
Research Article
J Bacteriol Mycol. 2014;1(2): 8.
Ecological Roles of Arc Signal Transduction System Revealed by Evolutionary Genetics Analysis
Yangyang Dong, Fen Wan, Jianhua Yin and Haichun Gao*
Institute of Microbiology and College of Life Sciences, Zhejiang University, China
*Corresponding author: Haichun Gao, Institute of Microbiology and College of Life Sciences, Zhejiang University, China
Zhang H, Z-BioMed, Inc., 15725 Crabbs Branch Way, Rockville, MD 20855, USA.
Received: August 14, 2014; Accepted: November 06, 2014; Published: November 12, 2014
Abstract
The Arc signal transduction system was studied in γ-proteobacteria to determine evolutionary mechanisms and ecological pressures responsible for the plasticity of this system. Phylogenic analysis of the arcA gene encoding the DNA-binding response regulator suggests that the gene has remained under strong purifying selection throughout its history while the arcB gene encoding the sensor histidine kinase has undergone extensive modification in various lineages. The major observed modification occurs in the Alteromonadaceae family, where the Hpt domain of ArcB has been uncoupled from the sensor component in several genera (Shewanella, Colwellia and Idiomarina). This phenomenon appears to be linked to the free-living lifestyles of the Alteromonads. In contrast to the prevailing view of the Arc system existing only in facultative anaerobes, several of the Alteromonad species which encode Arc systems are strict aerobes or aero-tolerant anaerobes, suggesting additional functions for the Arc system beyond facultative anaerobiosis. Molecular clock estimates using linearized phylogenetics trees for the arcA gene place the origins of the system at 600-1300 million years ago, a period roughly corresponding to the rise in atmospheric oxygen levels that occurred in the Precambrian Era. It is hypothesized that the Arc system originated as an early oxidative stress response mechanism in the γ-proteobacteria and was later co-opted for the facultative anaerobic lifestyle.
Keywords: Shewanella; Alteromonaceae; Arc System; Signal Transduction; Molecular
Introduction
The metabolic transition to anaerobiosis in γ-proteobacterial species, such as model microorganism Escherichia coli, is mediated in part by the aerobic respiratory control (Arc) phosphorelay twocomponent system (TCS) consisting of a hybrid sensor histidine kinase (ArcB) and a global OmpR-family winged-helix response regulator (ArcA) [1, 2] (Figure 1). Reduction of the quinone pool in response to anaerobiosis triggers auto-phosphorylation of ArcB, which initiates a phosphorelay from the histidine kinase dimerization (HisKA) domain (ArcB, H292) to the receiver domain (ArcB, D576) to the Hpt domain (ArcB, H717) and finally to the ArcA receiver domain (D54) [3,4]. Most ArcB proteins also contain a PAS domain of unknown function, though this domain has lost in the Pasteurellaceae [5]. ArcA serves primarily as a global repressor of aerobic metabolic pathways, but recent studies have shown that the ArcA regulon is significantly more varied than was previously believed [6-15].
A major exception to the canonical γ-proteobacterial Arc system was observed in Shewanella oneidensis [9]. Shewanella species are facultative anaerobes of the Altermonadaceae family that utilize numerous compounds for respiration, including nitrate, DMSO, insoluble iron oxides and a variety of heavy metal ions [16]. An analysis of the genomic sequence of S. oneidensis revealed a gene encoding a putative ArcA homolog with 81% amino acid sequence identity to the E. coli ArcA protein. Despite the presence of an experimentally verified ArcA, annotation of the genome has not revealed a full-length arcB gene. Instead, signal transduction to ArcA is initiated by ArcS, which displays substantial sequence and structure variation fromE. coli ArcB [17, 18] (Figure 1). The most striking difference betweenE. coli ArcB and ArcS with respect to phosphorelay is that the latter lacks the Hpt domain. To compensate for the loss, S. oneidensis employs a free HptA protein with homology to the Hpt domain of E. coli ArcB [9]. Consistent with this, the ArcA regulon of S. oneidensis is substantially different from that of E. coli although their DNA-binding motifs are similar [11, 19]. Therefore, the cumulative computational and experimental results show that the Arc system of Shewanella has undergone significant evolution since the divergence of the lineage from the Enterobacteria.
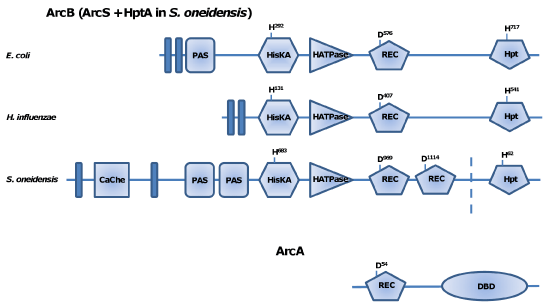
Figure 1: Domain organization of Shewanella ArcS compared to that of E. coli ArcB. The numbers in boxes mark the positions of the domains in the amino acid
sequence. The position of conserved amino acid residues putatively involved in phosphotransfer is marked within the corresponding domain. The numbers in
shaded areas between the domains of ArcS and ArcB display levels of identity/similarity between the domains (n/h, no homologies). Black vertical bars show the
positions of transmembrane domains. CaChe, CaChe-sensing domain; PAS, energy-sensing domain; HisKA, histidine kinase dimerization domain; HATPase_c,
histidine kinase ATPase domain; REC, receiver domain.
