the -308(a/g) tumor necrosis factoralpha genes and susceptibility to chronic periodontitis in a czech population. J Periodontal Res. 2003; 38: 394-399.
- Trombone APF, Cardoso CR, Repeke CE, Ferreira SB, Martins W, Campanelli AP, et al. Tumor necrosis factor-alpha -308g/a single nucleotide polymorphism and red-complex periodontopathogens are independently associated with increased levels of tumor necrosis factor-a in diseased periodontal tissues. J Periodontal Res. 2009; 44: 598-608.
- Holla LI, Fassmann A, Stejskalova A, Znojil V, Vanek J, Vacha J. Analysis of the interleukin-6 gene promoter polymorphisms in czech patients with chronic periodontitis. J Periodontol. 2004; 75: 30-36.
- Komatsu Y, Tai H, Galicia JC, Shimada Y, Endo M, Akazawa K, et al. Interleukin-6 (il-6)--373 a9t11 allele is associated with reduced susceptibility to chronic periodontitis in japanese subjects and decreased serum il-6 level. Tissue Antigens. 2005; 65: 110-114.
- Nibali L, D’Aiuto F, Donos N, Griffiths GS, Parkar M, Tonetti MS, et al. Association between periodontitis and common variants in the promoter of the interleukin-6 gene. Cytokine. 2009; 45: 50-54.
- D’Aiuto F, Parkar M, Brett PM, Ready D, Tonetti MS. Gene polymorphisms in pro-inflammatory cytokines are associated with systemic inflammation in patients with severe periodontal infections. Cytokine. 2004; 28: 29-34.
- D’Aiuto F, Ready D, Parkar M, Tonetti MS. Relative contribution of patient-, tooth-, and site-associated variability on the clinical outcomes of subgingival debridement. I. Probing depths. J Periodontol. 2005; 76: 398-405.
- Yamazaki K, Nakajima T, Kubota Y, Gemmell E, Seymour GJ, Hara K. Cytokine messenger rna expression in chronic inflammatory periodontal disease. Oral Microbiol Immunol. 1997; 12: 281-287.
- Cesar-Neto JB, Duarte PM, de Oliveira MC, Casati MZ, Tambeli CH, Parada CA, et al. Smoking modulates interferon-gamma expression in the gingival tissue of patients with chronic periodontitis. Eur J Oral Sci. 2006; 114: 403- 408.
- Pravica V, Asderakis A, Perrey C, Hajeer A, Sinnott PJ, Hutchinson IV. In vitro production of ifn-gamma correlates with ca repeat polymorphism in the human ifn-gamma gene. Eur J Immunogenet. 1999; 26: 1-3.
- Pravica V, Perrey C, Stevens A, Lee JH, Hutchinson IV. A single nucleotide polymorphism in the first intron of the human ifn-gamma gene: Absolute correlation with a polymorphic ca microsatellite marker of high ifn-gamma production. Hum Immunol. 2000; 61: 863-866.
- Sutherland GR, Baker E, Callen DF, Hyland VJ, Wong G, Clark S, et al. Interleukin 4 is at 5q31 and interleukin 6 is at 7p15. Hum Genet. 1988; 79: 335-337.
- McFarlane CG, Meikle MC. Interleukin-2, interleukin-2 receptor and interleukin-4 levels are elevated in the sera of patients with periodontal disease. J Periodontal Res. 1991; 26: 402-408.
- Mout R, Willemze R, Landegent JE. Repeat polymorphisms in the interleukin-4 gene (il4). Nucleic Acids Res. 1991; 19: 3763.
- Michel J, Gonzales JR, Wunderlich D, Diete A, Herrmann JM, Meyle J. Interleukin-4 polymorphisms in early onset periodontitis. J Clin Periodontol. 2001; 28: 483-488.
- Gonzales JR, Kobayashi T, Michel J, Mann M, Yoshie H, Meyle J. Interleukin-4 gene polymorphisms in japanese and caucasian patients with aggressive periodontitis. J Clin Periodontol. 2004; 31: 384-389.
- Holla LI, Fassmann A, Augustin P, Halabala T, Znojil V, Vanek J. The association of interleukin-4 haplotypes with chronic periodontitis in a czech population. J Periodontol. 2008; 79: 1927-1933.
- Kim JM, Brannan CI, Copeland NG, Jenkins NA, Khan TA, Moore KW. Structure of the mouse il-10 gene and chromosomal localization of the mouse and human genes. J Immunol. 1992; 148: 3618-3623.
