
Research Article
J Bacteriol Mycol. 2024; 11(2): 1222.
Biofilm Formation and Whole Genome Analysis of MDR Klebsiella Pneumoniae Strains Isolated from Hospital Acquired Infections in Tertiary Hospitals in Dakar, Senegal
Issa Ndiaye1,8*; Ousmane Sow1; Abdoulaye Cissé1; Bissoume Sambe Ba4; Farma Thiam5; Mouhamadou Moustapha Boye2; Baidy Dièye3; Cheikh Fall1; Yakhya Dieye1; Assane Dieng2; Amadou Diop3; Guillaume Constantin de Magny6,7; Abdoulaye Seck1,8
1Pôle de Microbiologie, Institut Pasteur de Dakar, Sénégal
2Laboratoire de Bactériologie et Virologie, Hôpital Aristide Le Dantec, Dakar, Sénégal
3Laboratoire de Bactériologie et Virologie, Hôpital Albert Royer, Dakar, Sénégal
4World Health Organization WCARO, Dakar, Senegal
5Laboratoire de Biologie Médicale, Hôpital régional de Pikine
6MIVEGEC, Université Montpellier, CNRS, IRD, Montpellier, France
7MEEDiN, Montpellier Ecology and Evolution of Disease Network
8Faculté de Médecine, Pharmacie et Odontostomatologie, Université Cheikh Anta Diop, Dakar, Sénégal
*Corresponding author: Issa Ndiaye, Pôle de Microbiologie, Institut Pasteur Dakar, 36, Avenue Pasteur, Dakar, BP: 220, Senegal. Email: seydi-na.indiaye14@gmail.com
Received: October 14, 2024; Accepted: November 01, 2024 Published: November 08, 2024
Abstract
Klebsiella pneumoniae is widely recognized as an opportunistic pathogen in both hospital and community settings. It is a key member of the ESKAPE group, which comprises priority microorganisms of major concern owing to their antibiotic resistance. The resistance of K. pneumoniae, particularly related to Extended-Spectrum β-Lactamases (ESBLs), poses a significant global public health challenge. The combination of its Multidrug Resistance (MDR) phenotype and various pathogenicity factors increases its potential to cause severe clinical infections. Biofilm formation was assessed via a semiquantitative microtiter technique. We employed various bioinformatics tools to analyze the Antimicrobial Resistance (AMR), virulence factors, plasmid replicons, and genomic diversity of the CRKP isolates. Overall, among the 24 K. pneumoniae isolates, most produced strong biofilms (n = 21), with some exhibiting moderate (n = 1) or weak (n = 2) biofilm production. An alarming level of resistance to multiple classes of antibiotics was correlated with the presence of various resistance genes, including those for β-lactams (blaOXA-48, blaOXA-181, blaCTX-M15, blaTEM and blaSHV), aminoglycosides (aph(6)-Id, aac(3)-IIe, aadA2, ant(3’’)-IIa, aph(3’)-Ia and aac(6’)-Ib-cr), and quinolones (qnrA, qnrB, qnrS, CRP, and emrR). Various efflux pumps, such as KpnGH, oqxAB, acrAB, acrD, and KpnEF, are ubiquitously distributed across MDR K. pneumoniae strains. Several virulenceassociated genes encoding type 1 fimbriae (fimH), type 3 fimbriae (mrkA), efflux pumps (acrAB, oqxAB), enterobactin (entA, entB, fepC), and yersiniabactin (irp1, irp2, ytbA, ybtE, ybtP, ybtQ, ytbT, ytbU, ytbX) have been identified. Genetically, the isolates presented high diversity, with 18 Sequence Types (STs) and an average of 70.1% accessory genes. On the basis of SNP distance and pairwise ANI analysis, the majority of K. pneumoniae isolates were grouped into one clade. The high plasticity of K. pneumoniae in the acquisition of an MDR phenotype, combined with the phenotypic and genotypic factors described in this report, underscores the challenges in achieving effective clinical therapy with the available antibiotics. The findings also emphasize the critical need for the surveillance of multidrug-resistant pathogens in clinical settings in Senegal, as well as the need to evaluate their prevalence, propagation, and impact on patient health outcomes.
Keywords: Klebsiella pneumoniae; MDR; Biofilm; Resistance genes; Virulence genes; Plasmid replicons; ST; WGS; Bionformatics
Introduction
Klebsiella pneumoniae is an opportunistic pathogen responsible for a broad spectrum of Healthcare-Associated Infections (HAIs), predominantly affecting immunocompromised individuals and is responsible for diverse diseases syndromes such as pneumonia, bacteremia, urinary tract infections, wound or soft tissue infections, and liver abscesses [1]. Klebsiella pneumoniae ranks among the priority pathogens categorized within the ESKAPE-E group and is classified as a critical organism on the WHO priority pathogens list for the research and development of novel antibiotics [2]. Recent studies have highlighted this organism as one of the top five pathogens contributing to global mortality, regardless of its susceptibility to antibiotics [3]. In the USA, K. pneumoniae has been identified as a predominant cause of HAIs, accounting for an estimated 8.0% of all HAIs, whereas in the UK, it has been implicated in 4.7%–6.0% of all bacterial infections [4]. Sparse data from sub-Saharan Africa (sSA) suggest that K. pneumoniae may be responsible for higher proportions of HAIs in this region than in industrialized countries, particularly among children under 5 years of age [5-7]. In South Africa, K. pneumoniae caused 22.0% of HAI bacteremia cases among neonates, whereas in Kenya, it was estimated to be responsible for 20.0% of HAI bacteremia cases [8,9]. The emergence of Multidrug- Resistant (MDR) strains in K. pneumoniae is largely attributed to the acquisition of Antimicrobial Resistance (AMR) genes, which are commonly found among globally disseminated clones and often contribute to hospital outbreaks. Presently, K. pneumoniae resistance is predominantly associated with molecules such as third-generation cephalosporins and carbapenems. Various Extended-Spectrum β-Lactamases (ESBLs) responsible for resistance to third-generation cephalosporins have been identified in Senegal, including those from major blaCTX-M groups such as blaCTX-M15, the most predominant [10- 12]; blaCTX-M109 [13]; blaSHV-derived enzymes (blaSHV-2 and blaSHV-12) [14]; and carbapenemases conferring resistance to carbapenems (e.g., blaKPC-2, blaNDM and blaOXA-48) [15,16]. Additionally, resistance to fluoroquinolones, which is primarily mediated by Plasmid-Mediated Quinolone Resistance (PMQR) mechanisms [17,18], as well as modification enzymes conferring resistance to aminoglycosides [17], has been reported. Owing to the high adaptability of this pathogen, ESBLs, carbapenemases, PMQRs, and aminoglycoside-modifying enzymes may coexist in the same clinical strain, posing challenges for treatment options for affected patients [19].
