
Research Article
Austin Immunol. 2016; 1(2): 1011.
Association between the Promoter -675 4G/5G Polymorphism of the Plasminogen Activator Inhibitor-1 Gene and Asthma: An Update of Meta-Analysis
Huang X, Yang M, Wang Y, Zhang X and Yang H*
Department of Preventive Medicine, Hubei University of Chinese Medicine, P. R. China
*Corresponding author: Haijun Yang, Department of Preventive Medicine, College of Basic Medical Sciences, Hubei University of Chinese Medicine, Wuhan 430065, Hubei, P. R. China
Received: October 12, 2016; Accepted: November 09, 2016; Published: November 10, 2016
Abstract
Background and Objectives: Several studies have explored the association between the promoter -675 4G/5G polymorphism of the Plasminogen Activator Inhibitor-1 (PAI-1) gene and asthma risk; however the results are inconsistent. The purpose of this study was to evaluate the genetic risk of this polymorphism for asthma using the method of meta-analysis.
Methods: Systemic electronic literature search was conducted on PAI-1 polymorphism and asthma risk in several databases. The data were pooled employing the meta-analysis method.
Results: Eight case-control studies involving 1551 asthmatics and 2339 healthy controls were included in this meta-analysis. In the overall population, our results showed that the PAI-1 -675 4G/5G polymorphism was significantly associated with elevated asthma risk in a dominant genetic model [odds ratio (OR)=1.716, 95% confidence interval (CI)=1.190-2.474]. Stratified analyses were conducted based on ethnicity, age and atopic status of asthmatic patients. We observed that the PAI-1 -675 4G allele carriers have increased risk of asthma in both Caucasian and Asian populations (OR=1.749, 95% CI=1.084- 2.823 and OR=1.456, 95% CI=1.019-2.081, respectively). Increased risk of asthma was also seen in adult and children populations of the PAI-1 -675 4G allele carriers (OR=1.558, 95% CI=1.028-2.360 and OR=2.380, 95% CI=1.486- 3.811, respectively). In the case of atopic asthma and non-atopic asthma, the PAI-1 4G/5G polymorphism was significantly associated with atopic asthma susceptibility (OR=2.436, 95% CI=1.783-3.328 for 4G4G+4G5G vs. 5G5G).
Conclusion: Data indicated that the PAI-1 -675 4G/5G polymorphism was associated with increased asthma risk. Recommendations for further studies include pooling of individual data to facilitate assessment of gene-gene and gene-environment interactions in asthma susceptibility.
Keywords: Asthma; Meta-analysis; Plasminogen activator inhibitor-1; Polymorphism
Abbreviations
BHR: Bronchial Hyperresponsiveness; PAI: Plasminogen Activator Inhibitor; OR: Odds Ratio; CI: Confidence Interval; CNKI: Chinese National Knowledge Infrastructure; HWE: Hardy-Weinberg Equilibrium
Introduction
Asthma is a common chronic inflammatory respiratory disease, which is characterized by chronic airway inflammation, Bronchial Hyperresponsiveness (BHR) and airway remodeling. The prevalence of asthma is high in developed countries and there is a concern that its prevalence is still rising in both developed and developing countries [1]. It is widely accepted that asthma is a complex polygenic disease whose pathogenesis involves complex interactions of environmental and genetic factors [2,3]. In the past decades, much effort has been made to explore the susceptible genes of asthma.
The Plasminogen Activator Inhibitor (PAI)-1, a 50 kD glycoprotein, belongs to the SERPIN family, which is abbreviated from the serine protease inhibitor. PAI-1 is the key inhibitor of the fibrinolytic system by hindering the activation of plasminogen and is known to play an essential role in tissue remodeling [4]. PAI-1 might play an important role in the pathogenesis of asthma [4]. For instance, Kowal, et al. and Xiao, et al. have reported that there are elevated PAI- 1 levels in the induced sputum or plasma of patients with asthma in comparison with that of healthy controls [5,6], and the plasma PAI-1 levels of asthmatics were pronouncedly up-regulated during allergen challenge [6]. A large number of mast cells, a predominant effector cell of asthma, expressing high level of PAI-1 were found in the lung tissue of severe asthmatics [7].