- Scarel-Caminaga RM, Trevilatto PC, Souza AP, Brito RB, Camargo LE, Line SR. Interleukin 10 gene promoter polymorphisms are associated with chronic periodontitis. J Clin Periodontol. 2004; 31: 443-448.
- Kube D, Platzer C, von Knethen A, Straub H, Bohlen H, Hafner M, et al. Isolation of the human interleukin 10 promoter. Characterization of the promoter activity in burkitt’s lymphoma cell lines. Cytokine. 1995; 7: 1-7.
- Eskdale J, Kube D, Tesch H, Gallagher G. Mapping of the human il10 gene and further characterization of the 5’ flanking sequence. Immunogenetics. 1997; 46: 120-128.
- Caton JG, Armitage G, Berglundh T, Chapple IL, Jepsen S, Kornman KS, et al. A new classification scheme for periodontal and peri-implant diseases and conditions–Introduction and key changes from the 1999 classification. Journal of periodontology. 2018; 89: S1-S8.
- Machin D, Campbell MJ, Tan SB, Tan SH. Sample size tables for clinical studies. John Wiley & Sons. 2011.
- Endo M, Tai H, Tabeta K, Kobayashi T, Yamazaki K, Yoshie H. Analysis of single nucleotide polymorphisms in the 5’-flanking region of tumor necrosis factor-alpha gene in japanese patients with early-onset periodontitis. J Periodontol. 2001; 72: 1554-1559.
- Trevilatto PC, Scarel-Caminaga RM, de Brito RB, de Souza AP, Line SR. Polymorphism at position -174 of il-6 gene is associated with susceptibility to chronic periodontitis in a caucasian brazilian population. J Clin Periodontol. 2003; 30: 438-442.
- Moreira PR, Costa JE, Gomez RS, Gollob KJ, Dutra WO. The il1a (-889) gene polymorphism is associated with chronic periodontal disease in a sample of brazilian individuals. J Periodontal Res. 2007; 42: 23-30.
- Raunio T, Nixdorf M, Knuuttila M, Karttunen R, Vainio O, Tervonen T. The extent of periodontal disease and the il-6 -174 genotype as determinants of serum il-6 level. J Clin Periodontol. 2007; 34: 1025-1030.
- Sumer AP, Kara N, Keles GC, Gunes S, Koprulu H, Bagci H. Association of interleukin-10 gene polymorphisms with severe generalized chronic periodontitis. J Periodontol. 2007; 78: 493-497.
- Nikolopoulos GK, Dimou NL, Hamodrakas SJ, Bagos PG. Cytokine gene polymorphisms in periodontal disease: A meta-analysis of 53 studies including 4178 cases and 4590 controls. J Clin Periodontol. 2008; 35: 754-767.
- Reichert S, Machulla HK, Klapproth J, Zimmermann U, Reichert Y, Glaser C, et al. Interferon-gamma and interleukin-12 gene polymorphisms and their relation to aggressive and chronic periodontitis and key periodontal pathogens. J Periodontol. 2008; 79: 1434-1443.
- Walker SJ, Van Dyke TE, Rich S, Kornman KS, di Giovine FS, Hart TC. Genetic polymorphisms of the il-1alpha and il-1beta genes in african-american ljp patients and an african-american control population. J Periodontol. 2000; 71: 723-728.
- Sugimoto M, Furuta T, Yamaoka Y. Influence of inflammatory cytokine polymorphisms on eradication rates of helicobacter pylori. J Gastroenterol Hepatol. 2009; 24: 1725-1732.
- Nakao F, Ihara K, Kusuhara K, Sasaki Y, Kinukawa N, Takabayashi A, et al. Association of ifn-gamma and ifn regulatory factor 1 polymorphisms with childhood atopic asthma. J Allergy Clin Immunol. 2001; 107: 499-504.
- Chin HJ, Na KY, Kim SJ, Oh KH, Kim YS, Lim CS, et al. Interleukin-10 promoter polymorphism is associated with the predisposition to the development of iga nephropathy and focal segmental glomerulosclerosis in korea. J Korean Med Sci. 2005; 20: 989-993.
- Loo WT, Jin LJ, Chow LW, Cheung MN, Wang M. Rhodiola algida improves chemotherapy-induced oral mucositis in breast cancer patients. Expert Opin Investig Drugs. 2010: S91-100.
- Belluco C, Olivieri F, Bonafe M, Giovagnetti S, Mammano E, Scalerta R, et al. -174 g>c polymorphism of interleukin 6 gene promoter affects interleukin 6 serum level in patients with colorectal cancer. Clin Cancer Res. 2003; 9: 2173-2176.