The Hpt domain of ArcB plays a significant role in signal transduction to ArcA. First, the Hpt domain represents the primary signal transfer point between the sensor and the response regulator and, as such, imparts specificity between the two components of the system [20]. This specificity helps to limit loss of signal between the two components and to reduce noise from unintended crosstalk with other signal transduction networks, which appears inevitable [21]. Second, the Hpt domain represents an important node for signal modulation. For instance, the ratio of phosphorylated to nonphosphorylated ArcA is mediated by forward and reverse phosphorelay reactions, both of which require the activity of the Hpt domain [22]. Signal transduction can also be modulated at the Hpt domain through the activity of additional components such as the phosphohistidine phosphatase SixA. In E. coli cells grown under anaerobic condition in the presence of nitrate, SixA interrupts the signal at the Hpt domain, allowing depression of the ArcA-regulated sdh operon [23].
The observation that S. oneidensis utilizes an Arc system substantially different from those observed in commensal/pathogens suggests a role other than for host interaction that maintains the canonical Arc system. With the recent release of additional Alteromonad genome sequences, this hypothesis may now be tested. Furthermore, the opportunity now presents itself to study the Arc system in a broader phylogenetics and ecological context beyond the commensal lifestyle. The following topics need to be addressed: 1) the distribution of the arcA, arcB , hptA genes in the γ-proteobacteria, 2) correlate this distribution to the particular ecological and physiological parameters of the organisms, if possible, i.e. determine whether evolutionary pressures imposed by the particular ecological niche of the organism affect the plasticity of the Arc system, 3) determine at what level(s) of the Arc system (i.e. regulon, regulator, phosphorelay, sensor) evolution of the system manifests and 4) correlate the accumulated data to specific temporal and geological events in the history of the γ-proteobacteria. To this end, the genomic sequences of multiple γ-proteobacterial species of the Alteromonaceae, Enterobacteraceae, Vibrionaceae, Pasteurellaceae and other families were analyzed to determine the distribution of the Arc system genes and these data were correlated to the particular ecology and physiology of each organism studied. The arcA genes were analyzed using maximum likelihood methods to determine the degree of selection acting upon this component. Finally, a linearized arcAphylogenetics tree was used to date major events in the evolution of the Arc system in relation to known geological and temporal phenomenon.
Experimental Procedures
Gene sequence selection
The publicly available γ-proteobacterial genomic sequences used in the computational analyses were obtained either from the NCBI or Joint Genome Institute Integrated Microbial Database (IMG). Using the E. coli ArcAB and S. oneidensis ArcA/ArcS/HptA amino acid sequences as queries, orthologs of Arc system genes were identified in the target genomes either by using the reciprocal best hit (RBH)- defined ortholog groupings of the IMG database or, when necessary, manual RBH using NCBI Blast 2.2.14 (tblastn, evalue 1e-10, defaults) [24]. Nucleotide sequences for arcA, arcB, arcS and hptA groups were imported into MEGA [25], translated to amino acid sequences, and aligned using ClustalW and reverse translated to codon alignments. Phylogenetics trees of the codon alignments were constructed in MEGA (Neighbor joining, Tamura-Nei model, variable rates along lineages) and compared to 16S rRNA and whole-genome alignments for the γ-proteobacteria [26]. Manual revision of trees was conducted in TreeView as indicated in the text. The resulting set of phylogenetic trees was used to perform evolution analyses in PAML [27].
Evolutionary analyses
Alignments and phylogenetics trees were analyzed using the codon models implemented PAML. The initial trees previously described were tested first under the one ratio model and the trees resulting from these analyses were used for all subsequent models. Standard branch test, site-specific and branch site models were conducted [28] and the complete set of analyses is listed in Supplemental Tables S1 and S2 with relevant results listed in Table 2. Evidence of positive selection was tested for statistical significance using the appropriate likelihood-ratio tests [27] (Table S3 and S4, Table 3). Tests of rate constancy of the phylogenies were conducted using the two-cluster and branch length tests implemented in the LINTREE program on arcA trees rooted with the paralogous E. coli ompR gene [29]. Lineages evolving faster or slower than the background at the 1% level were removed and the resulting dataset was reanalyzed until rate constancy was achieved. Lineages showing rate deviation at the 5% level were selectively removed if removal of the lineage improved or did not significantly impact the tree topology [29].
H0
H1
2Δl
df
Tree A
Tree B
One Ratio
2 Ratio (Sh)
2.60
*4.00
1
One Ratio
2 Ratio (P)
0.00
0.10
1
Nearly Neutral
Model A (Sh)
**17.70
**14.48
2
Model A (Sh, ω=1)
Model A (Sh, ω=1)
-1.14
0.32
1
Nearly Neutral
Model A (P)
**90.46
**86.20
2
Abbreviations are as follows: Sh – Shewanella oneidensis, P – Pasteurellaceae
Statistical significance was determined using the ?2 test with the indicated degrees of freedom (* 5%, **1%). Abbreviations are as defined in Table 2.
