
Research Article
J Bacteriol Mycol. 2014;1(2): 7.
Isolation and Characterization of Lactic Acid Bacteria Producing Bacteriocin from Newborn Infants Feces
Yalian Sun, Xiuyu Lou, Xuan Zhu, Han Jiang, Qing Gu*
Zhejiang Gongshang University, Key Laboratory for Food Microbial Technology of Zhejiang Province, China
*Corresponding author: Qing Gu, Zhejiang Gongshang University, Key Laboratory for Food Microbial Technology of Zhejiang Province, China.
Zhang H, Z-BioMed, Inc., 15725 Crabbs Branch Way, Rockville, MD 20855, USA.
Received: August 28, 2014; Accepted: October 20, 2014; Published: October 24, 2014
Abstract
Ninety-six strains of bacteria were isolated from newborn infants feces. After Antimicrobial activity was screened, these strains showed a broad inhibition spectrum. Particularly, twenty-one strains had antimicrobial abilities to inhibit Escherichia coli ATCC 35150, Micrococcus luteus CICC 10209, Salmonella typhimurium ZJJK 311. Regarding the physiological analysis of strains and BLAST analysis of 16S rDNA sequences, twenty-one strains were divided into six species: Enterococcus faecium (six isolates), Enterococcus faecalis (six isolates), Enterococcus durans (two isolates), Lactobacillus paracasei (one isolate), Lactobacillus rhamnosus (three isolates), and Lactobacillus plantarum (three isolates). All of the selected strains showed antimicrobial activity against six food-borne pathogens. After treatments with proteinase K or Trypsin, L. paracasei (LZ54), Enterococcus faecalis (LZ95), and L. plantarum (LZ222, LZ227) lost antibacterial activity, indicating that the proteinaceous nature of the antimicrobial compounds were produced by these strains. These strains could be introduced as preservatives in the food industry.
Keywords: Lactic acid bacteria; Bacteriocin; Antibacterial
Introduction
The lactic acid bacteria (LAB) constitute a large group of nonsporulating, gram-positive, catalase and oxidize-negative rods and cocci. These bacteria produce lactic acid as the major metabolite of the carbohydrate fermentation. All LAB are anaerobic and aero tolerant. They generally have complex nutritional requirements. The main genera of the LAB are Lactobacillus, Lactococcus, Streptococcus, Enterococcus, and Sporolactobacillus. LAB plays an important role in physiological functions and exists widely in the intestinal tracts of the human body. Lactobacilli, Bifidobacteria, and Enterococci are normally associated with infant gut microbiota [1].
In neonates, especially preterm babies, intestinal microbiota plays important roles in metabolism, nutrition, immune functions, and defense against pathogens. However, many of the microfloras that colonize the neonatal gut have different origins. Recent molecular studies have shown that LAB colonization is not significantly related to the method of delivery. Rather, it is also influenced by the infant diet during the first few days of life [2]. Formula-fed infants have greater numbers of more diverse enterobacteria and enterococci and maintain these populations longer [3, 4]. In contrast, breast milk seems to facilitate the growth of maternal lactobacilli in the infant gut [2, 5].
Recent research has revealed that the probiotic effects of LAB including inhibiting the growth of pathogenic bacteria, reduce therate of colon cancer, increase the immune response, and decrease serum cholesterol [6, 7, 8]. It has also been reported that they produce antimicrobial substances, including bacteriocins, organic acids, and low-molecular-weight substances that can inhibit the growth of undesirable pathogens that can cause diarrhea or other diseases of the human intestine [9, 10, 11]. We choose the feces of healthy newborn babies from Zhejiang Maternal and Child Health Hospital as the research objects for screening the strains capable of producing antibacterial substances. Special attention was paid to bacteriocins.
The aim of the present work was to characterize and identify the lactic acid bacteria isolated from feces of healthy newborn babies in Zhejiang Maternal and Child Health Hospital and to select bacteriocins-producing strains for use in the food industry.
Materials and Methods
Sample collection
Three hundred and forty-seven samples were collected from newborn infants in Zhejiang Maternal and Child Health Hospital. These infants defecated 72 h after birth, generally three or four pieces per infant. The samples were placed at 4 °C as soon as they were collected, during transporting way, and at the laboratory. After examination, they were disposed of immediately.
