Abstract
Exosomes have been extensively studied as diagnostic biomarkers and for monitoring disease progression. This study examined the role of exosomes in the pathogenesis of candidemia. Exosomes in the serum of patients with candidemia were analyzed by liquid chromatography-mass spectrometry/ mass spectrometry. Proteins differentially expressed in patient (n=4) vs. control (n=4) sera were confirmed by Enzyme-Linked Immunosorbent Assay (ELISA). Protein expression was evaluated using receiver operating characteristic curve analysis. In the serum samples of the patients, 4,055 fragment ions and 418 proteins were detected, including 11 proteins whose expression levels differed significantly from the controls. Three of these proteins (cathepsin b, pentraxin 3, and the serine protease inhibitor A3N) were shown by ELISA to be closely related to the development of candidemia. Our results suggest a role for serum exosomes as molecular markers in the diagnosis and monitoring of candidemia.
Keywords: Candidemia; Candida albicans; Plasma exosomes; Biomarkers; Proteomics
Introduction
Candida albicans is present on the skin and mucosal surfaces; it is the most common opportunistic fungal pathogen in humans, and is responsible for up to 50% of all cases of invasive candidiasis [1]. Due to the lack of innovative prophylactic and therapeutic strategies against fungal pathogens, invasive candidiasis has high rates of morbidity and mortality [2]. While blood cultures have a very high positive predictive value, due to the small fungal load and difficulties in culturing fungal pathogens their overall positivity rate for the diagnosis of bloodstream infections is only 20% [3]. Identification of serum biomarkers would allow early diagnosis of candidemia.
Exosomes are lipid bilayer vesicles with a diameter of 30-200 nm that are produced by endocytosis and secreted by multiple cell types. They are present in the supernatants of cell cultures and body fluids, including plasma, saliva, breast milk, urine, blood, cerebrospinal fluid, bile, and lymph [4]. Their role as functional organelles in a variety of biological processes, including the immune response, is evidenced by their protein, lipid, and RNA contents [5]. For example, using Liquid Chromatography-Mass Spectrometry/Mass Spectrometry (LC-MS/ MS), researchers have identified >100 phosphoproteins in plasma exosomes with significantly higher expression levels in breast cancer patients than healthy controls [6]. Another study demonstrated that the presence of the GPCI protein in exosomes can serve as a marker for early diagnosis of pancreatic cancer [7]. Similar roles for other exosome biomarkers (CD44, CD47, CXCR4, Del-1, HER2 and KDR) in the early diagnosis and prognosis of breast cancer have been reported [8].
In this study, LC-MS/MS was used for quantitative detection of fragment ions and peptides produced after the digestion of exosomes isolated from the serum of patients with candidemia. Our aim was to obtain a better understanding of the pathogenesis of candidemia based on comparison of exosome protein expression between patients with candidemia and healthy controls. A bioinformatics approach was then applied to analyze the cellular components, molecular functions, and potential signaling pathways of these proteins with respect to the pathogenesis of candidemia.
Materials and Methods
Patients and controls
Plasma samples for method optimization were contributed by ourselves, and by patients diagnosed with sepsis at The Fifth People’s Hospital of Shanghai (Shanghai, China), in accordance with the requirements of the Ethics Committee of the Fifth People’s Hospital of Shanghai. Candidemia was diagnosed by a clinician and confirmed by blood culture. Written informed consent was obtained from all patients. The plasma samples were stored in aliquots at -80oC prior to their use.
Serum exosome isolation and enrichment
Four 1mL aliquots of the plasma samples were quickly thawed, and then centrifuged for 30min at 3,000 × g and 4oC to remove cellular debris and impurities. The supernatants were diluted to 7mL with precooled 1× phosphate-buffered saline (PBS; Sangon Biotech, China), filtered (0.22mm; Millipore, USA), and then subjected to two rounds of ultracentrifugation for extraction and washing, respectively. Ultracentrifugation was carried out at 150,000 × g and 4oC using a SW41 rotor (Beckman Coulter, USA). After the first round of ultracentrifugation, the supernatant was removed and the pellet was resuspended in 1mL of 1× PBS for the second round. The washed, pelleted exosomes were resuspended in 200mL of 1× PBS.