Apart from AMR, the propensity of K. pneumoniae to cause severe infections is linked to virulence factors, biofilm formation and sequence types [20,21]. The determination of sequence types and clonal distributions is important, as certain clones, such as ST11 and ST258 or ST14, ST15, ST17, and ST37, are widely acknowledged for carrying MDR traits and have been linked to global outbreaks in human populations in recent years [22]. Virulence-associated genes, encompassing both fimbriae and nonfimbrial adhesins, ironscavenging systems, and surface polysaccharides, play pivotal roles in the pathogenicity of K. pneumoniae. They are responsible for processes such as colonization, invasion, and pathogenicity of the strains [23,24]. One of the key virulence traits of K. pneumoniae is its ability to form biofilms, which are composed of bacteria enclosed within a selfgenerated extracellular matrix adhering to either living or nonliving surfaces [25]. This matrix is composed of proteins, exopolysaccharides, DNA, and lipopeptides [26]. Also, several virulence factors, including capsule polysaccharides, lipopolysaccharides, type 1 and type 3 fimbriae, outer membrane proteins, and mechanisms for iron acquisition and nitrogen utilization enable K. pneumoniae to survive, evade the immune system during infection, and contribute to biofilm formation [27,28].
In Senegal, there is a lack of whole-genome studies on AMR, which are essential for better understanding the mechanisms of resistance, virulence, and clonal distribution of isolates. This knowledge is crucial to strengthening AMR surveillance and evaluating available therapeutic options. This, the aim of this study was to give first insight of biofilm formation, resistome and virome among MDR-producing K. pneumoniae strains isolated from HAIs in Senegal.
Materials and Methods
Sample Collection
MDR K. pneumoniae strains were collected and processed from Hospital Aristide Le Dantec and the Children’s Hospital Center Albert Royer of Fann Microbiology Laboratory from a previous study [29]. Bacterial strains resistant to at least three different antibiotic classes were classified as MDR, while those susceptible to only one or two antibiotic classes were categorized as XDR respectively, as previously described [30]. Infections were considered hospital-acquired if they developed at least 48 hours after hospital admission. Isolates were collected between January 2018 to February 2021. Antimicrobial susceptibility was evaluated by measuring strain growth zone diameters using the Kirby-Bauer method according to CA-SFM/ EUCAST guidelines (version 2023) during a previous study [29]. The phenotypic resistance of the strains is given in Table 1 supp.
Hypermucoviscosity Characterization and Biofilm Formation Assay
The Hypermucoviscous (HM) phenotype was assessed via the "string test," following established protocols [31]. Klebsiella pneumoniae cultures were incubated on agar plates overnight at 37°C. A colony from the plate was subsequently stretched using a loop. If a viscous string formed, exceeding a length of 5 mm, the strain was classified as exhibiting the HM phenotype.
Biofilm production of MDR K. pneumoniae was assessed via a crystal violet staining assay. Briefly, the strains were grown overnight in Luria Bertani (LB) broth at 37°C under static conditions. Initially, 20 μL of the 0.5 McFarland bacterial standard and 180 μL of Luria– Bertani broth were inoculated into each well of a 96-well microplate, with six wells per strain, followed by incubation at 37°C for 24 hours. The Luria–Bertani broth was subsequently aspirated, and the wells were washed three times with Phosphate-Buffered Saline (PBS). The plates were stained with a 1% crystal violet dye solution (150 μL/well) for 15 minutes. After staining, the wells were washed three times with sterile water to remove unbound dye and then air-dried. The stained biofilms were solubilized with 150 μL of 100% ethanol for 10 minutes, and quantification was performed by measuring the optical density at 570 nm (OD575). Each experiment was conducted in triplicate. The OD of the control wells with only media was used as the cutoff value (ODc). Using the ODc, the results of the biofilm formation assay were interpreted as follows: non biofilm producer (OD < ODc), weak producer (ODc < OD < 2ODc), moderate producer (2ODc < OD < 4ODc), and strong producer (4ODc < OD) [32]. Graphs and statistical analyses were performed via GraphPad Prism 9 (GraphPad Software, San Diego, CA, USA).
DNA Extraction and Whole-Genome Sequencing
The genomic DNA of the K. pneumoniae isolates was extracted via the PureLinkTM Genomic DNA Mini Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Nucleic acid concentrations were measured via a Nanodrop spectrophotometer, and samples were adjusted to concentrations between 100 and 300 ng/μL. Whole-genome sequencing (WGS) was conducted via the NextSeq platform Illumina®. Dual-index sequencing libraries were prepared via the NEBNext® library preparation kit, Multiplex Oligos for Illumina® (NEB, Boston, MA, USA), and pooled. Sequencing was performed on an Illumina® Next 500 cartridge (2 × 150 bp).
Genomic Analysis
Genome assembly, annotation and sequence analysis: The quality of reads was conducted using FastQC [33]. The adaptor trimming was executed with fastp v0.23.2 [34] and the genomes were assembled from draft genomes with SPAdes v3.15.5 [35]. Finally, annotation was performed using Prokka [36]. AMR genes, virulence genes and plasmid replicons were identified with the Comprehensive Antibiotic Resistance Database (CARD) [37], Virulence Factor Database (VFDB) [38], and PlasmidFinder [39], respectively, via Abricate v1.0.1 [40]. The approach proposed by Diancourt et al. [41] was employed for in silico Multilocus Sequence Typing (MLST), which involves evaluating allelic diversity across seven housekeeping genes (gapa, infb, mdh, pgi, phoe, rpob, and tonb). Whole-Genome Sequencing (WGS) data were utilized to identify the different sequence types (STs) and to determine the MLST profiles of the K. pneumoniae isolates via MLST v2.23.0 [42,43]. The K and O serotypes were determined via Kaptive 2.0 [44].