Bacterial strains
Lactic acid bacteria (LAB) were isolated from samples of newborn infants. Standard ten-fold dilutions of the feces were made in 0.9 % sterile physiological saline, and each sample was plated on both MRS (Becton Dickinson, U.S.) agar and M17 (Becton Dickinson, U.S.) agar containing CaCO3 (5 g/L). A volume of 0.1 mL of appropriate dilutions was spread and plated in triplicate on the following medium for isolation of LAB: M17 agar for lactococci and MRS agar for lactobacilli. All plates were incubated at 30 °C under anaerobic condition until colonies became visible. Colonies were randomly selected based on morphological differences (colony size and shape, clearance zone formed from hydrolysis of CaCO3 by lactic acid). Colonies were purified, again on the MRS and M17 agar plates. The isolates were routinely maintained in broth at 30 °C and stock culture of each isolate was kept for further study at -80 °C in broth supplemented with 30 % glycerol.
Indicator bacteria used for antimicrobial assays were cultured in Luria-Bertani (LB) agar under aerobic conditions at 37°C. These bacteria included Escherichia coli ATCC 35150, Micrococcus luteus CICC 10209, Staphylococcus citreus ZJJK 311, Vibrio parahaemolyticus ATCC 27519, Salmonella typhimurium CGMCC 1.1190, and Listeria monocytogenes ATCC 7644. These strains were provided by Microbiological Laboratory of Clinical Detection Center of Zhejiang (Hangzhou, China) or from our own supplies.
Antimicrobial assays
Cell-free culture supernatants (CFCS) were obtained by centrifugation (12,000×g, 4°C, 20 min) of LAB cultures grown in 20 mL broth at 30 °C for 24 h. The supernatant was filtered through a 0.22 µm filter to remove residual cells. The antimicrobial activity of all samples was tested using the agar-well assay [12]. Briefly, indicator bacteria were grown in LB broth overnight and spread onto the soft agar (0.75 %, w/v) plate of LB after diluting to 107 CFU/mL. Then wells 8 mm in diameter were punched onto the surface using a sterile borer. Then 200 µL sample prepared from filtrate was added to each well of the plate and stored at 4 °C for 30 min. Samples were then incubated at 37 °C for 18 h. Antimicrobial activity was recorded as growth-free inhibition zones (diameter) around the well. MRS and M17 adjusted to 4.5 as controls.
Sensitivity to proteolysis enzymes of the cell-free supernatants of bacteriocins producer strains was assessed through treatment with proteinase K and Trypsin (Sangon). All enzymes (10 mg/mL in sterile distilled water) were filter-sterilized and added to supernatants at a final concentration of 1 mg/mL in phosphate buffer (pH 6.5). Following incubation at 37 °C for 2 h, enzymes were denatured by heating at 100 °C for 5 min. Untreated samples were used as controls. The residual activity of enzyme-treated samples against Listeria monocytogenes was determined using the well-diffusion method [12].
The effect of temperature on cell-free supernatants was determined by treatment at 80 °C or 100 °C for 20 min in water bath and at 121 °C for 20 min (sterilization). A portion of each supernatant was treated at room temperature for 20 min. This served as a control. Samples retaining activity after heat treatment were identified through the well-diffusion method [12]. Regarding our previous work, Gramnegative bacteria Escherichia coli and Gram-positive bacteria Micrococcus luteus are more sensitive to bacteriocins. In this work, Escherichia coli ATCC 35150 and Micrococcus luteus CICC10209 were chosen as the indicator strains.
Biochemical characterization
Each isolated strain was propagated twice on a plate before use. We used bacterial trace biochemical reaction tubes produced by Hangzhou Microbial Reagent Co., Ltd.
Identification of LAB isolates
Identification of the twenty-one LAB isolates was performed using Phylogenetics analysis based on 16S rRNA sequence.
Total genomic DNA from each isolate was extracted from 3 ml pure overnight cultures grown in broth at 30 °C using a modified version of the CTAB (cetyltrimethylammonium bromide) method [13, 14]. Purified DNA was diluted to 100 ng/µL for further use.
Genomic DNA from each isolate was used as a template for PCR amplification of a segment of the 16S rDNA gene. Genomic DNA from each isolate was used as template for PCR amplification of 16S rRNA gene using the primers 27F (5'-AGAGTTTGATCCTGGCTCAG-3') and 1492R (5'-CTACGGCTACCTTGTTACGA-3') [15, 16]. The 25 µL reaction mixture contained 100 ng templates DNA, 2.5 µL 10×buffer, 1 µL 3 mM MgCl2, 1 µL 0.2 mM of dNTP, 0.5 µL 10 pmol of each primer, and 6.0 units of Taq DNA polymerase. PCR amplifying procedures were as follows: 5 min at 94°C, 30 cycles of 1 min at 94°C, 30 s at 54°C, 1 min at 72°C, and then 7 min at 72°C. It was carried out in an automatic thermal cycler. The sequencing of purified products was performed by Shanghai Sangon Biosciences Corporation of China (Shanghai, China).