Nanometer particle size analysis of exosomes
The enriched exosomes were suspended in 100μL of PBS. A 10μL aliquot was then diluted to 1mL with ultrapure water and injected into a S300 nanometer particle size analyzer (NanoSight, UK) to determine the size distribution and concentration of the particles according to the standard procedure. The measurement was performed in triplicate. NanoSight NTA (V3.2) software was used to analyze the results.
Proteolysis
FASP enzymolysis [9] was carried out as follows: 1) Exosome proteins were precipitated in five volumes of an organic solution composed of 50% ethanol, 50% acetone, and 0.1% acetic acid, and the obtained precipitate was dissolved in 6mol guanidine hydrochloride/L. After overnight freezing at -20oC, 2μL of the reducing agent DLdithiothreitol was added and the mixture was incubated for 1h at 60oC, followed by the addition of 10μL of the alkylating agent indole- 3-acetic acid and a 40min incubation in the dark at 4oC. The resulting protein solution was transferred to a 10kDa filter tube and centrifuged for 20min at 12,000 × g and 4oC to concentrate the proteins. Trypsin (Promega, USA) was then added at an enzyme:protein ratio of 1:20. After enzymatic hydrolysis, the peptides were desalted using a C18 cartridge, freeze-dried, and then reconstituted in 40μL of dissolution buffer [9].
LC-MS/MS data acquisition
Dried peptides were resuspended in 0.1% formic acid to a final concentration of 0.5mg/mL; 2mL of each sample was then analyzed in an Easy Nano-UPLC 1000 coupled with a QE Plus mass spectrometer (Thermo Scientific, USA). The peptides were eluted using a gradient of buffer B (0.1% TFA in acetonitrile), as follows: 0-65 min, 3-35% B; 65-68 min, 35-80% B; and 68-75 min, 80% B at a flow rate of 300nL/ min. The MS data files were acquired in data-dependent mode. The resolution of the full-scan MS spectra (m/z 300-1,500) was 70,000 (at m/z 200), with an Automatic Gain Control (AGC) of 2 × 105, a maximum injection time of 100ms, and a precursor charge of 2-6. The 20 most abundant precursors identified in the full scan were selected with an isolation window of 2.0m/z and fragmented by higher energy collision-induced dissociation at a normalized energy of 35%. The AGC was 1 × 104, the maximum injection time was 35ms, and the dynamic exclusion time was 60s.
Protein identification and quantitative analysis
The original files (raw files) were searched using MaxQuant software (version 1.5.5.1; www.maxquant.org). Protein identification and the results of the quantitative analysis were obtained using the following search criteria: missed trypsin cleavage site to allow two loci, oxidative and acetylated modifications as variables, quality of the precursor and fragment ion error of 6 ppm [10-6], 0.5-Da protein identification of standards: peptides based on at least two characteristics, p <0.05, a False Positive Rate (FDR) of <1%, and determination of the relative abundance of the proteins based on the values of at least two peptides. A significant difference between patients and controls was defined as a p value <0.01. A >2-fold change was considered to reflect significant up-regulation, and a <0.5-fold change significant down-regulation.
Enzyme-linked immunosorbent assay (ELISA)
Protein markers identified in the serum exosome proteomics analysis as potentially associated with candidemia were confirmed by ELISA, which was performed using commercially available kits (Biosh, China) in accordance with the manufacturer’s instructions.
Results
Enrichment and separation of serum exosomes
The exosomes isolated from serum were 100-150 nm (Figure 1A and B) in size, consistent with their known morphological characteristics. The size range was further confirmed by transmission electron microscopy, which also showed the bilayer goblet structure of the exosomes (Figure 1C). The efficiency of the exosome preparation method was assessed by immunoblot analysis of a known exosome marker, CD63 (Figure 1D). Together, these results demonstrated successful enrichment of serum exosomes and their suitability for use in the experiments.