- Scarel-Caminaga RM, Trevilatto PC, Souza AP, Brito RB, Line SR. Investigation of il4 gene polymorphism in individuals with different levels of chronic periodontitis in a brazilian population. J Clin Periodontol. 2003; 30: 341-345.
- Stashenko P, Fujiyoshi P, Obernesser MS, Prostak L, Haffajee AD, Socransky SS. Levels of interleukin 1 beta in tissue from sites of active periodontal disease. J Clin Periodontol. 1991; 18: 548-554.
- Alexander MB, Damoulis PD. The role of cytokines in the pathogenesis of periodontal disease. Curr Opin Periodontol: 1994; 39-53.
- Graves DT, Cochran D. The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J Periodontol. 2003; 74: 391-401.
- Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999; 340: 448-454.
- Costa PP, Trevisan GL, Macedo GO, Palioto DB, Souza SL, Grisi MF, et al. Salivary interleukin-6, matrix metalloproteinase-8, and osteoprotegerin in patients with periodontitis and diabetes. J Periodontol. 2010; 81: 384-391.
- Imamura Y, Fujigaki Y, Oomori Y, Kuno T, Ota N, Wang PL. Polymorphism of genes encoding toll-like receptors and inflammatory cytokines in periodontal disease in the japanese population. J Int Acad Periodontol. 2008; 10: 95-102.
- Laine ML, Loos BG, Crielaard W. Gene polymorphisms in chronic periodontitis. Int J Dent. 2010: 324719.
- Laine ML, Crielaard W, Loos BG. Genetic susceptibility to periodontitis. Periodontol. 2012; 2000; 58: 37-68.
- Lopez NJ, Valenzuela CY, Jara L. Interleukin-1 gene cluster polymorphisms associated with periodontal disease in type 2 diabetes. J Periodontol. 2009; 80: 1590-1598.
- Geismar K, Enevold C, Sorensen LK, Gyntelberg F, Bendtzen K, Sigurd B, et al. Involvement of interleukin-1 genotypes in the association of coronary heart disease with periodontitis. J Periodontol. 2008; 79: 2322-2330.
- Hou LT, Liu CM, Chang WK. Increased interleukin-1 beta levels in gingival crevicular fluid of chinese periodontal patients. J Formos Med Assoc. 1994; 93: 99-103.
- Li QY, Zhao HS, Meng HX, Zhang L, Xu L, Chen ZB, et al. Association analysis between interleukin-1 family polymorphisms and generalized aggressive periodontitis in a chinese population. J Periodontol. 2004; 75: 1627-1635.
- Hao L, Li JL, Yue Y, Tian Y, Wang M, Loo WT, et al. Application of interleukin-1 genes and proteins to monitor the status of chronic periodontitis. Int J Biol Markers. 2013; 28: e92-e99.
- Sheng WS, Hu S, Ni HT, Rowen TN, Lokensgard JR, Peterson PK. Tnf-alphainduced chemokine production and apoptosis in human neural precursor cells. J Leukoc Biol. 2005; 78: 1233-1241.
- Babel N, Cherepnev G, Babel D, Tropmann A, Hammer M, Volk HD, et al. Analysis of tumor necrosis factor-alpha, transforming growth factor-beta, interleukin-10, il-6, and interferon-gamma gene polymorphisms in patients with chronic periodontitis. J Periodontol. 2006; 77: 1978-1983.
- Pontes CC, Gonzales JR, Novaes AB, Taba Junior M, Grisi MF, Michel J, et al. Interleukin-4 gene polymorphism and its relation to periodontal disease in a brazilian population of african heritage. J Dent. 2004; 32: 241-246.
- Hooshmand B, Hajilooi M, Rafiei A, Mani-Kashani KH, Ghasemi R. Interleukin-4 (c-590t) and interferon-gamma (g5644a) gene polymorphisms in patients with periodontitis. J Periodontal Res. 2008; 43: 111-115.
- Anovazzi G, Kim YJ, Viana AC, Curtis KM, Orrico SR, Cirelli JA, et al. Polymorphisms and haplotypes in the interleukin-4 gene are associated with chronic periodontitis in a brazilian population. J Periodontol. 2010; 81: 392- 402.
- Berglundh T, Donati M, Hahn-Zoric M, Hanson LA, Padyukov L. Association of the -1087 il 10 gene polymorphism with severe chronic periodontitis in swedish caucasians. J Clin Periodontol. 2003; 30: 249-254.
- Parameter on periodontitis associated with systemic conditions. American academy of periodontology. J Periodontol. 2000; 71: 876-879.