Construction of phylogenetics trees
The 16S rDNA gene sequences of all twenty-one isolates were submitted to the National Center for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov) for BLAST search. The sequences of four bacteriocins-producing strains were imported into MEGA version 4.0 software, with which a sequence alignment and Phylogenetics tree were created on the basis of the neighbor-joining method [17]. The reliability of the groups was evaluated by bootstrap analysis with 500 resamplings. Bacillus subtilis DSM 10T was used as an outgroup.
Results
Detection of antimicrobial activity
Twenty-one isolates collected from infant feces were isolated on plates and screened for antimicrobial activity against Escherichia coli ATCC 35150 and Micrococcus luteus CICC10209 using ager well diffusion assay. Then the CFCS of the 21 isolates were analyzed and found to show wide-spectrum antibacterial activity, some of them can inhibit all the indicate bacteria in our study, including Staphylococcus citreus ZJJK 311, Vibrio parahaemolyticus ATCC 27519, Salmonella typhimuriumCGMCC 1.1190, and Listeria monocytogenes ATCC 7644. These isolates displayed antibacterial effects against at least three indicator strains (Table 1).
Strain
Indicator strains
Inhibition zone (mm)a
a
M. luteus
E. coli
S. typhimurium
V. parahaemolyticus monocytogenes
LZ3
+++
+++
++++
+
+
+
LZ4
+++
++
++++
-
+
-
LZ6
+
+++
++++
++
++
+++
LZ18
++
++++
++++
+
++
+++
LZ32
-
++
++
+
+
++
LZ37
-
+++
++
++
++
++
LZ42
-
++
++++
+++
-
+
LZ45
++
+++
+++
-
++
+++
LZ54
++++
++++
++++
+++
++
++++
LZ81
+++
+++
++++
-
++
-
LZ83
-
+++
+++
+
+
-
LZ92
-
+++
++++
+++
+
+
LZ95
+
++
+++
++
-
+
LZ200
++
++++
++
++
++
++++
LZ202
-
++++
+++
++
++
-
LZ206
++
++++
+++
+++
++
+
LZ215
+
+++
+++
+
+
-
LZ216
-
++++
++++
+
+++
++
LZ222
+++
++++
++++
+++
+++
++
LZ225
++
++++
+++
+
++
+
LZ227
+++
++++
+++
+++
++
++
aSymbols refer to the diameter of the inhibition zone observed among growing cells: +, 9.0–11.9 mm; ++, 12.0–14.9 mm; +++, 15.0–17.9 mm; ++++, over 18.0 mm; -, absence of an inhibitory zone or presence of a zone under 9.0 mm. Well diameter 8.0 mm.
Table 1: Inhibitory effects of the isolates against six indicator strains in the agar well diffusion assay.
The CFCS of twenty one strains were treated with proteinase K, Trypsin. LZ54, LZ200, LZ222, and LZ227 all became inactivated after digestion by proteinase K and trypsin (Table 2). The CFCS of these four strains were found to be resistant to heating at 80 °C for 20 min. The CFCS of four strains retained antibacterial activity after heating at 100 °C for 20 min. The inhibitory activity of LZ200 decreased relative to that before beating. The supernatants of strains LZ54, LZ222, and LZ227 remained stable even after treatment at 121 °C for 20 min (Table 3).
Phenotypic characterization
A total of twenty-one strains, including seven rods and fourteen cocci, were obtained from the newborn infant fecal samples using the pure culture method. The phenotypic characteristics of all isolates are shown in Tables 4 and 5. The strains were divided into six groups according to their morphological, physiological, and biochemical properties [19-21]. The seven rods were divided into three groups [1- 3] and the fourteen cocci were also divided into another three groups [4-6]. All rods were found to grow at pH 4.2 and pH 4.8 and exhibited growth in 2.0 % and 4 % NaCl solution. Most of the cocci were found to grow at pH 9.2 and pH 9.6 at 45 °C and in 6.5 % NaCl solution. None were able to grow at pH 4.2 at 50°C.