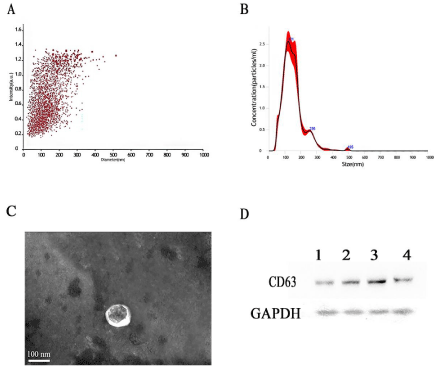
Figure 1: Extraction and characterization of exosomes. A,B) Exosome particle size distribution. C) Transmission electron microscopy images of exosomes. D)
Western blot analysis of CD63.
Differential protein expression and clustering analysis
The serum samples of the control (n=4) and candidemia (n=4) groups were analyzed by label-free proteomics. Based on a difference between the two groups of >1.5 or <0.667 at p <0.05, 91 differentially expressed proteins were obtained, including 32 and 59 that were upand down-regulated in the candidemia group, respectively (Figure 2A).
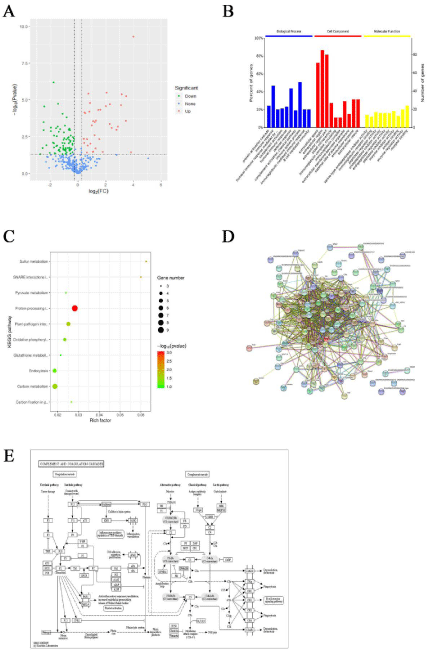
Figure 2: A) The dysregulated proteins in candidemia patients. B) Gene Ontology (GO) enrichment analysis of differentially expressed proteins. C) KEGG pathway
enrichment of differentially expressed proteins. D) Overview of the Protein-Protein Interaction (PPI) network analysis. E) Details of the PPI.
Gene Ontology (GO) enrichment analysis of differentially expressed proteins
Fisher’s exact test was used to conduct a GO enrichment analysis of the differentially expressed proteins (Figure 2B). Most were primarily involved in biological processes, such as protein activation cascade, immune response, humoral immune response by immunoglobulin, complement activation and humoral immune response, extracellular space, extracellular region, extracellular region part, blood microparticles and immunoglobulin complex, serinetype endopeptidase inhibitor activity, and immunoglobulin receptor binding. Significant changes in the expression of proteins involved in peptidase regulator activity, endopeptidase inhibitor activity, and endopeptidase regulator activity were also determined.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed proteins
The annotation of significantly differentially expressed proteins using the KEGG pathway can provide insights into metabolic and signaling pathways. In this study, KEGG pathway enrichment of the differentially expressed proteins of candidemia patients was analyzed using Fisher’s exact test. Significant changes in the complement and coagulation cascades, and in the Staphylococcus aureus infection, malaria, pertussis and prion disease pathways, were found (Figure 2C). Details of the complement and coagulation cascade pathways are shown as an example in Figure 2E.
Protein-protein interaction (PPI) network analysis
Investigations of both the interaction between proteins and the network formed by those interactions can provide information on protein function. The results of a PPI network analysis can then be combined with those of pathway annotation to obtain a comprehensive and systematic model of cell activity at the molecular level. The PPI network constructed based on the differentially expressed proteins identified in this study is shown in Figure 2D. Pathways related to the STAT cascade, ion transport, acute inflammatory response, and leukocyte-mediated immunity were enriched.