A polymorphism at position -675 in the 5’ terminal promoter region of the PAI-1 gene, consisting of two alleles 4G and 5G (PAI-1 -675 4G/5G, rs1799889), has been described to regulate transcription of the PAI-1 gene. For example, Dawson, et al. and Kowal, et al. have reported that the plasma levels of PAI-1 are higher in individuals with the 4G4G genotype than in those with the 5G5G genotype, whereas the 4G5G genotype has intermediate values [6,8]. Since Cho, et al. demonstrated that the PAI-1 4G allele is preferentially transmitted to asthmatic children based on the transmission disequilibrium test in nuclear families from the UK [9], several case-control studies have investigated the association between the PAI-1 4G/5G polymorphism and asthma risk [10-18]. However, the results of these studies were inconsistent and inconclusive. Because a single study may have low power to detect the effect of polymorphism, it is necessary to carry out a meta-analysis to summarize the effect size of PAI-1 4G/5G polymorphism on asthma risk. To date, one meta-analysis of association of this polymorphism with asthma risk has been reported [19], however, the results needed further evaluation for the following reasons: firstly, the included studies were not strictly checked in accordance with their inclusion criteria and some overlapped studies were included into the meta-analysis more than once; secondly, the previous meta-analysis was performed using a classical meta-analysis method. Considering a meta-analysis of genetic polymorphism association studies involving multiple comparisons with a classical method, this might increase the risk of type I error. Thirdly, one new case-control study concerning this polymorphism and asthma risk has been reported since the meta-analysis was published.
Based on above analysis, an updated meta-analysis was performed to summarize reported case-control studies concerning the PAI-1 -675 4G/5G polymorphism and asthma risk in all ethnic populations according to the framework for conducting a meta-analysis of molecular association studies [20]. To overcome the limitation of the classical meta-analysis involving multiple comparisons in genetic association studies, a logistic-regression based meta-analysis of genetic association case-control studies was applied to calculate the Odds Ratio (OR) and 95% Confidence Interval (CI) for PAI-1 -675 4G4G versus (vs.) 5G5G (OR4G4G vs. 5G5G) and 4G5G vs. 5G5G (OR4G5G vs. 5G5G) and to decipher the most plausible genetic action model [21].
Methods
Search strategy and inclusion criteria
A systematic literature search was carried out in Medline, Embase, Wanfang, Weipu and Chinese National Knowledge Infrastructure (CNKI) to indentify studies concerning the association between the PAI-1 polymorphism and asthma susceptibility. The following search terms were used: “asthma or asthmatic” in combination with “plasminogen activator inhitor-1 or PAI-1 or SERPIN-1”. We reviewed all related studies published before October 1, 2016.
The studies which meet the following criteria were incorporated into this meta-analysis: (1) the paper should include asthma risk and PAI-1 4G/5G polymorphism; (2) the article should be published in English or Chinese; (3) only case-control or cohort studies were considered, however, the study design should not be a family-based association or sibling pairs; (4) the study should clearly report the frequencies of each genotype; (5) when there were multiple publications from the same group, only the most recent or the publication with more complete information was included in the analysis.
Data extraction
The following information was extracted from each study: the name of the first author, publication year, country of origin, ethnicity and age of subjects, sample size and asthma definition and frequencies of each genotype.
Statistical analysis
The effect size of the PAI-1 -675 4G/5G polymorphism on asthma risk was evaluated using OR with corresponding 95% CI. Firstly, the 4G and 5G alleles of PAI-1 4G/5G promoter polymorphism were compared. Then the risks of a dominant model (4G4G+4G5G vs. 5G5G) and a recessive model (4G4G vs. 4G5G+5G5G) were estimated. Data were pooled using a fixed-effect model when there was no significant heterogeneity, otherwise a random-effect model (DerSimonian and Laird method) was used [22]. The statistical significance of summary ORs was analyzed by the Z test. A chisquare- based Cochran’s Q statistic and index of inconsistency (I2) were employed to assess heterogeneity among studies [23].
To calculate ORs for 4G4G vs. 5G5G genotypes and 4G5G vs. 5G5G genotypes, which involve multiple comparisons if using the classical meta-analysis method, a novel logistic-regression based meta-analysis of case-control genetic association studies was adopted [21]. It was reported that this methodology could avoid multiple comparisons and give the most plausible genetic model based on statistical results rather than empirical observations. The estimation algorisms are as follows: OR4G4G vs. 5G5G (OR1) and OR4G5G vs. 5G5G (OR2) are calculated using the logic-regression-based method and then compared, if OR1=OR2=1, no statistically significant association was indicated; if OR1>1 and OR2=1 (the difference between OR1 and OR2 are statistically significant), then a recessive genetic model is proposed; if OR1>OR2>1 (statistically significantly), then a codominant model is suggested; if OR1=OR2>1, then a dominant model is indicated.