Table 3: Likelihood ratio test of results from Table 2.
Results
Phylogenetic distribution of Arc system components
The distribution of the arcA, arcB, arcS, and hptA orthologs in the γ-proteobacteria was determined and the results were summarized with corresponding ecological data in Table 1. The immediate observation is that the Enterobacteracaea, Vibrionacaea and Pasteurellacaea families maintain strictly canonical Arc systems whereas the Alteromonacaea family members display a far greater variety in the system. Within the Alteromonaceae family, the following distribution was observed: canonical ArcAB systems (Alteromonas, Pseudoalteromonas, and Psychromonas); Shewanellalike ArcA/ArcS/HptA systems (Shewanella, Colwellia, Ferrimonas, and Idiomarina); no Arc system (Marinobacter and Saccharophagus). This distribution of arcB vs. arcS/hptA within the Alteromonads could not be definitively linked to the phylogeny of the lineage, suggesting that the uncoupling of the Hpt domain from ArcB may have occurred multiple times during the radiation of the Alteromonaceae. All of the Alteromonad species having Arc system genes represent freeliving, non-commensal organisms in contrast to those of the other lineages studied. This distribution suggests that maintenance of the canonical Arc system is important in organisms requiring host interaction while increased plasticity in the system is more common in free-living organisms. Otherwise, the Arc system is rather stable in evolution evidenced by the lack of obvious correlation between Arc system gene distribution and other environmental or physiological factors (temperature, oxygen metabolism, pathogen vs. commensal/ symbionts).
Organism
Order
Genome Size (Mb)
Ecology
Arc System Components
O2a
Lifestyle
ArcA
ArcB
ArcS
HptA
Actinobacillus pleuropneumoniae serovar
Pasteurellaceae
2.2
F
Symbiotic
y
y
n
n
Aeromonas salmonicida A449
Aeromonadaceae
5.0
F
Free-Living
y
y
n
n
Aestuariibacter salexigens DSM15300
Alteromonadaceae
3.8
A
Free-Living
y
y
n
n
Algicola sagamiensis DSM14643
Aeromonadaceae
4.3
A
Free-Living
y
n
y
y
Aliagarivorans marinus DSM23064
Alteromonadaceae
4.9
A
Free-Living
y
y
n
n
Alishewanella aestuarii B11
Alteromonadaceae
3.6
A
Free-Living
y
y
n
n
Alteromonadaceae bacterium Bs12
Alteromonadaceae
4.9
F
Commensal
n
n
n
n
Alteromonas macleodii Deep Ecotype
Alteromonadaceae
4.2
A
Free-Living
y
y
n
n
Catenovulum agarivoransDS-2
Alteromonadaceae
4.6
A
Free-Living
y
y
n
n
Citrobacter koseri ATCC27026
Enterobacteriaceae
4.7
F
Free-Living
y
y
n
n
Colwellia psychrerythraea 34H
Alteromonadaceae
5.4
F
Free-Living
y
n
y
y
Dickeya zeae Ech1591
Enterobacteriaceae
4.8
A
Free-Living
y
y
n
n
Edwardsiella tarda FL6-60
Enterobacteriaceae
3.7
F
Free-Living
y
y
n
n
Enterobacter cloacae EcWSU1
Enterobacteriaceae
4.8
N
Free-Living
y
y
n
n
Escherichia coli K12
Enterobacteriaceae
4.6
F
Commensal
y
y
n
n
Ferrimonas balearica PAT
Alteromonadales
4.3
F
Free-Living
y
n
y
y
Gayadomonas joobiniege G7
Alteromonadaceae
3.9
A
Free-Living
y
y
n
n
Gilvimarinus chinensis DSM19667
Alteromonadaceae
4.1
A
Free-Living
n
n
n
n
Glaciecola mesophila KMM241
Alteromonadaceae
5.1
F
Commensal
y
y
n
n
Haemophilus influenzae RdKW20
Pasteurellaceae
1.8
F
Commensal
y
y
n
n
Haliea salexigens DSM19537
Alteromonadaceae
4.3
A
Free-Living
n
n
n
n
Idiomarina baltica OS145
Alteromonadaceae
2.7
A
Free-Living
y
n
y
y
Klebsiella neumonia KCTC
Enterobacteriaceae
5.5
A
Commensal
y
y
n
n
Mannheimia succiniciproducens MBEL55E
Pasteurellaceae
2.3
F
Commensal
y
y
n
n
Marinimicrobium agarilyticum DSM16975
Alteromonadaceae
4.5
A
Free-Living
n
n
n
n
Marinobacter adhaerens HP15
Alteromonadaceae
4.7
A
Free-Living
n
n
n
n
Marinobacterium stanieri S30
Alteromonadaceae
4.4
A
Free-Living
n
n
n
n
Melitea salexigens DSM19753
Alteromonadaceae
3.4
A
Free-Living
n
n
n
n
Microbulbifer variablilis ATCC700307
Alteromonadaceae
4.8
F
Commensal
n
n
n
n
Moritella dasanensis ArB0140
Alteromonadaceae
4.9
A
Free-Living
y
y
n
n
Pantoea ananatis AJ13355
Enterobacteriaceae
4.9
A
Free-Living
y
y
n
n
Pasteurella multocida Pm70 Type A
Pasteurellaceae
2.2
F
Commensal
y
y
n
n
Pectobacterium wasabiae WPP163
Enterobacteriaceae
5.1
F
Free-Living
y
y
n
n
Photobacterium profundum SS9
Vibrionaceae
6.4
F
Free/Comb
y
y
n
n
Photorhabdus luminescens laumondi TT01
Enterobacteriaceae