Phenotypic characteristic
Strains of groups
1
2
3
4
5
6
Number of isolates
3
1
3
6
2
6
Shape a
R
R
R
C
C
C
Catalase reaction
0 b/3 c
0/1
0/3
0/6
0/2
0/6
CO2 from glucose
3/3
0/1
0/3
0/6
0/2
0/6
NH3 from arginine
0/3
0/1
0/3
0/6
0/2
6/6
Growth at pH
3.0
1/3
0/1
0/3
NT
NT
NT
4.2
3/3
1/1
3/3
0/6
0/2
1/6
4.8
3/3
1/1
3/3
6/6
2/2
6/6
9.2
NTd
NT
NT
6/6
2/2
5/6
9.6
NT
NT
NT
6/6
2/2
5/6
Growth in NaCl (w/v)
2.0 %
3/3
1/1
3/3
NT
NT
NT
4.0 %
3/3
1/1
3/3
NT
NT
NT
5.0 %
NT
NT
NT
6/6
2/2
6/6
6.5 %
1/3
0/1
3/3
6/6
2/2
6/6
10.0 %
NT
NT
NT
0/6
0/2
0/6
Growth at temperature
10 °C
3/3
1/1
3/3
6/6
2/2
6/6
45 °C
2/3
1/1
3/3
6/6
2/2
6/6
55 °C
NT
NT
NT
0/6
0/2
0/6
Growth in glycolcholate sodium
10 %
NT
NT
NT
6/6
2/2
6/6
40 %
NT
NT
NT
6/6
2/2
6/6
Hydrolysis arginine
Nitrate reduction
H2S from tryptone
0/3
0/3
0/3
0/3
0/1
0/1
0/1
0/1
0/3
0/3
0/3
0/3
6/6
4/6
0/6
0/6
2/2
2/2
0/2
0/2
6/6
6/6
0/6
0/6
aR: rods, C: cocci. bNumber of positive strains. cTotal number of strains. dNT= not tested.
Table 4: Phenotypic characteristics of 21 strains of LAB isolated from newborn infants in Zhejiang Maternal and Child Health Hospital.
Acid from
Strains of groups
1
2
3
4
5
6
Glucose
3a/3b
1/1
3/3
2/2
6/6
Lactose
3/3
1/1
3/3
6/6
2/2
5/6
Maltose
3/3
1/1
3/3
6/6
2/2
6/6
Mannitol
3/3
1/1
3/3
1/6
1/2
0/6
Sucrose
3/3
1/1
3/3
6/6
1/2
5/6
Arabinose
2/3
0/1
1/3
1/6
(2/2)
6/6
Xylose
0/3
0/1
2/3
0/6
0/2
0/6
Raffinose
0/3
0/1
3/3
0/6
0/2
0/6
Fructose
3/3
1/1
3/3
6/6
2/2
6/6
Rhamnose
3/3
0/1
0/3
0/6
0/2
0/6
Galactose
3/3
1/1
3/3
6/6
2/2
6/6
Sorbose
3/3
1/1
3/3
0/6
0/2
0/6
Cellobiose
3/3
1/1
3/3
6/6
2/2
5/6
Melibiose
0/3
0/1
3/3
0/6
0/2
6/6
Melezitose
3/3
1/1
3/3
(6/6)c
0/2
0/6
Mannose
3/3
1/1
3/3
(6/6)
2/2
(6/6)
Salicin
3/3
1/1
3/3
6/6
2/2
6/6
Esculin
3/3
1/1
3/3
6/6
2/2
6/6
Amygdalin
3/3
1/1
3/3
0/6
2/2
0/6
Starch
0/3
0/1
0/3
0/6
0/2
0/6
Gluconate
0/3
0/1
0/3
0/6
0/2
0/6
a Number of positive strains. b Total number of strains. c Weakly positive.
Table 5: Biochemical characteristics of 21 strains of LAB isolated from newborn infants in Zhejiang Maternal and Child Health Hospital.
Group 1 included LZ202, LZ216, and LZ225. All strains were found to produce CO2 from glucose and were able to ferment lactose, melezitose, rhamnose, galactose, glucose, cellolezitose, mannitol, and other sugars. However, LZ202 was not able to use arabinose and none could ferment xylose, raffinose, melibiose, or starch. In this way, isolates of group 1 were identified as Lactobacillus rhamnosus.
Strain LZ54 was found to belong to group 2. It grew well at 10 °C and 45 °C and showed positive results for most sugars except arabinose, raffinose, xylose, rhamnose, melibiose, and starch. This group was identified as L. casei.