Deviations from Hardy-Weinberg Equilibrium (HWE) of the genotype distribution of each control group were assessed by Pearson’s chi-squared test. Publication bias was examined using Egger’s regression test and Begg’s rank correlation method [24,25]. All statistical analyses were performed using STATA of version 10.1 (STATA Corporation, College Station, Texas, USA). All tests were two-sided, and a P value of less than 0.05 was considered to be statistically significant, with the exception of heterogeneity tests where a P value less than 0.10 and I2 value of more than 50.0% were used.
Results
Characteristics of studies included in the meta-analysis
A total of 57 articles were identified after the initial search, including 46 papers written in English and 11 in Chinese. Based on the abstract of each article, 11 studies were enrolled for full-text review. After reading the full texts, three studies were excluded from the meta-analysis for the following reasons: one for repeated publication [12], one for a family-based association study design [9] and one for being unrelated to asthma risk and PAI-1 4G/5G polymorphism [26]. Accordingly, eight case-control studies were summarized in this meta-analysis, including 1551 asthmatics and 2339 controls [10,11,13-18]. Among all studies, 6 were conducted in Caucasians [10,13-15,17,18] and 2 were performed in Asians [11,16]. Six studies were conducted in adults [10,11,13,14,16,17] and 2 were performed in children [15,18]. Four studies involved atopic asthmatics [10,13,14,18] and 2 studies clearly reported non-atopic asthmatics [14,18]. The characteristics of the included studies are listed in (Table 1) and the detailed allele and genotype frequencies of the 4G/5G polymorphism of PAI-1 in each study are shown in (Table 2). The genotype frequency distributions of control groups were all in consistent with HWE (Table 2).
First author
Year
Country
Ethnicity
Case age (year)
Control age (year)
Asthmatic category
Asthma definition
Buckova D [10]
2002
Czech
Caucasian
26.9±12.8
32.6±10.4
Atopic
Questionnaire with physician diagnosed asthma
Hizawa N [11]
2006
Japan
Asian
45 (16-81)
32 (18-72)
Mixed
Asthma diagnosed by a physician
Kowal K [13]
2008
Poland
Caucasian
25 (23-26)
24 (23-26)
Atopic
Guidelines of the Global Initiative for Asthma
Cosan D [14]
2009
Turkey
Caucasian
42.8±1.05
41.8±1.90
Mixed
American Thoracic Society
Ozbek OY [15]
2009
Turkey
Caucasian
9.47±2.79
10.80±3.30
NA
American Thoracic Society
Zhang XY [16]
2009
China
Asian
50±15
47±15
NA
Chinese asthma diagnosis criteria (2003)
Dijkstra A [17]
2011
Holland
Caucasian
50 (35-75)
52 (35–79)
NA
Published algorithm [35]
Bora E [18]
2012
Turkey
Caucasian
9.24±2.92
10.84±3.15
Mixed
Guidelines of the Global Initiative for Asthma
Table 1: Characteristics of the eight case-control studies included in the meta-analysis.
Studies
Case
Control
Case
Control
HWE
No.
5G5G
5G4G
4G4G
No.
5G5G
5G4G
4G4G
5G
4G
5G
4G
?2
P
Overall
Buckova D [10]
159
27
75
57
186
50
83
53
129
189
183
189
2.414
0.143
Hizawa N [11]
374
49
194
131
374
59
185
130
292
456
303
445
0.259
0.611
Kowal K [13]
372
38
154
180
160
43
70
47
230
514
156
164
2.478
0.115
Cosan D [14]
98
29
43
26
67
19
29
19
101
95
67
67
1.209
0.272
Ozbek Y [15]
106
23
39
44
83
27
41
15
85
127
95
71
0.007
0.934
Zhang XY [16]
99
13
49
37
101
27
46
28
75
123
100
102
0.800
0.371
Dijkstra A [17]
241
54
117
70
1267
275
627
365
225
257
1177
1357
0.035
0.852
Bora E [18]
102
15
63
24
101
37
43
21
93
111
117
85
1.619
0.203
Total
1551
248
734
569
2339
537
1124
678
1230
1872
2198
2480
2.931
0.087
Atopic asthma
BuckovaD [10]
159
27
75
57
186
50
83
53
129
189
183
189
2.414
0.143
Kowal K [13]
372
38
154
180
160
43
70
47
230
514
156
164
2.478
0.115
Cosan D [14]
19
5
9
5
67
19
29
19
19
19
67
67
1.209
0.272
Bora E [18]
67
10
40
17
101
37
43
21
60
74
117
85
1.619
0.203
Total
617
70
278
259
514
149
225
140
438
796
523
505
7.935
0.005
Non-atopic asthma
Cosan D [14]
79
24
34
21
67
19
29
19
72
76
67
67
1.209
0.272
Bora E [18]
35
5
23
7
101
37
43
21
33
37
117
85
1.619
0.203
Total
114
29
57
28
168
56
72
40
105
113
184
152
3.062
0.080
HWE: Hardy-Weinberg Equilibrium
Table 2: PAI-1 -675 4G/5G polymorphism genotype and allele frequencies among asthmatics and controls.