5.7
F
Commensal
y
y
n
n
Proteus mirabilis HI4320
Enterobacteriaceae
4.1
F
Free-Living
y
y
n
n
Pseudoalteromonas haloplanktis TAC125
Alteromonadaceae
3.8
A
Free-Living
y
y
n
n
Pseudomonas aeruginosa PAO1
Pseudomonadaceae
6.2
A
Commensal
n
n
n
n
Psychromonas aquimarina ATCC1526
Alteromonadaceae
5.5
A
Free-Living
y
y
n
n
Saccharophagus degradans 2-40
Alteromonadaceae
5.1
A
Free-Living
n
n
n
n
Salinimonas chungwhensisDSM16280
Alteromonadaceae
4.0
A
Free-Living
y
y
n
n
Salmonella typhimurium LT2
Enterobacteriaceae
4.9
F
Commensal
y
y
n
n
Shewanella oneidensis MR-1
Alteromonadaceae
5.0
F
Free-Living
y
n
y
y
Shigella boydii Sb227
Enterobacteriaceae
4.7
F
Free-Living
y
y
n
n
Sodalis glossinidius str. moristans
Enterobacteriaceae
4.1
F
Endosymbiont
y
y
n
n
Vibrio cholerae O1
Vibrionaceae
4
F
Free/Comb
y
y
n
n
Xenorhabdus bovienii SS-2004
Enterobacteriaceae
4.2
F
Free-Living
y
y
n
n
Yersinia pestis KIM
Enterobacteriaceae
4.6
F
Commensal
y
y
n
n
aOxygen tolerance. Abbreviations are as follows: A – strict aerobe, N – strict anaerobe, F – facultative anaerobe
bB free-living and Commensal
Table 1: Distribution of Arc system genes in representative γ-proteobacteria and correlation to ecological conditions.
Several lines of evidence suggest that the hptA gene is descended from an ancestral arcB gene. First, the HptA protein of S. oneidensis shows greater homology to the Hpt domains of the E. coli ArcB and the Vibrio cholerae FexB than to other Hpt domains and vice versa when analyzed using Blast (data not shown). Second, it is experimentally confirmed that HptA is a component in the Shewanella Arc signal transduction pathways [9, 17, 18]. Third, most species studied show common synteny in the region of arcB/hptA, even across broad taxonomic groupings (Figure 2). Finally, hptA and arcB orthologs are found in a variety of Alteromonad species but never together in the same organism. The cumulative results suggest that the hptA gene descended from a common ancestral arcB gene that predated the divergence of families Alteromonaceae and Enterobacteraceae-Vibrionaceae-Pasteurellaceae.
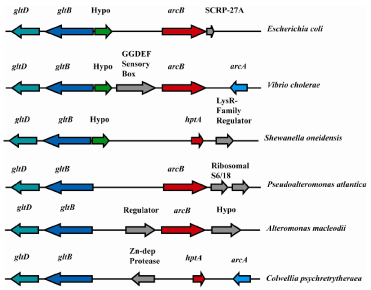
Figure 2: Synteny of the arcB/hptA regions of representative γ-proteobacterial species. Orthologous genes are colored similarly.
Evolutionary analysis of the arcA gene
Transcriptomic analysis of global regulator mutants of S. oneidensis, including ArcA, has revealed a significant degree of shuffling between regulon members, including global regulators Crp and Fnr [11, 13, 30]. These results, coupled with the previous observations regarding HptA, show that evolution of the Arc system occurs (at least in Shewanella) on at least 2 levels, that of the regulon and of the signal transduction network. To complement these analyses, evolutionary analyses were conducted to determine if the arcA gene itself has been subject to positive selection or neutral evolution within its history. To conduct these analyses, phylogenetics trees were constructed with microorganisms listed in Table 1 (Figure 3 and Figure S1). An immediate observation was that the Pasteurellacaea lineage was misplaced in the original tree; in response, a second tree topology was constructed with the Pasteurellaceae lineage placed according to the 16S rRNA phylogeny. The complete set of results for both tree topologies are listed in Supplementary Tables S1-2 and the relevant results are listed in Table 2. While the original tree gave slightly better likelihood values than the corrected tree, changing the tree topology had only negligible effects on the final results. The branch-site Model A test (free ω for Shewanella lineage, variable rates at site classes) suggested positive selection along that lineage but could not be shown statistically to be greater than the neutral condition. The Model A test for the P lineage revealed 7 sites potentially under positive selection, but again, the results could not be distinguished from the neutral condition, suggesting that the arcA gene has remained under strong purifying selection throughout time and space. These data, collectively, conclude that ArcA acts as an evolutionary foundation for the Arc system, with the major modifications to the system occurring at the level of the regulon and the phosphorelay network topology.