Validation of the identified dysregulated proteins
According to a significance criterion of a >2-fold change, 11 proteins that differed significantly among the candidemia samples were identified (Table 1), with the largest differences being seen in cathepsin B, pentraxin 3, and the serine protease inhibitor A3N. To validate our findings, serum was collected from 10 healthy individuals (control group) and 10 patients diagnosed with candidemia. The levels of the three proteins in serum samples from both groups were then determined by ELISA. The cathepsin B expression level in the control and candidemia groups was 232.07 ± 21.80 and 282.65 ± 39.76 ng/mL, respectively; the difference was significant (t = 3.53, P <0.01; Figure 3A and 3B). The difference in expression of pentraxin 3 between the control (40.53 ± 9.55 ng/mL) and candidemia (61.34 ± 10.17 ng/mL) groups was also significant (t = 4.72, P <0.01), as was the difference in expression of A3N (51.678 ± 19.80 and 95.351 ± 23.11 ng/mL, respectively; t = 4.608, P <0.01), as shown in Figure 3C. A Receiver Operating Characteristic (ROC) diagnostic model was established using GraphPad Prism 6.0 software. The Area under the Curve (AUC) for cathepsin B was 0.9100, with a 95% Confidence Interval (CI) of 0.78-1.03. For pentraxin 3 and A3N, the AUC was 0.930 and 0.940, and the 95% CI was 0.819-1.041 and 0.843-1.037, respectively, as shown in Figure 3D.
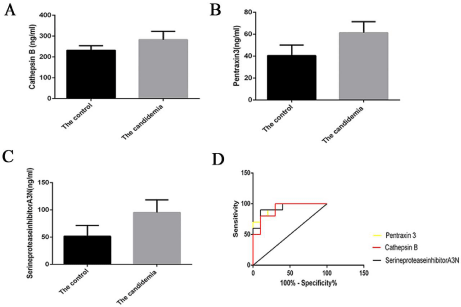
Figure 3: A) Cathepsin B expression in the control group (232.07 ± 21.80 ng/mL) was significantly higher than in the candidemia group (282.65 ± 39.76 ng/mL; t
= 3.53, P <0.01). B) Pentraxin 3 expression in the control group (40.53 ± 9.55 ng/mL) was significantly higher than in the candidemia group (61.34 ± 10.17 ng/mL;
t = 4.72, P <0.01). C) Expression of the serine protease inhibitor A3N in the control group (51.678 ± 19.80 ng/mL) was significantly higher than in the candidemia
group (95.351 ± 23.11 ng/mL; t = 4.608, P <0.01). D) Receiver Operating Characteristic (ROC) curve analysis of cathepsin B, pentraxin 3, and A3N: controls vs.
candidemia patients. The Area under the Curve (AUC) of cathepsin B was 0.9100 and the 95% Confidence Interval (CI) was 0.78–1.03; for pentraxin 3, the AUC
was 0.930 and the 95% CI was 0.819-1.041, and for A3N, the AUC was 0.940 and the 95% CI was 0.843-1.037.
Gene names
Peptides
Ratio (The patients/The healthy)
p value (The patients/The healthy)
Q5BKC4
C9
28
2.748338
4.88E-05
Q6IN22
Cathepsin B
Ctsb
3
3.42543
0.034993
P27139
Ca2
6
8.901092
4.17E-06
A0A0H2UHM3
Hp
27
2.117521
0.000288
P06238
Alpha-2-macroglobulin
A2m
82
11.16629
5.72E-06
A0A0H2UHF8
Orm1
7
16.03046
5.07E-10
D3ZT94
Ptx3
9
8.546177
2.25E-05
H6X2X0
C-reactive protein
Crp
7
2.333257
0.000325
A0A0H2UHI5
Serpina3n
28
4.053251
2.71E-05
P20059
Hpx
27
2.442018
0.000254
Q7TQ70
Fga
26
3.043354
4.64E-05
Table 1: Eleven proteins differentially expressed between healthy controls and patients with candidemia.
Application of exosome proteome profiling to a temporal study of candidemia
We applied our novel exosome proteome profiling approach to study temporal changes in the exosomal proteomes of patients diagnosed with candidemia. Plasma samples were collected from patients on days 1-3 after the diagnosis of candidemia. Candidemia progression was monitored by measuring three clinical indicators: the levels of C - Reactive Protein (CRP) and Interleukin (IL)-6 and the APACHE-II score (Figure 4A-4C). All three indicators increased between days 1 and 3, suggesting a dynamic acute phase response. Whether these changes were reflected in the exosome proteome of the patients was then examined. Cathepsin B, pentraxin 3, and A3N levels were higher in the candidemia group than in the control group on days 1-3 (Figure 4D-4F). These results demonstrated that cathepsin B, pentraxin 3, and A3N are predictors of candidemia, with good sensitivity and specificity.