Heterogeneity test
(Table 3) shows the relationship between the PAI-1 -675 4G/5G polymorphism and asthma risk. The heterogeneity of PAI-1 4G/5G polymorphism: 4G versus 5G (allele), 4G4G+4G5G vs. 5G5G (dominant model), and 4G4G vs. 4G5G+5G5G (recessive model), were analyzed in eight case-control studies. The results indicate that both the dominant and recessive comparisons in Asians, all comparisons in atopic asthmatics, allele and recessive model comparisons in nonatopic asthmatics and allele and dominant model comparisons in childhood asthmatics had no significant heterogeneity, therefore those ORs were calculated with a fixed-effect model. A Random-effect model was used to examine the other ORs.
Comparisons
Sample size
No. of studies
Hypothesis test
Heterogeneity test
Publication bias test (P)
Case/control
OR (95% CI)
P
?2 (df)
P
Begg’ test
Egger’test
Overall
4G vs. 5G
3102/4678
8
1.396 (1.108-1.760)
0.005
32.97 (7)
<0.001
0.368
0.279
4G4G vs. 4G5G+5G5G
8
1.394 (1.047-1.856)
0.023
21.07 (7)
0.004
0.368
0.252
4G4G+4G5G vs. 5G5G
1551/2339
8
1.716 (1.190-2.474)
0.004
26.78 (7)
<0.001
0.548
0.291
Caucasians
4G vs. 5G
2156/3728
6
1.445 (1.068-1.954)
0.017
27.75 (5)
<0.001
1.000
0.559
4G4G vs. 4G5G+5G5G
1078/1864
6
1.479 (1.015-2.156)
0.041
17.44 (5)
0.004
1.000
0.524
4G4G+4G5G vs. 5G5G
1078/1864
6
1.749 (1.084-2.823)
0.078
24.25 (5)
<0.001
0.806
0.552
Asians
4G vs. 5G
946/950
2
1.261 (0.846-1.879)
0.255
3.26 (1)
0.071
1.000
NA
4G4G vs. 4G5G+5G5G
473/475
2
1.104 (0.844-1.444)
0.470
1.59 (1)
0.207
1.000
NA
4G4G+4G5G vs. 5G5G
473/475
2
1.456 (1.019-2.081)
0.039
2.41 (1)
0.120
1.000
NA
Atopic asthma
4G vs. 5G
1234/1028
4
1.705 (1.428-2.035)
<0.001
6.12 (3)
0.106
0.308
0.342
4G4G vs. 4G5G+5G5G
617/514
4
1.685 (1.288-2.204)
<0.001
4.33 (3)
0.228
0.308
0.236
4G4G+4G5G vs. 5G5G
617/514
4
2.436 (1.783-3.328)
<0.001
4.96 (3)
0.175
0.734
0.569
Non-atopic asthma
4G vs. 5G
228/336
2
1.252 (0.880-1.780)
0.211
0.96 (1)
0.326
1.000
NA
4G4G vs. 4G5G+5G5G
114/168
2
0.928 (0.520-1.658)
0.802
<0.01 (1)
0.948
1.000
NA
4G4G+4G5G vs. 5G5G
114/168
2
1.682 (0.454-6.235)
0.437
4.40 (1)
0.036
1.000
NA
Adulthood asthma
4G vs. 5G
2686/4310
6
1.297 ( 0.998-1.686)
0.052
26.56 (5)
<0.001
0.452
0.476
4G4G vs. 4G5G+5G5G
1343/2155
6
1.289 (0.967-1.716)
0.083
13.80 (5)
0.017
0.452
0.574
4G4G+4G5G vs. 5G5G
1343/2155
6
1.558 (1.028-2.360)
0.037
21.01 (5)
0.001
0.452
0.351
Childhood asthma
4G vs. 5G
416/368
2
1.804 (1.357-2.396)
<0.001
0.46 (1)
0.499
1.000
NA
4G4G vs. 4G5G+5G5G
208/184
2
1.937 (0.720-5.209)
0.190
4.34 (1)
0.037
1.000
NA
4G4G+4G5G vs. 5G5G
208/184
2
2.380 (1.486-3.811)
<0.001
1.86 (1)
0.172
1.000
NA
OR: Odds Ratio; CI: Confidence Interval; df: degree of freedom
Table 3: Summary odds ratios of the association between PAI-1 -675 4G/5G polymorphism and asthma risk.