Model
Parameters
l
dN/dS
Positively Selected Sites
Tree A
One Ratio
ω = 0.0400
-8710.17
ω
NA
2 Ratio (Sh)
ω = 0.0394
-8708.87
0.0959
NA
2 Ratio (P)
ω = 0.0398
-8710.17
0.0404
NA
Model A (Sh)
p0 = 0.81, p1 = 0.15
(p2 + p3) = 0.05, ω2 = 3.0367
-8455.49
-
-
Model A (Sh, ω=1)
p0 = 0.79, p1 = 0.14
(p2 + p3) = 0.06, ω2 = 1
-8456.06
-
-
Model A (P)
p0 = 0.80, p1 = 0.13
(p2 + p3) = 0.08, ω2 = 1
-8419.11
-
2, 130, 131, 136, 148, 150, 152
Tree B
One Ratio
ω = 0.0404
-8727.26
ω
-
2 Ratio (Sh)
ω = 0.0396
-8725.26
0.1107
-
2 Ratio (P)
ω = 0.0396
-8727.20
0.0424
Model A (Sh)
p0 = 0.81, p1 = 0.15
(p2 + p3) = 0.04, ω2 = 1.958
-8460.80
-
-
Model A (Sh, ω=1)
p0 = 0.80, p1 = 0.15
(p2 + p3) = 0.06, ω2 = 1
-8460.96
-
-
Model A (P)
p0 = 0.80, p1 = 0.13
(p2 + p3) = 0.07, ω2 = 1
-8424.94
-
2, 130, 131, 136, 148, 150, 152
Abbreviations are as follows: Sh – Shewanella oneidensis, P – Pasteurellaceae
Table 2: Representative results of PAML analysis of arcA.
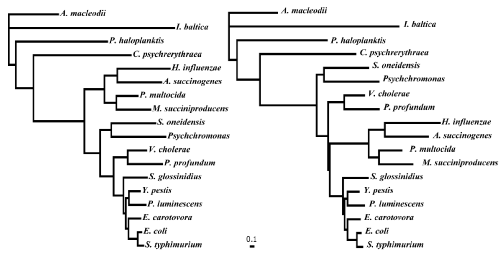
Figure 3: The arcA gene phylogenies used in PAML analyses. Microorganisms for the analysis listed in Table 1. The simplified tree is shown and the full tree is
given in Figure S1. A) Original unaltered tree derived from the codon alignment of arcA genes. B) Modified version of Tree A with Pasteurellacaea branch placed
according to the 16S rRNA phylogeny.
Molecular clock tests
A third arcA phylogenetics tree rooted with the E. coli ompR gene sequence was used to test the molecular clock hypothesis for this gene family (Figure 4). The first pass suggested an accelerated rate of evolution in the Sodalis lineage at the 1% significance level; the lineage was removed for subsequent LINTREE analyses. Further passes saw in the removal of Alteromonas macleodii (5%) and the Pasteurellacaea lineage (to provide a more accurate phylogeny), resulting in a final linearized tree. Using this tree and a previously determined estimate of the divergence time of E. coli and Salmonella typhimurium of 160 million years ago (MYA) as the calibration point [31], divergence times for the lineages were determined (Figure 4). These estimates give a divergence time between the Vibrionacaea and Enterobacteracaea of 480 MYA, consistent with previous estimates [31], and a divergence time between Enterobacteracaea-Vibrionacaea and Alteromonacaea (Shewanella) of 680 MYA. For comparison, divergence times were calculated based on a linearized tree with the same lineages removed and the same calibration point was constructed in MEGA, with similar results (Figure 4). Recent estimates place the divergence of the γ-proteobacteria from the β-proteobacteria at 1.89 billion years ago (GYA) [32], thus divergence times derived from arcA suggest that the Arc system appeared early in the history of the γ-proteobacteria.
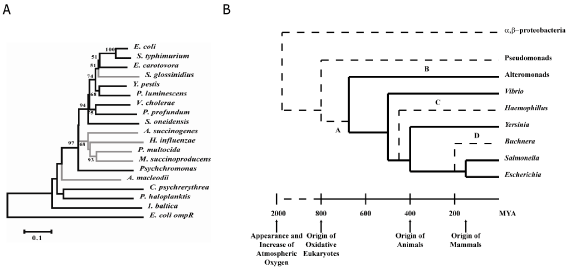
Figure 4: Evolution analysis of arcA genes. (A) Rooted arcA tree used to produce a linearized tree. Lineages in grey were removed based on successive LINTREE
runs to produce the final tree topology. (B) Linearized tree of arcA genes with scale depicting major geological and environmental events. Lineages in bold are
those used to construct the linearized tree while dashed lines represent lineages predicted from other sources. Letters indicate major events in the evolution of the
Arc system: A) origin of Arc system, B) uncoupling of HPt domain from ArcB in Alteromonad genera, C) loss of PAS domain in Pasteurellaceae ArcB, D) loss of
Arc system in Buchnera and other P-symbionts.