Phylogenomic analysis: To investigate the genetic diversity of the 24 MDR K. pneumoniae isolates in this study, pairwise Single Nucleotide Polymorphism (SNP) distances and pairwise Average Nucleotide Identity (ANI) values were analyzed via snp-dists v0.8.2 (https://github.com/tseemann/snp-dists) and FastANI v1.32 [45], respectively. Core genome alignment was performed via Roary [46] with a 95% minimum identity for BLASTX and a 99% core definition threshold. SNPs for each isolate were called from core genes using SNP sites v2.4.1 [47]. The phylogenetic tree was subsequently constructed employing gubbins [48], with the RAXML option for tree builder, and branch support was subsequently assessed using the neighbor-joining method with 500 bootstrap replicates. The resulting tree was visualized using Geneious [49]. The number of pangenomes was obtained, and a phylogenetic tree was visualized against a presence and absence matrix of the pangenomes via the roary_plots script (https://github. com/sanger-pathogens/Roary/tree/master/contrib/roary_plots).
Results
String Test and Biofilm Formation
The string test was negative, and none of the isolates were considered hypermucoviscous. Biofilm formation was detected in all strains, the majority of which were categorized as strong producers (n = 21), moderate producers (n = 1), or weak producers (n = 2) (Figure 1).
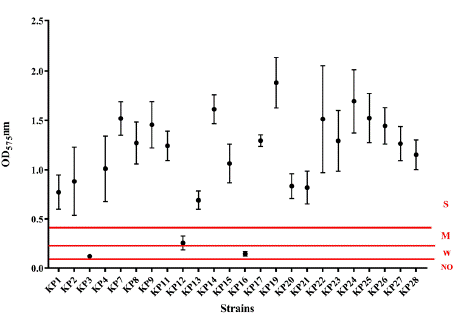
Figure 1: Biofilm formation of the 24 K. pneumoniae strains studied. The
graphic shows the values (mean and standard deviation) of the optical
density at 575 nm (OD575 nm) of crystal violet obtained for each strain. The
dashed lines at 0.1, 0.21, and 0.42 represent the threshold value for each
biofilm capability formation category: no biofilm Producer (NO), Weak (W),
and Moderate (M) biofilm, respectively. Values above 0.42 were considered
Strong (S) biofilm formation.
Resistome, Virulome and Plasmid Analysis
For the resistome analysis, we identified carbapenem resistance genes, oxacillinases or class D β-lactamases, with blaOXA-48 found in 2 strains and its variant blaOXA-181 in 3 strains. For the ESBL genes, blaCTX-M15 was detected in 22 out of 24 strains. The blaOXA-1 ESBL gene was detected in 15 out of 24 strains. Multiple strains simultaneously harbored ESBL genes such as blaTEM and blaSHV. Additionally, the ESBL gene blaPKP-A-6 was detected in one strain. Tetracycline resistance genes (tetA) and macrolide resistance genes (mphA) were detected in 14 and 2 strains, respectively. Various aminoglycoside resistance genes were also found: aph(6)-Id (20 strains), aac(3)-IIe (15 strains), aadA2 (2 strains), ant(3’’)-Iia (2 strains) and pph(3’)-Ia (2 strains). The aac(6’)- Ib-cr gene, which confers resistance to both aminoglycoside and fluoroquinolones, was found in 15 out of 24 strains. Other important resistance determinants, such as quinolone resistance genes (qnrA (1/24), qnrB (14/24), qnrS (8/24), crp (24/24), and emrR (24/24)), efflux pump genes (oqxAB (23/24)), macrolide resistance genes (mphA (2/24)), trimethoprim resistance genes (dfrA12 (2/24), dfrA14 (17/24), dfrA15 (2/24), and cpxA (24/24)), sulfonamide resistance genes (sul1 (3/24) and sul2 23/24)), phenicol resistance genes (catI (4/24) and catII (1/24)), and fosfomycin resistance genes (fosA5 (3/24) and fosA6 (21/24)), were detected (Figure 2A, and Table S3).
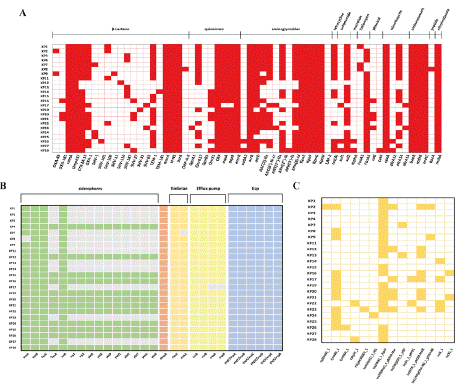
Figure 2: Distribution of A) Resistance genes. The white square indicates
that the gene was not detected. B) Virulence genes of interest are shown in
gray. ECP: E. coli common pilus. C) Plasmid replicons of 24 K. pneumoniae
strains isolated from HAIs in Dakar.
Various types of efflux pumps are ubiquitously distributed across MDR K. pneumoniae strains, including major facilitator superfamily (MFS) antibiotic efflux pump genes (KpnGH), Resistance-Nodulationcell Division (RND) antibiotic efflux pump genes (oqxAB, acrAB, and acrD (22 out of 24 strains)), and small multidrug resistance (SMR) antibiotic efflux pump genes (KpnEF). The porin OmpK37, which confers reduced susceptibility to β-lactams and carbapenems, was also detected in all the strains (Figure 2A).
Regarding virulence genes, those related to adherence, specifically the E. coli Common Pilus (ECP) genes (ecpA, ecpB, ecpC, ecpD, ecpE, and ecpR), type 1 (fimH), and outer membrane protein A (ompA), were present in all strains. Type 3 fimbriae (mrkA) were present in 23 out of 24 strains. The siderophore genes entB and fepC were also found in all strains, whereas entA, irp1, irp2, ytbA, ybtE, ybtP, ybtQ, ytbT, ytbU, and ytbX were present in some strains (Figure 2B and Table S4).
Fourteen plasmid replicons were detected in the studied isolates. They comprised five replicons from the IncF group, four from the Col group, and one each from the IncC, IncH, IncL, IncR, and IncX groups. Up to 6 plasmid replicons were found within the isolates. The IncFIB(K)_1_Kpn3 (n=20) and Col440I_1 (n=12) plasmid replicons were the most common in the K. pneumoniae isolates analyzed in this study, followed by IncHI1B_1_Pndm-MAR, IncFIB(Mar)_1_ Pndm-Mar, and IncR_1, which were detected in 8, 6 and 4 isolates, respectively (Figure 2C and Table S5).