Genetic model decipherment
Table 4 shows the ORs for PAI-1 -675 4G4G vs. 5G5G and 4G5G vs. 5G5G based on a logistic-regression method and genetic model decipherment results. The heterogeneity of genetic effect i.e. OR and genetic model i.e. the inferred dominant, co-dominant or recessive model is also listed in (Table 4). Our results indicated that the PAI-1 675 4G/5G polymorphism was associated with asthma risk in the overall population in a dominant genetic model. When stratified analyses based on ethnicity, age and atopic status, the data suggested that the PAI-1 -675 4G/5G polymorphism might be linked with asthma in a co-dominant model in Caucasians and atopic populations; the polymorphism might be complicated in asthma risk in a dominant model in Asian, children and adult populations. Our results also indicated that the PAI-1 4G/5G polymorphism might not be associated with non-atopic asthma risk.
Group
Sample size
(case/control)
OR
Logistic-regression
Heterogeneity test
Genetic model test
Genetic model selection
Genetic effect
Genetic model
OR(95% CI)
P
?2 (df)
P
I2
?2 (df)
P
?2
P
Overall
1551/2339
OR1
1.888 (1.337-2.666)
<0.001
39.987(14)
<0.001
0.650
13.866(7)
<0.001
3.544
0.060
Dominant
OR2
1.520 (1.196-1.932)
0.001
Asians
473/475
OR1
1.475 (0.996-2.185)
0.052
2.909(2)
0.233
0.313
0.499(1)
0.480
0.006
0.941
Dominant
OR2
1.459 (1.004-2.120)
0.047
Caucasians
1078/1864
OR1
1.986 (1.311-3.009)
0.001
35.528(10)
<0.001
0.719
11.957(5)
0.035
4.187
0.041
Co-dominant
OR2
1.527 (1.137-2.050)
0.005
Atopic
617/514
OR1
2.911 (2.038-4.159)
<0.001
8.161(6)
0.227
0.265
3.393(3)
0.335
3.893
0.049
Co-dominant
OR2
2.187 (1.560-3.066)
<0.001
Non-atopic
114/168
OR1
1.248 (0.628-2.484)
0.527
4.654(2)
0.570
0.098
0.427(1)
0.513
0.692
0.405
No association
OR2
1.628 (0.899-2.905)
0.101
Adulthood
1343/2155
OR1
1.669 (1.119-2.490)
0.012
25.930(10)
0.004
0.614
5.687(5)
0.338
1.777
0.183
Dominant
OR2
1.420 (1.080-1.868)
0.012
Childhood
208/184
OR1
3.097 (1.627-5.893)
0.001
9.253(2)
0.010
0.784
7.054(4)
0.008
0.705
0.401
Dominant
OR2
2.018 (0.939-4.336)
0.072
OR: Odds Ratio; OR1: Odds Ratio for 4G4G versus 5G5G comparison; OR2: Odds Ratio for 4G5G versus 5G5G comparison; df: degree of freedom.
Table 4: Summary Odds ratios of 4G4G vs. 5G5G and 4G5G vs. 5G5G comparisons and genetic model decipherment results.
Quantitative data synthesis
Table 3 lists the summary ORs of the PAI-1 4G/5G polymorphism in the form of allele (4G vs. 5G), recessive (4G4G vs. 4G5G+5G5G) and dominant (4G4G+5G5G vs. 5G5G) genetic model in overall and various stratified populations. Our results revealed that the PAI-1 -675 4G4G or 4G5G carriers have 71.6% increased risk of asthma compared with 5G5G homozygote in the overall population [OR (95% CI)=1.716 (1.190-2.474)]. As illustrated in (Figure 1), the 4G variant was associated with elevated risk of asthma in both Caucasians and Asians, with OR (95% CI)=1.749 (1.084-2.823) and 1.456 (1.019- 2.081), respectively. When stratified analysis was conducted by atopic status of asthmatic patients, we observed that the 4G allele was correlated with increased risk of atopic asthma with OR (95% CI) = 2.436 (1.783-3.328). Our data also indicated that the PAI-1 -675 4G variant was involved in increased risk of asthma in populations of both children and adults, as shown in (Figure 2) and (Table 4).