Group 3 comprised three bacteria LZ206, LZ222, and LZ227. The strains in group 3 were found to grow at 10 °C and 45 °C and grew well at 6.5 % NaCl. The strains in this group were found to utilize most sugars except rhamnose and starch. However, LZ222 was not found to ferment arabinose or xylose and LZ206 was not found to ferment arabinose. The three isolates were preliminarily assigned to the L. plantarum group according to their characteristics.
The strains of group 4, LZ3, LZ4, LZ32, LZ81, LZ83, and LZ95, were identified asEnterococcus faecalis based on physiological and biochemical properties. The strains were found to grow at pH 4.8 and pH 9.6 at 45 °C in 6.5 % NaCl and to hydrolyze arginine. LZ3, LZ4, LZ81, and LZ95 were also found to hydrolyze hippurate. All strains could ferment glucose, lactose, maltose, sucrose, fructose, galactose, cellobiose, salicin, and esculin and showed weakly activity on melezitose and mannose, but only LZ81 could ferment mannitol and arabinose. None could ferment xylose, raffinose, rhamnose, sorbose, melibiose, amygdalin, or starch.
Both group 5 strains, LZ42 and LZ92, were classified as E. durans. The strains could not produce NH3 from arginine but could hydrolyze arginine and hippurate. Both strains fermented glucose, lactose, maltose, arabinose, fructose, rhamnose, galactose, cellobiose, melibiose, mannose, salicin, esculin, and amygdalin. LZ42 was also able to ferment sucrose and mannitol. Neither could use xylose, raffinose, sorbose, melibiose, melezitose, or starch.
All strains in group 6 were found to be E. faecium. This group contained 6 isolates, LZ6, LZ18, LZ37, LZ45, LZ200, and LZ215. These strains could produce NH3 from arginine and hydrolyze arginine and hippurate. They were able to ferment glucose, maltose, arabinose, fructose, galactose, melibiose, salicin, and esculin and had a weakly effect on mannose. Only LZ200 could not ferment lactose, sucrose, or cellobiose. None were able to use mannitol, xylose, raffinose, rhamnose, sorbose, melezitose, amygdalin, or starch.
16S rDNA sequence analysis
Genotypic methods were used to corroborate the phenotypic classification. Specifically, 16S rDNA gene sequencing was used to classify the twenty-one antimicrobial-producing isolates. The 16S rDNA gene sequences of 4 bacteriocins-producing strains were obtained and then a Phylogenetics tree was constructed using MEGA software (version 4.0) and the neighbor-joining method. As shown in Figure 1., LZ54 showed more than 99.7 % homology to L. paracasei JCM 8130T. LZ227 appeared to be equally linked to both L. plantarum JCM 1149T and L. pentosus JCM 1558, and its 16S rDNA gene sequence showed 99.7 % similarity to L. plantarum and 99.6 % similarity to L. pentosus. LZ222 was found to have a situation similar to that of LZ227 when examined under the16S rDNA sequence. LZ95 was placed in the cluster of genus Enterococcus, which was recovered in 100 % of bootstrap analyses. It showed a similarity of over 99.8 % to these strains.
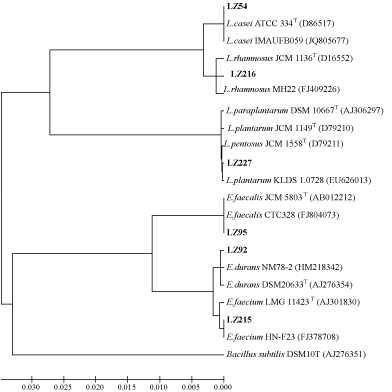
Figure 1: Neighbor-joining tree based on 16S rDNA gene sequences of the four bacteriocin-producing strains and the reference strains. Bacillus subtilis was used as the outgroup. L.: Lactobacillus; E.: Enterococcus.
Discussion
In this work, we attempted to screen lactic acid bacteria, specifically those with antimicrobial and bacreiocin-producing capacities, from the feces of healthy newborn babies recruited from Zhejiang Maternal and Child Health Hospital. To our knowledge many studies regarding intestinal microflora and the screening of lactic acid bacteria from yogurt, steamed buns, fermented vegetables, and other materials, but there have been few attempts to screen antibacterial LAB from the feces of newborn infants in Zhejiang Province.