Discussion
With the availability of new Alteromonad genomic sequences, computational studies of the Arc system (and by extension other γ-proteobacterial regulatory systems) may now be extended beyond the scope of commensal organisms and known pathogens with the goal of characterizing the function of the network in a broader ecological sense. Of particular interest are the plasticity of the phosphorelay network in the Alteromonads in contrast to the commensals and the modifications of the regulon across family boundaries. As the topic of regulon evolution is beyond the scope of this work, the discussion will focus on the evolutionary origins of the Arc system and the distribution, plasticity and ecological significance of the Arc system components.
While OmpR-family regulators and hybrid sensor histidine kinase proteins are common in bacteria, the Arc system is only known to exist in four γ-proteobacteria families (Alteromonaceae, Enterobacteraceae, Vibrionaceae and Pasteurellaceae). Unlike most of TCSs, the components of all Arc systems are never encoded by a single operon [33]. Arc systems studied to date employ a conserved ArcA regulator but show significant diversity at the level of the regulon and the phosphorelay network. The Arc system without a full length ArcB as in Shewanella is actually quite common among the Alteromonads and may in fact represent the dominant form of the system in nature. As such, the following classifications for the Arc system are proposed: Class I, encompassing a full-length hybrid sensor histidine kinase (typified by E. coli ArcB) and Class II, encompassing a free Hpt domain protein and one or more unknown sensor/signal transduction components (typified by S.oneidensis HptA). Further sub-classifications are possible based in lineage-specific modifications to the system (i.e. modified ArcB of Pasteurellaceae). Based on the current genomic sequence library, Class I systems are common throughout all four studied lineages while Class II systems are exclusive to the Alteromonads. At this time it is unknown whether Class II systems share common sensor/signal transduction mechanisms or whether each genera employs a unique network topology.
Having established the distribution of the Arc system components within the γ-proteobacteria, the next question to be addressed is the origin of the hptA gene from the arcB gene. The most likely scenario for the uncoupling of the Hpt domain from the sensor histidine kinase component is gene duplication followed by sub fictionalization [34, 35]. Under this scenario, the Arc system would initially remain biochemically intact as a 3-component system (ArcA, ArcB, and HptA). Uncoupled from the Hpt domain, the sensor could then be replaced as necessary as an adaptation to changing environmental conditions. The conserved synteny of the arcB/hptA regions across the γ-proteobacteria and the lack of a canonical ArcB sensor component in hptA-encoding organisms would seem to support this hypothesis. Furthermore, there is no obvious pattern as to which Alteromonad genera utilize Class I vs. Class II systems, suggesting that there is no common ancestor to the hptA genes and that the uncoupling of the Hpt domain has likely occurred multiple times in multiple lineages. If these hypotheses are correct, one would predict the existence of an Alteromonad species that is in the process of transitioning from Class I to Class II. Such a hypothetical system would be expected to encode independent ArcB and HptA.
Comparison of the distribution of Arc system components to the ecological data of the studied organisms revealed a few correlations (Table 1). It was assumed prior to the sequencing of the Alteromonad genomes that the Arc system was only employed by facultative anaerobes and that facultative an aerobiosis was the selective pressure maintaining the existence of the system [5], but this does not in fact appear to be the case. At least three strictly aerobic (Alteromonas, Pseudoalteromonas and Idiomarina) and one aerotolerant anaerobic (Psychromonas) Alteromonad species employ Arc systems, which would imply that the traditional role of the Arc system in facilitating the transition to an aerobic lifestyle likely does not apply to these organisms.
The most striking correlation between environment and Arc system component distribution is host interaction. Class II Arc systems are only found in completely free-living Alteromonad species while all commensal, pathogenic and secondary symbiotic species encode Class I systems (Table 1). This distribution strongly suggests that, even in commensal/pathogenic species capable of a freeliving lifestyle such as Vibrio cholerae, host interaction is a selective pressure that helps maintain the integrity of the Arc system in these organisms. All of the commensal species are facultative anaerobes, in which the Arc system is presumed to serve the same general function, repression of aerobic metabolic pathways [3, 4]. Conceivably, such a system would be useful for an organism that transitions between the free-living and commensal lifestyle. Additionally, the Arc systems are involved in many other biological processes independent of metabolism, especially oxidative stress response. Many plant and animal species are known to initiate an “oxidative burst” (increased production of reactive oxygen species) in response to invasion by a bacterial species, an environmental change to which the bacteria must respond [36, 37]. In a number of well-studied bacteria, including E. coli, Salmonella enterica, V. cholerae, and Haemophilus influenzae, the Arc systems have been shown to play important role in combating reactive oxygen species, with different mechanisms, depending on the species [10, 38-41]. Thus, the accumulated evidence would seem to suggest that oxidative stress response, adaptation to changing oxygen conditions and/or regulation of virulence factors all contribute to maintaining the integrity of the Class I Arc system in commensal and pathogenic organisms.