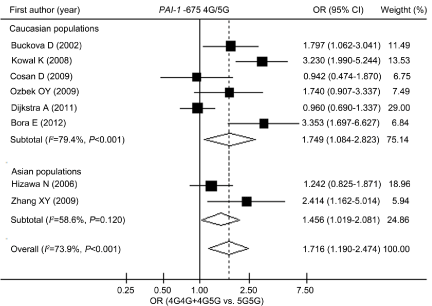
Figure 1: Overall odds ratios from the meta-analysis and odds ratios
from individual studies for the association between PAI-1 -675 4G/5G
polymorphism and risk of asthma (stratified by ethnicity). 95% CI: 95%
Confidence Interval; OR: Odds Ratio.
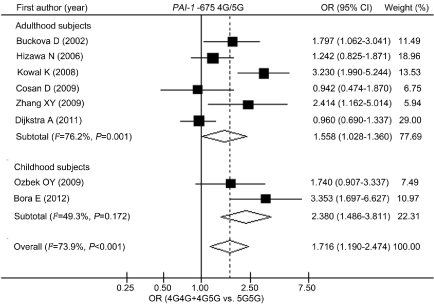
Figure 2: Overall odds ratios from the meta-analysis and odds ratios
from individual studies for the association between PAI-1 -675 4G/5G
polymorphism and risk of asthma (stratified by age). 95% CI: 95% Confidence
Interval; OR: Odds Ratio.
Publication bias diagnosis
Publication bias in this study was assessed by Begg’s rank correlation test and Egger’s regression method. As shown in (Table 4), the results of these two tests both indicated that publication bias in the current meta-analysis was not statistically significant (P>0.05).
Discussion
Due to the important role of PAI-1 in the pathogenesis of asthma, many studies have investigated the association between polymorphisms in the PAI-1 gene and asthma risk. To address the inconsistent results among those studies, a meta-analysis was conducted using a standard procedure of meta-analysis of genetic association studies advocated by Thakkinstian, et al. [20]. Our results indicated that the PAI-1 -675 4G/5G polymorphism was associated with increased asthma risk in the overall population in a dominant genetic model. When stratified analysis was performed based on ethnicity, age and atopic status of asthmatic patients, we observed that the PAI-1 -675 4G/5G polymorphism was associated with elevated asthma risk in both Asians and Caucasians. However, the genetic model in these two types of populations might be different, and a co-dominant genetic model in Caucasians and a dominant genetic model in Asians were indicated, respectively. Our results also suggested that the PAI-1 -675 4G/5G polymorphism was associated with enhanced atopic asthma risk in a co-dominant genetic model, with OR (95% CI) for 4G4G vs. 5G5G and 4G5G vs. 5G5G being equal to 2.911 (2.038-4.159) and 2.187 (1.560-3.066), respectively, using logistic-regression-based meta-analysis method. The difference between the two ORs was statistically significant, indicating a doseresponse relationship effect of the PAI-1 -675 4G/5G polymorphism on atopic asthma risk. With respect to the stratified analysis by age of asthmatic patients, our results showed that the PAI-1 -675 4G/5G polymorphism was associated with increased risk of asthma in populations of both children and adults in a dominant genetic model.
In general, the findings of our study were similar to that of Nie, et al. [19] although several discrepancies also occur. Firstly, we observed that the PAI-1 -675 4G/5G polymorphism is associated with increased asthma risk in Caucasians which was not found in the study of Nie, et al. This might be due to a different ethnic classification. In our study, we define Turkish subjects as Caucasian referring to other well-conducted meta-analyses [27,28]. Secondly, significant associations of the PAI-1 4G/5G polymorphism with elevated asthma risk were seen in both adolescent and adult populations; however Nie, et al. reported no significant association of this polymorphism with adolescent asthma risk [19]. Our analysis showed several advantages compared to the previous meta-analysis [19]: (1) excluding overlapped studies from the analysis; (2) including new publications; (3) overcoming the limitation of classical meta-analysis of molecular genetic studies involving multiple comparisons, reducing the risk of type I error; (4) giving the most plausible genetic model based on statistical analysis rather than empirical observation. Thus the result of our meta-analysis is more reliable and comprehensive compared with the previous one.
There are several lines of evidence suggesting the involvement of the PAI-1 -675 4G/5G polymorphism in the pathogenesis of asthma. For instance, in the stimulated human MC line, Ma, et al. demonstrated that the PAI-1 -675 4G allele has higher promoter activity by binding to the upstream stimulatory factor 1 with greater affinity compared with the 5G allele [29]. Kowal, et al. reported that in allergic asthma patients the PAI-1 gene -675 4G allele corresponded to higher increase in plasma PAI-1 levels and strongly correlated with BHR compared with the 5G allele [6]. The findings of our metaanalysis lend supports to the experimental evidence that the PAI- 1 gene -675 4G allele is associated with higher expression levels of PAI-1 protein [6,29] and the coagulation system such as fibrin was involved in the pathogenesis of asthma [30,31].