Twenty-one strains were originally isolated from the feces of newborn infants. All strains were divided into 6 groups by conventional methods using physiological properties. Strains in groups 1 through 6 were identified as L. rhamnosus, L. casei, L. plantarum, E. faecalis, E. durans, and E. faecium, respectively. Genotypic analysis confirmed the results of the physiological analysis, excepting only strain LZ54 in group 2. BLAST showed LZ54 to be L. paracasei, a species of L. casei.
As shown in Table 2, LZ54, LZ200, LZ222, and LZ227 become inactivated after digestion with proteinase K or trypsin. This suggests that these inhibitory compounds are proteinaceous, so they can be considered bacteriocins [22]. As indicated by 16S rDNA sequence analysis, the four bacteriocin-producing strains belong to L. paracasei (LZ54), L. plantarum (LZ222, LZ227), and E. faecalis (LZ95). However, strains LZ222 and LZ227 were shown to be highly similar to the L. plantarum group (L. plantaum, L. paraplantarum, L. pentosun). Corsetti and Settanni L [23] reported that both phenotypic determination tools, such as carbohydrate fermentation patterns, and genetic methods, such as 16S rDNA gene sequence, were not able
Strain
Indicator strains (L. monocytog)
Inhibition zone (mm)a
a
control
Proteinase K
trypsin
LZ3
++++
+++
++++
LZ4
++++
++++
+++
LZ6
++++
++++
++++
LZ18
++++
+++
++++
LZ32
++
++
++
LZ37
++
++
++
LZ42
++++
++++
++++
LZ45
+++
+++
+++
LZ54
++++
-
-
LZ81
++++
++++
++++
LZ83
+++
+++
+++
LZ92
++++
++++
+++
LZ95
+++
+++
+++
LZ200
++
-
-
LZ202
+++
+++
+++
LZ206
+++
++
+++
LZ215
+++
+++
+++
LZ216
++++
+++
+++
LZ222
++++
-
-
LZ225
+++
+++
+++
LZ227
+++
-
-
aSymbols refer to the diameter of the inhibition zone observed among growing cells: +, 9.0–11.9 mm; ++, 12.0–14.9 mm; +++, 15.0–17.9 mm; ++++, over 18.0 mm; -, absence of an inhibitory zone or presence of zone under the 9.0 mm. Well diameter: 8.0 mm.
Table 2: Effects of enzymes on inhibitory activity of LAB as shown in agar well diffusion assay.
to distinguish between members of the L. plantarum group species, primarily because they differ by only 2 bp [24]. Tanasupawat et al. [25] reported that the production of acid from melezitose and xylose could be used to differentiate between L. plantarum and L. pentosus species, even though these two strains were similar to each other. The ability of each strain to produce acid from carbohydrates is summarized in Table 3. Strains LZ222 and LZ227 showed the same trends as L. plantarum due to the absence of acid production from glycerol and xylose, but they were also able to produce acid from melezitose.
Strain
Indicator strains (L. monocytog)
Inhibition zone(mm)a
a
control
80 °C for 20 min
100 °C for 20 min
121 °C for 20 min
LZ54
++++
+++
++++
++++
LZ200
++++
++++
+
-
LZ222
++++
++++
+++
+++
LZ227
++++
+++
++++
++++
aSymbols refer to the diameter of the inhibition zone diameter observed among growing cells: +, 9.0–11.9 mm; ++, 12.0–14.9 mm; +++, 15.0–17.9 mm; ++++, over 18.0 mm; -, absence of an inhibitory zone or presence of a zone under 9.0 mm. Well diameter: 8.0 mm.
Table 3: Effects of heat treatment on inhibitory activity of LAB as shown in agar well diffusion assay.
The strains we screened were mainly enterocci and lactobacillus. This is consistent with the fact that enterocci are typical inhabitants of the intestine and frequently isolated from infant feces soon after birth [26]. Several studies have indicated that enterococci are as the most commonly isolated species during the first two months of infancy, with Lactobacilli being the second most common early colonizer [27].
As shown in Table 1, all strains showed different levels of ability to inhibit indicator bacteria. Some results proposed that the Lactobacilli that we screened could inhibit S. citreus and L. monocytogenes, though the effects differed across different strains. Generally, the inhibitive effects were stronger against Gram-positive bacteria than against Gram-negative bacteria. Lactic acid bacteria showed an important synergetic effect on the production of certain biogenic amines by food-borne pathogenic bacteria. However, the effects of certain lactic acid bacteria strains on biogenic amine production were straindependent under in vitro conditions [28].