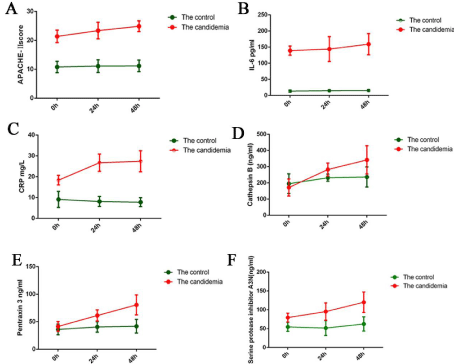
Figure 4: A-F) C-Reactive Protein (CRP) levels, interleukin (IL)-6 levels, and the APACHE-II score according to the changes in cathepsin B, pentraxin-3, and A3N
on days 1-3 after diagnosis of candidemia.
Discussion
This study showed differences in the expression of plasma exocrine proteins between candidemia patients and healthy controls. Therefore, our method, combining ultracentrifugation and MS, can be used to diagnose and monitor candidemia. Among the 11 plasma exosomes proteins with significantly increased expression in the candidemia group, 3 were further assessed in terms of their potential as molecular markers: cathepsin B, pentraxin 3, and A3N4. Cathepsin B is a cysteine proteolytic lysosomal enzyme of the papain family widely distributed in a variety of tissue cells. Candida albicansinduced NLRP3 inflammasome activation is thought to be regulated by at least two pathways: in the first, phagocytosis of extracellular bacteria results in lysosomal destabilization and the release of cathepsin B; in the second, extracellular exposure to a bacterial poreforming toxin or ATP results in potassium efflux and a decrease in intracellular potassium levels [10]. One of the main pathogenic toxins of C. albicans is a serine protease, which is inhibited by A3N [11]. In a study of NK92 cells, A3N was shown to block the inflammation induced by lipopolysaccharide, by regulating endoplasmic reticulum stress signaling, thus affecting granzyme B activity [12]. However, the role of A3N in C. albicans infection remains unknown. Pentraxin 3, a soluble pattern recognition receptor of the humoral innate immune system, is an antibody precursor [13] that recognizes foreign microorganisms and modulates the inflammatory response. The protein, which is produced by phagocytes and stored in neutrophils, has a nonredundant role in mediating resistance to fungal pathogens [14]. Pentraxin 3 is also produced by vascular endothelial cells and macrophages; its expression levels directly reflect inflammatory status. Moreover, because of its extrahepatic synthesis, pentraxin 3 levels are superior as an independent indicator of disease activity compared to CRP [15]. Increased pentraxin 3 plasma levels have been demonstrated in patients with pulmonary aspergillosis, tuberculosis, dengue virus infection, meningococcal disease, leptospirosis, and shigellosis, and were shown to correlate with disease severity and predict an unfavorable outcome [16]. Although the role of pentraxin 3 in C. albicans infection has yet to be determined, our exosome proteomics analysis showed that it was highly specifically expressed during the infection thus may serve a useful marker of candidemia.
Conclusion
Our study of the proteome of serum exosomes in patients with candidemia revealed differential expression of exocrine proteins. While the functions of these proteins remain to be elucidated, our results demonstrated that they are potential targets for the diagnosis and treatment of candidemia.
Acknowledgements
We thank the department of Trauma-Emergency & Critical Care medicine of Shanghai Fifth People’s Hospital, particularly Dr. Jianguo Tang, for help in disseminating the survey to the group.
Funding
This study was funded by Shanghai Health and Family Planning Commission Research Project Youth Project (20174Y0061); Special fund for high-level talents construction of Shanghai Fifth People’s Hospital, Fudan University (9-1).
Availability of Data and Material
We declared that materials described in the manuscript, including all relevant raw data, will be freely available to any scientist wishing to use them for non-commercial purposes, without breaching participant confidentiality.
Authors’ Contributions
JGT conceived of the study. ZTW prepared samples and drafted the initial manuscript. ZTW participated in data analysis. All authors participated in data synthesis and interpretation of results. All authors read and approved the final manuscript.