It has been reported that IL-13 promotes the deposition of coagulation proteins such as fibrinogen and fibrin on the airway surface [31]. Inflammatory cytokines, in particular TNF-alpha and TGF-beta1, activate the PAI-1 gene transcription [4]. Our previous and other meta-analyses indicated that certain polymorphisms in those genes were associated with susceptibility to asthma [32-34]. In combination with the results of our present study, future studies to clarify the gene-gene interaction among these genes would be of great significance.
There were several limitations, which should be taken into account when interpreting the results of our meta-analysis. Firstly, summary ORs were derived from heterogeneous individual studies, albeit a random-effect model was used to combine data. Subgroup analysis according to ethnicity, heterogeneity was absent in Asians, indicating that ethnicity is a source of heterogeneity. Secondly, all incorporated studies were published in Chinese and English from selected databases, thus some related studies written in other languages or unpublished data might be missing. Thirdly, all existing studies were performed in Caucasians and Asians. No studies were conducted in Africans, thus the results might not apply to Africans. Fourthly, other stratified factors, such us severity of asthma patients, were not considered in this meta-analysis, because most of the included casecontrol studies did not provide information on category severity.
Conclusion
This meta-analysis revealed that the PAI-1 -675 4G/5G polymorphism is associated with increased asthma risk. Further studies should be conducted to ascertain whether the association applies to African populations and to clarify the potential gene-gene interaction such as with IL13 and TGF-beta1 or gene-environment interaction in the susceptibility to asthma.
Acknowledgement
This work was financially supported by the Scientific Research Project of Hubei Provincial Department of Education (No. Q20152008).
References
- Eder W, Ege MJ, Von Mutius E. The asthma epidemic. N Engl J Med. 2006; 355: 2226-2235.
- Holgate ST. Genetic and environmental interaction in allergy and asthma. J Allergy Clin Immunol. 1999; 104: 1139-1146.
- Cho SH, Ryu CH, Oh CK. Plasminogen activator inhibitor-1 in the pathogenesis of asthma. Exp Biol Med (Maywood). 2004; 229: 138-146.
- Xiao W, Hsu YP, Ishizaka A, Kirikae T, Moss RB. Sputum cathelicidin, urokinase plasminogen activation system components, and cytokines discriminate cystic fibrosis, COPD, and asthma inflammation. Chest. 2005; 128: 2316-2326.
- Kowal K, Bodzenta-Lukaszyk A, Pampuch A, Szmitkowski M, Donati MB, Iacoviello L. Plasminogen activator inhibitor-1 plasma concentration in allergic asthma patients during allergen challenge. Int Arch Allergy Immunol. 2007; 144: 240-246.
- Cho SH, Tam SW, Demissie-Sanders S, Filler SA, Oh CK. Production of plasminogen activator inhibitor-1 by human mast cells and its possible role in asthma. J Immunol. 2000; 165: 3154-3161.
- Dawson SJ, Wiman B, Hamsten A, Green F, Humphries S, Henney AM. The two allele sequences of a common polymorphism in the promoter of the plasminogen activator inhibitor-1 (PAI-1) gene respond differently to interleukin-1 in HepG2 cells. J Biol Chem. 1993; 268: 10739-10745.
- Cho SH, Hall IP, Wheatley A, Dewar J, Abraha D, Del Mundo J, et al. Possible role of the 4G/5G polymorphism of the plasminogen activator inhibitor 1 gene in the development of asthma. J Allergy Clin Immunol. 2001; 108: 212-214.
- Buckova D, Izakovicova Holla L, Vacha J. Polymorphism 4G/5G in the plasminogen activator inhibitor-1 (PAI-1) gene is associated with IgEmediated allergic diseases and asthma in the Czech population. Allergy. 2002; 57: 446-448.
- Hizawa N, Maeda Y, Konno S, Fukui Y, Takahashi D, Nishimura M. Genetic polymorphisms at FCER1B and PAI-1 and asthma susceptibility. Clin Exp Allergy. 2006; 36: 872-876.
- Pampuch A, Kowal K, Bodzenta-Lukaszyk A, Di Castelnuovo A, Chyczewski L, Donati MB, et al. The -675 4G/5G plasminogen activator inhibitor-1 promoter polymorphism in house dust mite-sensitive allergic asthma patients. Allergy. 2006; 61: 234-238.
- Kowal K, Bodzenta-Lukaszyk A, Pampuch A, Szmitkowski M, Zukowski S, Donati MB, et al. Analysis of -675 4 g/5 G SERPINE1 and C-159T CD14 polymorphisms in house dust mite-allergic asthma patients. J Investig Allergol Clin Immunol. 2008; 18: 284-292.