While it is possible that the Arc system originated prior to the divergence of the γ-lineage from the other proteobacterial lineages, it is more likely that the system represents lineage-specific gene expansions in the γ-proteobacteria. Consistent with this view is that facultative anaerobes outside of the γ-proteobacteria have evolved novel systems for making the oxic-anoxic transition [33]. If this is the case, this places the upper temporal boundary on the origins of the system at ~1.89 GYA [32]. Assuming that the arcA orthologs studied are in fact descended from a common ancestor, this ancestor must have predated the divergence of the Alteromonacaea and Enterobacteraceae-Vibrionaceae-Pasteurellacaea lineages, approximately 600-1300 MYA based on the linearized arcA tree (Figure 4). This places the emergence of the Arc system in the late Pre-Cambrian Era at the time that oxygen was becoming a major component of the atmosphere [42]. While the above dates are technically only applicable to arcA, the same reasoning would suggest similar dates for arcB. This timeline is convenient in that it ties a modern phenotype (ability to transition between oxic and anoxic environments) to an ancient selective pressure (increasing atmospheric oxygen). This suggests that the Arc system may have originally evolved as an oxidative stress response mechanism that was later co-opted by emerging facultative anaerobes to facilitate the exploitation of aerobic environments. If this is the case, it also suggests a mechanism for the evolution of aerobiosis, at least withinthe γ-proteobacteria. It has been proposed that the proteobacteria originally evolved from an anaerobic photosynthetic ancestor [43], though evidence has been presented to show that some oxygen respiration systems predate the archaeal-bacterial divergence [44]. Regardless of whether oxygen respiration coupled to growth existed in the proteobacterial ancestor, increasing atmospheric oxygen represented an intense environmental pressure that promoted the emergence of novel oxygen defense mechanisms. Thus, it is attractive to suppose that the Arc system (and by extension, other oxidative stress defense mechanisms) evolved to provide aero tolerance to strict anaerobes, followed by the transition to facultative an aerobiosis and finally to strict aerobiosis.
Several directions about the Arc system are worth mentioning for future research : 1) addressing the question of whether the Shewanella Arc sensor/histidine kinase component is shared between all Alteromonad genera or whether each genus employs a unique sensor, 2) identification of the driving mechanistic force behind the plasticity of the Class II Arc system, i.e. addressing whether evolution of the regulon drives rearrangement of the phosphorelay network, or vice versa, or both, 3) given that the Arc regulon overlaps significantly with other global regulons (i.e. Fnr), determine whether modification of one system induces changes in other systems, 4) elucidation of the role of the Arc system in strictly aerobic and/or aero-tolerant anaerobic species and 5) determining if specific environmental conditions beyond the free-living lifestyle promote the formation of Class II Arc systems. It is clear from the presented results that the functional diversity of the Arc system is far greater than previously anticipated.
Acknowledgement
This research was supported by Major State Basic Research Development Program (973 Program: 2010CB833803), National Natural Science Foundation of China (31270097), and Doctoral Fund of the Ministry of Education of China (20130101110142) to H.G.
References
- Iuchi S, Lin EC . Purification and phosphorylation of the Arc regulatory components of Escherichia coli. J Bacteriol. 1992; 174: 5617-5623.
- Krell T, Lacal J, Busch A, Silva-Jiménez H, Guazzaroni ME, Ramos JL . Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol. 2010; 64: 539-559.
- Georgellis D, Kwon O, Lin EC . Quinones as the redox signal for the arc two-component system of bacteria. Science. 2001; 292: 2314-2316.
- Kwon O, Georgellis D, Lin EC . Phosphorelay as the sole physiological route of signal transmission by the arc two-component system of Escherichia coli. J Bacteriol. 2000; 182: 3858-3862.
- Georgellis D1, Kwon O, Lin EC, Wong SM, Akerley BJ . Redox signal transduction by the ArcB sensor kinase of Haemophilus influenzae lacking the PAS domain. J Bacteriol. 2001; 183: 7206-7212.
- Sengupta N, Paul K, Chowdhury R . The global regulator ArcA modulates expression of virulence factors in Vibrio cholerae. Infect Immun. 2003; 71: 5583-5589.
- Green J, Paget MS (2004) Bacterial redox sensors. Nat Rev Micro 2: 954-966.
- Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP . Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J Biol Chem. 2005; 280: 15084-15096.
- Gralnick JA, Brown CT, Newman DK . Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol Microbiol. 2005; 56: 1347-1357.
- Wong SM1, Alugupalli KR, Ram S, Akerley BJ . The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol Microbiol. 2007; 64: 1375-1390.
- Gao H, Wang X, Yang ZK, Palzkill T, Zhou J . Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics. 2008; 9: 42.
- Rolfe MD, Ter Beek A, Graham AI, Trotter EW, Asif HM, Sanguinetti G, et al . Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J Biol Chem. 2011; 286: 10147-10154.