As shown in Table 1, the species classified as Enterococci possessed a large difference ability to restrain the same indicator. LZ3, LZ4, LZ32, LZ81, LZ83, and LZ95 were found to belong to E. faecalis. LZ3, LZ4, and LZ95 showed stronger inhibition than LZ81. LZ32 and LZ83 showed no ability to inhibit M. luteus. LZ95 showed a particularly visible ability to inhibit L. monocytogenes.Enterococcus can be associated with pathogenicity, but these bacteria also have many benefits. They have been found in traditional artisanal fermented products, are they are used as probiotic cultures and are currently extensively studied for the production of bacteriocins, particularly enterocins. Many of these enterocins have been found to be active against L. monocytogenes [29]. LZ54, LZ200, LZ222, LZ227 all showed strong inhibition of V. parahaemolyticus, and a few have also been reported to be active against Gram-negative bacteria, which is unusual for bacteriocins produced by LAB [30]. As shown in Table 3, the activity of bacteriocins produced by LZ54, LZ200, LZ222, and LZ227 could not be destroyed by heat treatment at 100 °C lasting 20 min. These properties have been the subject of many studies describing the use of bacteriocins as preservatives in foodsof both animal and vegetable origin [31]. Many of the bacteriocins produced by bacteria have been fully characterized at the biochemical and genetic levels. This has allowed researchers to divide them into three major classes [32]. Strains in different groups show different levels of activity against different indicators. Bacteriocin-producing strains have attracted considerable interest regarding their potential for use as natural and non-toxic food preservatives because they can inhibit not only closely related species but also Gram-positive foodspoilage and food-borne pathogens [33]. Enterocins with high levels of anti-listerial activity show promise as antimicrobial agents and may be suitable for use in food preservation [31].
The twenty-one strains isolated from the feces of newborn infants were evaluated for antibacterial activity, physiology, and 16S rDNA sequences. Strains capable of inhibiting the growth of Escherichia coli and Micrococcus luteus were identified during preliminary screening. The CFCS of these twenty-one strains showed various levels of antagonistic effects against food-borne pathogens. LZ54 and LZ227 showed broad, pronounced antibacterial activity. This study suggests that the feces of newborn infants may serve as sources of probiotic lactic acid bacteria. The advantages of these particular twenty-one strains merit further investigation in vivo studies to determine which antimicrobial substances they produce. Some of these strains could be exploited in the development of infant formulas or novel food products. Bacteriocin-producing strains may play an important role in food fermentation and preservation. The procedure here developed for isolation of LAB may permit researchers to isolate unique and unknown novel LABs in nature, increasing the resources available for bacteriocins production.
Acknowledgments
This project was supported by the National High Technology Research and Development Program (“863” Program) of China (2012AA022105B), the National Natural Science Foundation of China (No. 31071513 and 31271821), the Natural Science Foundation of Zhejiang Province (No. Z3110399), and the International Science and Technology Cooperation Program of China (2013DFA32330).
References
- Eva R, Arqués JL, Rodríguez R, Ángela P, Landete MJ, Medina M. Antimicrobial properties of probiotic strains isolated from breast-fed infants. J of Functional Foods.2012; 4: 542-551.
- Matsumiya Y, Kato N, Watanabe K, Kato H. Molecular epidemiological study of vertical transmission of vaginal Lactobacillus species from mothers to newborn infants in Japanese, by arbitrarily primed polymerase chain reaction. J Infect Chemother.2002; 8: 43-49.
- Stark PL, Lee A . The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol. 1982; 15: 189-203.
- Yoshioka H, Iseki K, Fujita K . Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983; 72: 317-321.
- Heikkilä MP, Saris PE . Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol. 2003; 95: 471-478.
- Hartemink R, Domenech VR, Rombouts FM. A new selective medium for the isolation of lactobacilli from faeces. J Microbiol Meth.1997; 29: 77-84.
- Tahri K, Grill JP, Schneider F . Involvement of trihydroxyconjugated bile salts in cholesterol assimilation by bifidobacteria. Curr Microbiol. 1997; 34: 79-84.
- Mombelli B, Gismondo MR . The use of probiotics in medical practice. Int J Antimicrob Agents. 2000; 16: 531-536.
- Aasen IM, Møretrø T, Katla T, Axelsson L, Storrø I . Influence of complex nutrients, temperature and pH on bacteriocin production by Lactobacillus sakei CCUG 42687. Appl Microbiol Biotechnol. 2000; 53: 159-166.