Genomic Diversity and Phylogenetic Tree
A matrix of pairwise SNP distances and pairwise ANI values is illustrated in Figure 3. The 23 K. pneumoniae isolates are grouped into one clade, with KP8 being relatively distant. The number of core SNPs within the clade varied from 42-882. Additionally, ANI values of 98.92-100% were found among all pairs of isolates, except for KP8, which presented an ANI of 93.83% compared with the other isolates. We identified 18 distinct STs (ST6, ST13, ST15, ST17, ST20, ST37, ST39, ST70, ST234, ST307, ST392, ST502, ST584, ST867, ST870, ST967, ST1077, and ST1243) among the 24 strains. The phylo-genetic tree analysis delineated separate clades on the basis of these STs (Figure 4 and Table S2). The comparison revealed 19,993 pan genes consisting of 5,988 (29.9%) core and 14,005 (70.1%) accessory genes. Among the accessory genes, 10,406 (74.3%) encoded hypothetical proteins (Figure 5 and Table S6).
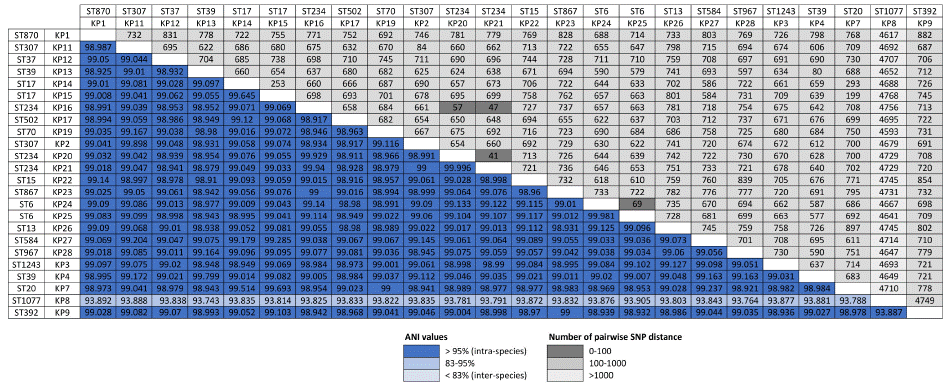
Figure 3: Matrix of pairwise Single Nucleotide Polymorphism (SNP) distances and pairwise Average Nucleotide Identity (ANI) values among MDR Klebsiella
pneumoniae clinical isolates.
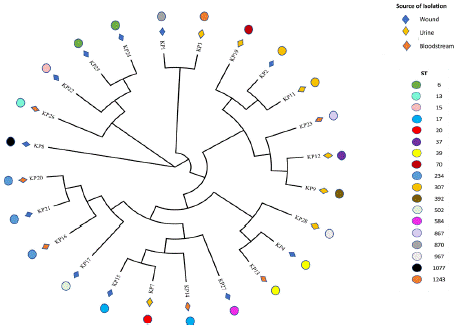
Figure 4: Phylogenetic tree of MDR Klebsiella pneumoniae clinical isolates
based on single nucleotide polymorphisms.
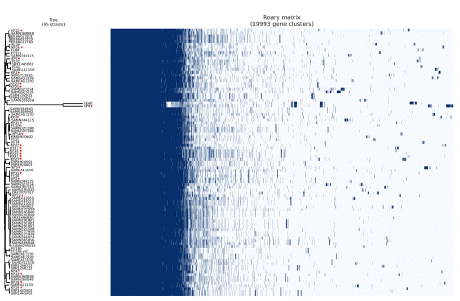
Figure 5: The single nucleotide polymorphism-based phylogenetic tree
is shown against the pangenome matrix of MDR Klebsiella pneumoniae
genomes compared with other published genomes.
Discussion
Gram-negative bacilli infections pose significant threats to hospitalized patients, with the potential to become life-threatening [50,51]. Klebsiella pneumoniae, an opportunistic pathogen, is associated with both community-acquired and nosocomial infections, causing pneumonia, abscesses, bacteremia, and urinary tract infections [52]. Its rapid acquisition of antimicrobial resistance has escalated K. pneumoniae into a global concern, prompting efforts to curb the spread of multidrug-resistant strains [53]. A primary advantage of WGS lies in its ability to characterize the genomic content of clinically relevant bacteria, linking them to antimicrobial resistance and virulence-associated phenotypes. This enhances our understanding of their transmission within healthcare settings, facilitates accurate diagnostics, and enables prompt therapeutic interventions. This study aimed to characterize K. pneumoniae isolates from HAIs that occurred between 2018 and 2020 in Dakar, Senegal. Antibiotic exposure is a key driver of antimicrobial resistance, influenced by factors such as antibiotic use in healthcare, communities, agriculture, and the environment. Overuse, often due to easy access without prescription, contributes to resistance. In healthcare settings, prolonged and intensive antibiotic use is a major cause of the spread of resistant healthcare associated infections [54]. All Multidrug Resistance (MDR) K. pneumoniae strains in our study exhibited high resistance rates to commonly prescribed antibiotics, either individually or in combination. The observed resistance rates were as follows: β-lactams (100%), aminoglycosides (83.3%), fluoroquinolones (91.6%), cyclins (100%), fosfomycin (33.3%), and trimethoprim-sulfamethoxazole (91.6%). This finding is consistent with previous studies by Nirwati et al. [55] and Moini et al. [56], which reported that MDR K. pneumoniae isolates exhibited high levels of resistance to penicillins, cephalosporins, fluoroquinolones, aminoglycosides, and sulfonamides. A global meta-analysis of 47 studies estimated the prevalence of antibiotic resistance in healthcareassociated MDR K. pneumoniae. According to this meta-analysis, the resistance rates were as follows: β-lactams (91.5%), aminoglycosides (85.1%), quinolones (87.2%), cyclins (34%), sulfonamides (51%), polymyxins (14.9%), and other classes of antibiotics (38.3%) [57].