Ethics Approval and Consent to Participate
The study protocol was approved by the Ethical and Protocol Review Committee of The Fifth People’s Hospital of Shanghai, Shanghai, China. All procedures performed in the study were in accordance with the ethical standards of The Fifth People’s Hospital of Shanghai. Written informed consents were obtained from all study participants.
Patient Consent for Publication
All authors have read and approved the content, and agree to submit for consideration for publication in the journal. There are no any ethical/legal conflicts involved in the article.
References
- Panahi, Jafar, Havasian, et al. In Vitro Inhibitory Effect Of Alcoholic Extract Of Inner Stratum Of Oak Fruit (Jaft) On Candida Albicans. Iranian journal of public health. 2014; 43: 277-277.
- Nami S, Mohammadi R, Vakili M, Khezripour K, Mirzaei H and Morovati H. Fungal vaccines, mechanism of actions and immunology: A comprehensive review. Biomed Pharmacother. 2019; 109: 333-344.
- Mohon AN, Oberding L, Hundt J, et al. Optimization and clinical validation of dual-target RT-LAMP for SARS-CoV-2. J Virol Methods. 2020; 286: 113972.
- Zhang Y, Chen B, Xu N, et al. Exosomes Promote the Transition of Androgen- Dependent Prostate Cancer Cells into Androgen-Independent Manner through Up-Regulating the Heme Oxygenase-1. Int J Nanomedicine. 2021; 16: 315-327.
- Zhang W, Ou X and Wu X. Proteomics profiling of plasma exosomes in epithelial ovarian cancer: A potential role in the coagulation cascade, diagnosis and prognosis. Int J Oncol. 2019; 54: 1719-1733.
- Li S and Wang X. The potential roles of exosomal noncoding RNAs in osteosarcoma. J Cell Physiol. 2020.
- Li J, Li B, Ren C, et al. The clinical significance of circulating GPC1 positive exosomes and its regulative miRNAs in colon cancer patients. Oncotarget. 2017; 8: 101189-101202.
- Wang M, Ji S, Shao G, et al. Effect of exosome biomarkers for diagnosis and prognosis of breast cancer patients. Clin Transl Oncol. 2018; 20: 906-911.
- Doellinger J, Schneider A, Hoeller M and Lasch P. Sample Preparation by Easy Extraction and Digestion (SPEED) - A Universal, Rapid, and Detergentfree Protocol for Proteomics Based on Acid Extraction. Mol Cell Proteomics. 2020; 19: 209-222.
- Wang Y, Jia L, Shen J, et al. Cathepsin B aggravates coxsackievirus B3- induced myocarditis through activating the inflammasome and promoting pyroptosis. PLoS Pathog. 2018; 14: e1006872.
- Dietrich MA, Slowinska M, Karol H, et al. Serine protease inhibitor Kazal-type 2 is expressed in the male reproductive tract of carp with a possible role in antimicrobial protection. Fish Shellfish Immunol. 2017; 60: 150-163.
- Wang L, Jiang S, Xiao L, Chen L, Zhang Y and Tong J. Inhibition of granzyme B activity blocks inflammation induced by lipopolysaccharide through regulation of endoplasmic reticulum stress signaling in NK92 cells. Mol Med Rep. 2018; 18: 580-586.
- Cui X, Zhang H, Cao A, Cao L and Hu X. Cytokine TNF-alpha promotes invasion and metastasis of gastric cancer by down-regulating Pentraxin3. J Cancer. 2020; 11: 1800-1807.
- Sugano A, Seo Y, Ishizu T, et al. Soluble ST2 and brain natriuretic peptide predict different mode of death in patients with heart failure and preserved ejection fraction. J Cardiol. 2019; 73: 326-332.
- Ramirez GA, Rovere-Querini P, Blasi M, et al. Corrigendum: PTX3 Intercepts Vascular Inflammation in Systemic Immune-Mediated Diseases. Front Immunol. 2019; 10: 1755.
- Porte R, Davoudian S, Asgari F, et al. The Long Pentraxin PTX3 as a Humoral Innate Immunity Functional Player and Biomarker of Infections and Sepsis. Front Immunol. 2019; 10: 794.