- Cosan D, Kurt E, Kurt H, Degirmenci I, Kucukarabaci B, Metintas M, et al. Plasminogen activator inhibitor type-1 gene 4G/5G polymorphism in Turkish adult patients with asthma. Genet Test Mol Biomarkers. 2009; 13: 543-546.
- Ozbek OY, Atac FB, Ogus E, Ozbek N. Plasminogen activator inhibitor-1 gene 4G/5G polymorphism in Turkish children with asthma and allergic rhinitis. Allergy Asthma Proc. 2009; 30: 41-46.
- Zhang XY, Lin JT, Liu CL, Su N, Chen X, Cai Z, et al. An association study between PAI-1 gene polymorphism and bronchial asthma. Chin J Tuberc Respir Dis. 2009; 32: 210-212.
- Dijkstra A, Postma DS, Bruinenberg M, Van Diemen CC, Boezen HM, Koppelman GH, et al. SERPINE1 -675 4G/5G polymorphism is associated with asthma severity and inhaled corticosteroid response. Eur Respir J. 2011; 38: 1036-1043.
- Bora E, Soylar R, Arikan-Ayyildiz Z, Uzuner N, Giray-Bozkaya O, Ercal D, et al. Plasminogen activator inhibitor-1 and angiotensin converting enzyme gene polymorphisms in Turkish asthmatic children. Allergol Immunopathol (Madr). 2012; 41: 11-16.
- Nie W, Li B, Xiu QY. The -675 4G/5G Polymorphism in Plasminogen Activator Inhibitor-1 Gene Is Associated with Risk of Asthma: A Meta-Analysis. PLoS One. 2012; 7: 34385.
- Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J. A method for metaanalysis of molecular association studies. Stat Med. 2005; 24: 1291-1306.
- Bagos PG, Nikolopoulos GK. A method for meta-analysis of case-control genetic association studies using logistic regression. Stat Appl Genet Mol Biol. 2007; 6: 17.
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986; 7: 177-188.
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003; 327: 557-560.
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994; 50: 1088-1101.
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997; 315: 629-634.
- De Alarcon A, Steinke JW, Caughey R, Barekzi E, Hise K, Gross CW, et al. Expression of leukotriene C4 synthase and plasminogen activator inhibitor 1 gene promoter polymorphisms in sinusitis. Am J Rhinol. 2006; 20: 545-549.
- Bagos PG. Plasminogen activator inhibitor-1 4G/5G and 5,10-methylenetetrahydrofolate reductase C677T polymorphisms in polycystic ovary syndrome. Mol Hum Reprod. 2009; 15: 19-26.
- Qian X, Lu Z, Tan M, Liu H, Lu D. A meta-analysis of association between C677T polymorphism in the methylenetetrahydrofolate reductase gene and hypertension. Eur J Hum Genet. 2007; 15: 1239-1245.
- Ma Z, Jhun B, Jung SY, Oh CK. Binding of upstream stimulatory factor 1 to the E-box regulates the 4G/5G polymorphism-dependent plasminogen activator inhibitor 1 expression in mast cells. J Allergy Clin Immunol. 2008; 121: 1006-1012.
- Matthay MA, Clements JA. Coagulation-dependent mechanisms and asthma. J Clin Invest. 2004; 114: 20-23.
- Wagers SS, Norton RJ, Rinaldi LM, Bates JH, Sobel BE, Irvin CG. Extra vascular fibrin, plasminogen activator, plasminogen activator inhibitors, and airway hyper responsiveness. J Clin Invest. 2004; 114: 104-111.
- Zhang Y, Zhang J, Huang J, Li X, He C, Tian C, et al. Polymorphisms in the transforming growth factor-beta1 gene and the risk of asthma: A metaanalysis. Respirology. 2010; 15: 643-650.
- Yang H, Dong H, Dai Y, Zheng Y. Association of interleukin-13 C-1112T and G+2044A polymorphisms with asthma: A meta-analysis. Respirology. 2011; 16: 1127-1135.
- Zhang Y, Zhang J, Tian C, Xiao Y, He C, Li X, et al. The -308 G/A polymorphism in TNF-a gene is associated with asthma risk: an update by meta-analysis. J Clin Immunol. 2011; 31: 174-185.
- Panhuysen CI, Bleecker ER, Koeter GH, Meyers DA, Postma DS. Characterization of obstructive airway disease in family members of probands with asthma. An algorithm for the diagnosis of asthma. Am J Respir Crit Care Med. 1998; 157: 1734-1742.