All of the strains were ESLB-producers and blaCTX-M15 was found as the main ESBL gene (22 out of 24, 91.6%) in our K. pneumoniae isolates confirms that CTX-M-15 is currently the most widely distributed CTX-M enzyme in Senegal [58,59] and worldwide [60]. A study conducted in medical biology laboratory at Institut Pasteur Dakar showed that a majority of ESBL-producing E. coli strains were sensitive to cefoxitin and piperacillin-tazobactam suggesting that these antibiotics can be used as alternatives to carbapenems in the treatment of ESBL-secreting Enterobacteriaceae infection [58]. Additionally, the presence of various ESBL genes, such as blaTEM (18 out of 24, 75%) and SHV (23 out of 24, 95.8%), underscores the potential role of K. pneumoniae as a reservoir for beta-lactam and non-betalactam resistance determinants, posing major concerns in countries with inadequate antibiotic resistance surveillance, prevention, and containment measures [61].
Among them, 8 K. pneumoniae strains were resistant to carbapenems, with the blaOXA-48 gene detected in 2 strains and blaOXA-181 in 3 strains. For the remaining carbapenem-resistant strains, the presence of various efflux pumps (ramA-acrAB) and a porin system (OmpK37) was observed, which can decrease susceptibility to betalactam antibiotics, including carbapenems [62,63]. The widespread of multidrug resistance mechanisms of K. pneumoniae, especially the global spread of carbapenemases, combined with the rapid increase in carbapenem consumption in LMICs are driving increased carbapenem resistance especially in the ICU where they are the leading causes of invasive HAIs [64].
In nearly all the isolates investigated, genes associated with resistance to aminoglycoside, trimethoprim, sulfonamide, tetracycline, and chloramphenicol were found, which correlates with antibiotic susceptibility results.
In addition to AMR genes, our K. pneumoniae isolates harbored several virulence-associated genes, including those encoding type 1 fimbriae (fimH), type 3 fimbriae (mrkA), efflux pumps (acrAB, oqxAB), enterobactins (entA, entB, fepC), and yersiniabactin (irp1, irp2, ytbA, ybtE, ybtP, ybtQ, ytbT, ytbU, ytbX). Previous studies have demonstrated a significant association between mrkA and biofilm formation in K. pneumoniae [65,66], while fimH has been strongly linked to the MDR phenotype [65,67].
Biofilms are microbial communities that are encased in a matrix that maintain bacterial structural integrity, can attach to both biotic and abiotic surfaces and protect bacterial cells against antibiotics and the host’s immune system [68]. The majority of the strains exhibited a strong biofilm formation phenotype (22/24, 91.6%) and several studies have reported strong biofilm production in the majority of clinical K. pneumoniae strains [55,69,70]. In our study, biofilm production correlates with the presence of type 1 (fimH) and type 3 fimbriae (mrkA), both of which are crucial for adhesion to host cells [71]. These fimbriae also play a significant role in biofilm formation across many species [72]. fimH has been implicated in adhesion to epithelial cells, colonization, biofilm formation, and immune evasion [73], whereas mrkA is crucial for binding to host cells and extracellular matrix proteins, promoting biofilm formation on both biotic and abiotic surfaces [74]. The Minimum Inhibitory Concentrations (MICs) of conventional antibiotics for biofilm bacteria are 100–1000 times higher than those for planktonic bacteria [75] and biofilmproducing ability have been shown to correlate with extensively drug resistant (XDR) K. pneumoniae antibiotic resistance profile [76]. This inherent tolerance to antimicrobial agents, can then lead to severe, persistent infections that are particularly difficult to treat particularly in hospital settings [77].
The overexpression of the multidrug efflux pump acrAB in gramnegative bacteria not only confers resistance against antibiotics such as fluoroquinolones, β-lactams, and tigecycline but also provides virulence factors, such as resistance to antimicrobial peptides produced by the innate immune system in the lungs [78,79]. The oqxAB multidrug efflux pump mediates resistance in various bacteria, especially K. pneumoniae and E. coli, and can be found on both chromosomes and plasmids [80,81]. Previous studies have demonstrated that oqxAB confers resistance to fluoroquinolone, olaquindox, tigecycline, nalidixic acid, and chloramphenicol [80-83]. Siderophores such as enterobactin and yersiniabactin facilitate iron uptake and protect microorganisms against oxidative stress from host innate immune cells, thereby promoting infection [84,85].
The persistence of carbapenem resistance genes is driven primarily by the clonal dissemination of isolates and the spread of these genes via conjugative or mobile plasmids [86]. In this study, we identified five types of Inc. plasmids known to facilitate the spread of the blaOXA-48 and blaNDM genes in K. pneumoniae isolates. All the detected IncL plasmids harbored blaOXA-48 resistance genes, while two plasmid replicons carrying blaNDM genes were identified. Although these particular plasmids did not carry the blaNDM gene, their presence suggests the potential for these strains to acquire it. IncFIB and Col were the most frequently detected plasmids. The IncFIB gene is a conjugative plasmid previously associated with the dissemination of the blaTEM gene in E. coli isolates from Africa and is responsible for the spread of the blaNDM-1, blaSHV-12, blaCTX-M15, and blaOXA-1 genes in K. pneumoniae in Europe [87,88]. This study is the first to report these plasmid replicons in clinical K. pneumoniae isolates from Senegal. The detection of these multiple plasmid profiles in our strains raises concerns about the rapid spread of antibiotic resistance in hospital settings, with the likelihood of these plasmid types becoming more prevalent in the future.
Among the 24 strains, 18 different sequence types were identified, indicating significant diversity among the clinical HAIs in Dakar. The variation in ST distribution across different study locations highlights the genetic diversity of this pathogen, with majority of the STs being widely dispersed [89,90]. Additionally, our analysis of the genetic relatedness of the strains revealed moderate pairwise SNP distances in pairs of different STs, whereas > 1000 SNPs were identified in ST1077 compared with the other STs. The commonly applied
ANI threshold of 95-96% supports species assignments [91]. Our study revealed that all MDR strains shared greater than 97% ANI, suggesting that they were closely related strains of K. pneumoniae with similar gene presence/absence patterns [92]. However, KP8 (ST1077) shared less than 94% ANI with other K. pneumoniae isolates, confirming that it is a relatively distinct ST from its closest phylogenetic neighbors. K. pneumoniae ST1077 is closely related to ST1224 that was isolated from dairy products and chicken meat, respectively, in libya and Western Algeria, suggesting that this Klebsiella isolate is not host-specific and could be easily transmitted to humans from food animals and their products [93,94]. Pangenome analysis revealed substantial diversity, with a high percentage of accessory genes (70.1%) and a significant proportion of hypothetical proteins (74.3%) among these accessory genes
K. pneumoniae is thought to possess an open pangenome due to its ubiquity across diverse environments, including mammalian guts, soils, and surfaces, where it can potentially exchange genetic material with other bacterial species [95]. These findings suggest the broad diversity, widespread dissemination, and rapid adaptive evolution potential of MDR clinical K. pneumoniae strains in Senegal.