- Bergogne-Bérézin E . Treatment and prevention of antibiotic associated diarrhea. Int J Antimicrob Agents. 2000; 16: 521-526.
- Martín R, Jiménez E, Olivares M, Marín ML, Fernández L, Xaus J, et al. Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from intant feces and breast milk of a mother-child. Int J Food Microbiol. 2006; 112: 35-43.
- Kumar M, Srivastava S . Antilisterial activity of a broad-spectrum bacteriocin, enterocin LR/6 from Enterococcus faecium LR/6. Appl Biochem Biotechnol. 2010; 162: 698-706.
- Rammelsberg M, Radler F. Antibacterial polypeptides of Lactobacillus species. J Appl Bacteriol.1990; 69:177-184.
- Liu W, Bao Q, Jirimutu, Qing M, Siriguleng, Chen X, Sun T . Isolation and identification of lactic acid bacteria from Tarag in Eastern Inner Mongolia of China by 16S rRNA sequences and DGGE analysis. Microbiol Res. 2012; 167: 110-115.
- Scarpellini M, Mora D, Colombo S, Franzetti L . Development of genus/species-specific PCR analysis for identification of Carnobacterium strains. Curr Microbiol. 2002; 45: 24-29.
- Liu W, Sun Z, Zhang J, Gao W, Wang W, Wu L, Sun T . Analysis of microbial composition in acid whey for dairy fan making in Yunnan by conventional method and 16S rRNA sequencing. Curr Microbiol. 2009; 59: 199-205.
- Saitou N, Nei M . The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4: 406-425.
- Sharpe ME, Fryer TF. Media for lactic acid bacteria. Laboratory Practice.1956; 14: 697.
- Greco M, Mazzette R, De Santis EP, Corona A, Cosseddu AM . Evolution and identification of lactic acid bacteria isolated during the ripening of Sardinian sausages. Meat Sci. 2005; 69: 733-739.
- Hugas M, Garriga M, Aymerich T, Monfort JM . Biochemical characterization of lactobacilli from dry fermented sausages. Int J Food Microbiol. 1993; 18: 107-113.
- Curk MC, Hubert JC, Bringel F . Lactobacillus paraplantarum sp. now., a new species related to Lactobacillus plantarum. Int J Syst Bacteriol. 1996; 46: 595-598.
- Fan L, Feng S, Sun Q, Xiang WL, Zhao J, Zhang J, et al. Screening for bacteriocin-producing lactic acid bacteria from kurut, a traditional naturally-fermented yak milk from Qinghai-Tibet plateau. Food Control.2011; 22: 50-53.
- Settanni L, Corsetti A . The use of multiplex PCR to detect and differentiate food- and beverage-associated microorganisms: a review. J Microbiol Methods. 2007; 69: 1-22.
- Ennahar S, Cai Y, Fujita Y . Phylogenetic diversity of lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl Environ Microbiol. 2003; 69: 444-451.
- Tanasupawat S, Ezaki T, Suzuki K, Okada S, Komagata K, Kozaki M. Characterization and identification of Lactobacillus pentosus and Lactobacillus plantarum strains from fermented food in Thailand. J Gen Appl Microbiol.1992; 38:121-134.
- Mitsou EK1, Kirtzalidou E, Oikonomou I, Liosis G, Kyriacou A . Fecal microflora of Greek healthy neonates. Anaerobe. 2008; 14: 94-101.
- Ahrné S, Lönnermark E, Wold AE, Aberg N, Hesselmar B, Saalman R, Strannegård IL . Lactobacilli in the intestinal microbiota of Swedish infants. Microbes Infect. 2005; 7: 1256-1262.
- Kuley E, Ozogul F2 . Synergistic and antagonistic effect of lactic acid bacteria on tyramine production by food-borne pathogenic bacteria in tyrosine decarboxylase broth. Food Chem. 2011; 127: 1163-1168.
- De Vuyst L, Foulquié Moreno MR, Revets H . Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int J Food Microbiol. 2003; 84: 299-318.
- Delves-Broughton J. Nisin as a food preserva tive. Food Australia.2005; 57:525-527.
- Khan H, Flint S, Yu PL . Enterocins in food preservation. Int J Food Microbiol. 2010; 141: 1-10.
- Franz CM, van Belkum MJ, Holzapfel WH, Abriouel H, Gálvez A . Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol Rev. 2007; 31: 293-310.
- Cotter PD, Hill C, Ross RP . Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005; 3: 777-788.