- Yuan J, Wei B, Lipton MS, Gao H . Impact of ArcA loss in Shewanella oneidensis revealed by comparative proteomics under aerobic and anaerobic conditions. Proteomics. 2012; 12: 1957-1969.
- Yun S, Shin JM, Kim OC, Jung YR, Oh DB, Lee SY, et al . Probing the ArcA regulon in the rumen bacterium Mannheimia succiniciproducens by genome-wide expression profiling. J Microbiol. 2012; 50: 665-672.
- Morales EH, Collao B, Desai PT, Calderón IL, Gil F, Luraschi R, et al . Probing the ArcA regulon under aerobic/ROS conditions in Salmonella enterica serovar Typhimurium. BMC Genomics. 2013; 14: 626.
- Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, et al . Towards environmental systems biology of Shewanella. Nat Rev Microbiol. 2008; 6: 592-603.
- Lassak J, Henche AL, Binnenkade L, Thormann KM . ArcS, the cognate sensor kinase in an atypical Arc system of Shewanella oneidensis MR-1. Appl Environ Microbiol. 2010; 76: 3263-3274.
- Shroff NP, Charania MA, Saffarini DA . ArcB1, a homolog of Escherichia coli ArcB, regulates dimethyl sulfoxide reduction in Shewanella oneidensis MR-1. J Bacteriol. 2010; 192: 3227-3230.
- Perraud AL, Kimmel B, Weiss V, Gross R . Specificity of the BvgAS and EvgAS phosphorelay is mediated by the C-terminal HPt domains of the sensor proteins. Mol Microbiol. 1998; 27: 875-887.
- Yin J, Gao H . Stress responses of shewanella. Int J Microbiol. 2011; 2011: 863623.
- Capra EJ, Laub MT . Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012; 66: 325-347.
- Peña-Sandoval GR, Kwon O, Georgellis D . Requirement of the receiver and phosphotransfer domains of ArcB for efficient dephosphorylation of phosphorylated ArcA in vivo. J Bacteriol. 2005; 187: 3267-3272.
- Matsubara M, Mizuno T . The SixA phospho-histidine phosphatase modulates the ArcB phosphorelay signal transduction in Escherichia coli. FEBS Lett. 2000; 470: 118-124.
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL . NCBI BLAST: a better web interface. Nucleic Acids Res. 2008; 36: W5-9.
- Kumar S, Nei M, Dudley J, Tamura K . MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008; 9: 299-306.
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, et al . Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013; 41: W597-600.
- Yang Z (2007) PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586-1591.
- Nickel GC, Tefft DL, Goglin K, Adams MD . An empirical test for branch-specific positive selection. Genetics. 2008; 179: 2183-2193.
- Takezaki N, Rzhetsky A, Nei M . Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 1995; 12: 823-833.
- Gao H, Wang X, Yang ZK, Chen J, Liang Y, Chen H, et al . Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis. PLoS One. 2010; 5: e15295.
- Ochman H, Wilson AC . Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987; 26: 74-86.
- Sheridan PP, Freeman KH, Brenchley JE (2003) Estimated minimal divergence times of the major Bacterial and Archaeal phyla. Geomicrobiol J 20: 1-14.
- Capra EJ, Laub MT . Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012; 66: 325-347.
- Ohta T . Time for spreading of compensatory mutations under gene duplication. Genetics. 1989; 123: 579-584.
- Rajon E, Masel J . Compensatory evolution and the origins of innovations. Genetics. 2013; 193: 1209-1220.
- Babior BM . NADPH oxidase: an update. Blood. 1999; 93: 1464-1476.
- Marino D, Dunand C, Puppo A, Pauly N . A burst of plant NADPH oxidases. Trends Plant Sci. 2012; 17: 9-15.
- Lu S, Killoran PB, Fang FC, Riley LW . The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar Enteritidis. Infect Immun. 2002; 70: 451-461.
- Calderón IL, Morales E, Caro NJ, Chahúan CA, Collao B, Gil F, et al . Response regulator ArcA of Salmonella enterica serovar Typhimurium downregulates expression of OmpD, a porin facilitating uptake of hydrogen peroxide. Res Microbiol. 2011; 162: 214-222.
- Loui C, Chang AC, Lu S . Role of the ArcAB two-component system in the resistance of Escherichia coli to reactive oxygen stress. BMC Microbiol. 2009; 9: 183.
- Alvarez AF, Malpica R, Contreras M, Escamilla E, Georgellis D . Cytochrome d but not cytochrome o rescues the toluidine blue growth sensitivity of arc mutants of Escherichia coli. J Bacteriol. 2010; 192: 391-399.
- Canfield DE, Habicht KS, Thamdrup B . The Archean sulfur cycle and the early history of atmospheric oxygen. Science. 2000; 288: 658-661.
- Gupta RS . Evolutionary relationships among photosynthetic bacteria. Photosynth Res. 2003; 76: 173-183.
- Castresana J, Lübben M, Saraste M . New archaebacterial genes coding for redox proteins: implications for the evolution of aerobic metabolism. J Mol Biol. 1995; 250: 202-210.