Conclusions
In this study, we present genomic insights into MDR K. pneumoniae clinical isolates from tertiary university hospitals in Dakar. These isolates exhibit strong biofilm formation ability, which could contribute to their persistence in hospital environments. They also belong to various sequence types and carry multiple antimicrobial resistance genes, virulence genes, and plasmid replicons. Through analysis of SNPs, ANI, and phylogenetic data, the isolates were found to be primarily clustered into a single major clade. Our findings elucidate the genomic characteristics and pathogenic traits of these clinical isolates. Pangenome analysis revealed significant genomic plasticity, suggesting the potential for the evolution and dissemination of these pathogens. We recommend search for alternative antibiotic treatment options, especially for carbapenem-resistant Enterobacteriaceae among clinicals isolates in Senegal, as well as novel therapeutic approaches such as bacteriophages for difficult-totreat and biofilm-associated infections. We also advocate reinforcing AMR surveillance, implementing antimicrobial stewardship policies, and enhancing infection control measures in hospitals to reduce the selective pressure driving the emergence and spread of MDR strains.
Author Statements
Author Contributions
Conceptualization: IN, GCDM and AS; Methodology, IN, OS, AC, BSB; Software, IN; Validation: GDCM and AS, Formal Analysis: IN; Bioinformatic and phylogenetic analysis, IN; Validation, GCDM and AS; Investigation: IN, MMB, AD and AD; Resources: MMB, AD, AD, BD, FT, OS, AC, BSB, CF, YD; Data Curation: IN; Writing – Original Draft Preparation: IN.; Writing – Review & Editing: BD, FT, OS, AC, BSB, CF, YD, GCDM and AS; Visualization: IN; Supervision: GCDM and AS; Project Administration: GCDM and AS.
Funding
This study was supported by GCDM at the Institut de Recherche pour le Développement (IRD), France.
Data Availability Statement: The genome raw reads were deposited in European Nucleotide Archive (ENA) and are also available at the National Center for Biotechnology Information (NCBI) under the project number PRJEB78995.
Acknowledgments
The authors thank all the members of the Pole of Microbiology, Pasteur Institute of Dakar, for their valuable assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 2016; 24: 944-956.
- Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Inf dis. 2018; 18: 318-327.
- Ikuta KS, Swetschinski LR, Aguilar GR, Sharara F, Mestrovic T, Gray AP, et al. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2022; 400: 2221-2248.
- Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013; 11: 297-308.
- Shi T, Denouel A, Tietjen AK, Lee JW, Falsey AR, Demont C, et al. Global and regional burden of hospital admissions for pneumonia in older adults: a systematic review and meta-analysis. The Journal of infectious diseases. 2020; 222: S570-S576.
- Verani JR, Blau DM, Gurley ES, Akelo V, Assefa N, Baillie V, et al. Child deaths caused by Klebsiella pneumoniae in sub-Saharan Africa and south Asia: a secondary analysis of Child Health and Mortality Prevention Surveillance (CHAMPS) data. The Lancet Microbe. 2024; 5: e131-e141.
- Abubakar U, Amir O, Rodríguez-Baño J. Healthcare-associated infections in Africa: a systematic review and meta-analysis of point prevalence studies. Journal of Pharmaceutical Policy and Practice. 2022; 15: 99.
- Morkel G, Bekker A, Marais BJ, Kirsten G, Van Wyk J, Dramowski A. Bloodstream infections and antimicrobial resistance patterns in a South African neonatal intensive care unit. Paediatrics and international child health. 2014; 34: 108-114.
- Henson SP, Boinett CJ, Ellington MJ, Kagia N, Mwarumba S, Nyongesa S, et al. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int J Med Microbiol. 2017; 307: 422-429.
- Abdallah R, Kuete Yimagou E, Hadjadj L, Mediannikov O, Ibrahim A, Davoust B, et al. Population diversity of antibiotic resistant Enterobacterales in samples from wildlife origin in Senegal: Identification of a multidrug resistance transposon carrying bla CTX–M–15 in Escherichia coli. Front Microbiol. 2022; 13: 838392.
- Ruppé E, Woerther PL, Diop A, Sene AM, Da Costa A, Arlet G, et al. Carriage of CTX-M-15-producing Escherichia coli isolates among children living in a remote village in Senegal. Antimicrob. Agents Chemother. 2009; 53: 3135- 3137.
- Dia ML, Ngom B, Diagne R, Ka R, Lo S, Cisse M, et al. Molecular detection of CTX-M-15-type β-lactamases in Escherichia coli strains from Senegal. New Microbes and New Infections. 2016; 9: 45-46.
- Diop A, Sambe-Ba B, Seck A, Dia ML, Timbine LG, Niang AA, et al. First Description of the Extended Spectrum-Beta-Lactamase Gene bla CTX-M-109 in Salmonella Grumpensis Strains Isolated from Neonatal Nosocomial Infections in Dakar, Senegal. PLoS One. 2016; 11: e0157683.
- Harrois D, Breurec S, Seck A, Delauné A, Le Hello S, de La Gandara MP, et al. Prevalence and characterization of extended-spectrum β-lactamaseproducing clinical Salmonella enterica isolates in Dakar, Senegal, from 1999 to 2009. Clinical Microbiology and Infection. 2014; 20: O109-O116.
- Baron SA, Mediannikov O, Abdallah R, Kuete Yimagou E, Medkour H, Dubourg G, et al. Multidrug-resistant Klebsiella pneumoniae clones from wild chimpanzees and termites in senegal. Antimicrob. Agents Chemother. 2021; 65: 02557-02520.
- Sarr H, Niang AA, Diop A, Mediannikov O, Zerrouki H, Diene SM, et al. The emergence of carbapenem-and colistin-resistant enterobacteria in Senegal. Pathogens. 2023; 12: 974.
- Breurec S, Bouchiat C, Sire JM, Moquet O, Bercion R, Cisse MF, et al. High third-generation cephalosporin resistant Enterobacteriaceae prevalence rate among neonatal infections in Dakar, Senegal. BMC Infect Dis. 2016; 16: 1-7.
- Diagne R, Diallo AA, Ka R, Ngom B, Lo S, Dia ML. Antimicrobial Resistance and Characterization of Broad-Spectrum Batalactamases and Quinolone Resistance Genes of Urinary E. Coli Isolated in Senegal between 2009 and 2017. J Bacteriol Mycol. 2019; 6: 1106.
- Papa-Ezdra R, Caiata L, Palacio R, Outeda M, Cabezas L, Bálsamo A, et al. Prevalence and molecular characterisation of carbapenemase-producing Enterobacterales in an outbreak-free setting in a single hospital in Uruguay. Journ of Glob Antimicrob Resist. 2021; 24: 58-62.
- Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Aguilar GR, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022.
- Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016; 80: 629-661.
- Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017; 41: 252-275.
- Aher T, Roy A, Kumar P. Molecular detection of virulence genes associated with pathogenicity of Klebsiella spp. isolated from the respiratory tract of apparently healthy as well as sick goats. Israel Journ of Vet Med. 2012; 67: 249-252.
- Hossain S, De Silva BCJ, Dahanayake PS, Heo GJ. Phylogenetic relationships, virulence and antimicrobial resistance properties of Klebsiella sp. isolated from pet turtles in Korea. Lett Appl Microbiol. 2020; 70: 71-78.
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the Natural environment to infectious diseases. Nat Rev Microbiol. 2004; 2: 95-108.
- Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002; 8: 881-890.
- Li B, Zhao Y, Liu C, Chen Z, Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014; 9: 1071-1081.
- Murphy CN, Clegg S. Klebsiella pneumoniae and type 3 fimbriae: nosocomial infection, regulation and biofilm formation. Future Microbiol. 2012; 7: 991- 1002.
- Ndiaye I, Bissoume Sambe BA, Thiam F, Boye MM, Sow O, Cissé A, et al. Antibiotic Resistance and Virulence Factors of Extended-Spectrum Beta- Lactamase-Producing Klebsiella Pneumoniae Involved in Healthcare- Associated Infections in Dakar, Senegal. Archiv Microbiol Immunol. 2023; 7: 65-75.
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012; 18: 268-281.
- Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013; 4: 107- 118.
- Angel Villegas N, Baronetti J, Albesa I, Polifroni R, Parma A, Etcheverría A, et al. Relevance of biofilms in the pathogenesis of Shiga-toxin-producing Escherichia coli infection. The scientific world journal 2013. 2013.
- Andrews S. Babraham bioinformatics-FastQC a quality control tool for high throughput sequence data. URL: 2010.
- Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34: i884-i890.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012; 19: 455-477.
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014; 30: 2068-2069.
- Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023; 51: D690-D699.
- Liu B, Zheng D, Zhou S, Chen L, Yang J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022; 50: D912-D917.
- Carattoli A, Hasman H. PlasmidFinder and in silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Horizontal gene transfer: methods and protocols. 2020; 2075: 285-294.
- Seemann T. ABRicate: mass screening of contigs for antimicrobial and virulence genes. Department of Microbiology and Immunology, The University of Melbourne, Melbourne, Australia. Available online: https://github.com/ tseemann/abricate (accessed on 28 February 2019). 2018.
- Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005; 43: 4178-4182.
- Grant JR, Arantes AS, Stothard P. Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genomics. 2012; 13: 1-8.
- Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010; 11: 1-11.
- Lam MM Wick RR, Judd LM, Holt KE, Wyres KL. Kaptive 2.0: updated capsule and lipopolysaccharide locus typing for the Klebsiella pneumoniae species complex. Microb genom. 2022: 8.
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nature communications. 2018; 9: 5114.
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015; 31: 3691- 3693.
- Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microbial genomics. 2016; 2: e000056.
- Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015; 43: e15-e15.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012; 28: 1647-1649.
- Ares MA, Fernández-Vázquez JL, Rosales-Reyes R, Jarillo-Quijada MD, Von Bargen K, Torres J, et al. H-NS nucleoid protein controls virulence features of Klebsiella pneumoniae by regulating the expression of type 3 pili and the capsule polysaccharide. Frontiers in cellular and infection microbiology. 2016; 6: 13.
- Joram N, de Saint Blanquat L, Stamm D, Launay E, Gras-Le Guen C. Healthcare-associated infection prevention in pediatric intensive care units: a review. Eur J Clin Microbiol Infect Dis. 2012; 31: 2481-2490.
- Gassama-Sow A, Diallo MH, Wane AA, Seck A, Samb-Ba B, Sow PS, et al. Genetic determinants of antibiotic resistance in diarrheagenic Klebsiella pneumoniae subspecies ozaenae: an emerging enteropathogen in Senegal. Clin Infect Dis. 2010; 50: 453-454.
- Jansen KU, Knirsch C, Anderson AS. The role of vaccines in preventing bacterial antimicrobial resistance. Nat Med. 2018; 24: 10-19.
- Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathogens and global health. 2015; 109: 309-318.
- Nirwati H, Sinanjung K, Fahrunissa F, Wijaya F, Napitupulu S, Hati VP, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. In Proceedings of the BMC Proc. 2019; 1-8.
- Moini AS, Soltani B, Ardakani AT, Moravveji A, Erami M, Rezaei MH, et al. Multidrug-resistant Escherichia coli and Klebsiella pneumoniae isolated from patients in Kashan, Iran. Jundishapur J Microbiol. 2015; 8: e27517.
- Mohd Asri NA, Ahmad S, Mohamud R, Mohd Hanafi N, Mohd Zaidi NF, Irekeola AA, et al. Global Prevalence of Nosocomial Multidrug-Resistant Klebsiella pneumoniae: A Systematic Review and Meta-Analysis. Antibiot. 2021; 10: 1508.
- Seck A, Ndiaye B, Diop A, Ndao M, Fall C, Dieng A, et al. Prevalence and Resistance Profile of Muenchen Cefotaximase (CTX-M) Group 1 Extended Spectrum Beta-Lactamase (ESBL)-Producing Uropathogenic Escherichia coli Strains in Dakar, Senegal. Open Journ of Med Microbiol. 2023; 13: 137- 145.
- Dossouvi KM, Ba BS, Lo G, Cissé A, Ba-Diallo A, Ndiaye I, et al. Molecular characterization of extended-spectrum beta-lactamase-producing extraintestinal pathogenic Escherichia coli isolated in a university teaching hospital Dakar-Senegal. BioRxiv. 2022; 2022.2007. 2020.500880.
- Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016; 6: 1-11.
- Moussa B, Hmami F, Arhoun B, El Fakir S, Massik AM, Belchkar S, et al. Intense Intestinal Carriage of Carbapenemase-Producing Klebsiella pneumoniae Co-harboring OXA-48, KPC, VIM, and NDM Among Preterm Neonates in a Moroccan Neonatal Intensive Care Unit. Cureus. 2023: 15.
- Doménech-Sánchez A, Hernández-Allés S, Martinez-Martinez L, Benedí VJ, Albertí S. Identification and characterization of a new porin gene of Klebsiella pneumoniae: its role in β-lactam antibiotic resistance. J. Bacteriol. 1999; 181: 2726-2732.
- Fonseca éL, Morgado SM, Freitas FS, Bighi NS, Cipriano R, Vicente ACP. Genomic characterization of a pandrug-resistant Klebsiella pneumoniae belonging to the high-risk ST11 in the Brazilian Amazon region. bioRxiv 2023, 2023.2004. 2025.538267.
- Ayobami O, Brinkwirth S, Eckmanns T, Markwart R. Antibiotic resistance in hospital-acquired ESKAPE-E infections in low-and lower-middle-income countries: a systematic review and meta-analysis. Emerg microb & infect. 2022; 11: 443-451.
- Swedan SF, Aldakhily DB. Antimicrobial resistance, biofilm formation, and molecular detection of efflux pump and biofilm genes among Klebsiella pneumoniae clinical isolates from Northern Jordan. Heliyon. 2024; 10: e34370.
- Gual-de-Torrella A, Delgado-Valverde M, Pérez-Palacios P, Oteo-Iglesias J, Rojo-Molinero E, Macià MD, et al. Prevalence of the fimbrial operon mrkABCD, mrkA expression, biofilm formation and effect of biocides on biofilm formation in carbapenemase-producing Klebsiella pneumoniae isolates belonging or not belonging to high-risk clones. Int J Antimicrob Agents. 2022; 60: 106663.
- Pourmohammad-Hosseini G, Ghandehari F, Hoveida L. The abundance of capsule (wabG) and fimbria (fimH) coding genes in multidrug-resistant Klebsiella pneumoniae strains isolated from patients admitted to Isfahan hospitals. Microbiology, Metabolites and Biotechnology. 2023; 6: 27-34.
- O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. An Rev in Microbiol. 2000; 54: 49-79.
- Seifi K, Kazemian H, Heidari H, Rezagholizadeh F, Saee Y, Shirvani F, Houri H. Evaluation of biofilm formation among Klebsiella pneumoniae isolates and molecular characterization by ERIC-PCR. Jundishapur J Microbiol. 2016: 9.
- Cepas V, López Y, Muñoz E, Rolo D, Ardanuy C, Martí S, et al. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microb. Drug Resist. 2019; 25: 72-79.
- Struve C, Bojer M, Krogfelt KA. Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect and im. 2009; 77: 5016-5024.
- Schroll C, Barken KB, Krogfelt KA, Struve C. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 2010; 10: 1-10.
- Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. Type 1 pilusmediated bacterial invasion of bladder epithelial cells. The EMBO journal. 2000.
- Langstraat J, Bohse M, Clegg S. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infection and immunity. 2001; 69: 5805-5812.
- Drago L, Mattina R, Vecchi Ed, Toscano M. Phenotypic and genotypic antibiotic resistance in some probiotics proposed for medical use. Inter Journ of Antimicrob Agents. 2013.
- Vuotto C, Longo F, Pascolini C, Donelli G, Balice MP, Libori MF, et al. Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J Appl Microbiol. 2017; 123: 1003-1018.
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004; 2: 95-108.
- Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 2010; 54: 177-183.
- Ruzin A, Keeney D, Bradford PA, AcrAB efflux pump plays a role in decreased susceptibility to tigecycline in Morganella morganii. Antimicrob. Agents Chemother. 2005; 49: 791-793.
- Rodríguez-Martínez JM, Díaz de Alba P, Briales A, Machuca J, Lossa M, Fernández-Cuenca F, et al. Contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-β-lactamase-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2013; 68: 68-73.
- Li J, Zhang H, Ning J, Sajid A, Cheng G, Yuan Z, et al. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob Resist & Infect Contr. 2019; 8: 1-13.
- Li X, Ma W, Qin Q, Liu S, Ye L, Yang J, et al. Nosocomial spread of OXA- 232-producing Klebsiella pneumoniae ST15 in a teaching hospital, Shanghai, China. BMC Microbiol. 2019; 19: 1-6.
- Wong MHY, Chan EWC, Chen S. Evolution and dissemination of OqxABlike efflux pumps, an emerging quinolone resistance determinant among members of Enterobacteriaceae. Antimicrob. Agents Chemother. 2015; 59: 3290-3297.
- El Fertas-Aissani R, Messai Y, Alouache S, Bakour R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol Biol. 2013; 61: 209-216.
- Peralta DR, Adler C, Corbalán NS, Paz García EC, Pomares MF, Vincent PA. Enterobactin as part of the oxidative stress response repertoire. PLoS One. 2016; 11: e0157799.
- Kessler C. Characterising AMR Plasmid Transmission in the Gut Microbiome. University of Birmingham. 2021.
- Paskova V, Medvecky M, Skalova A, Chudejova K, Bitar I, Jakubu V, et al. Characterization of NDM-encoding plasmids from Enterobacteriaceae recovered from Czech hospitals. Front Microbiol. 2018; 9: 1549.
- Ferjani S, Saidani M, Maamar E, Harbaoui S, Hamzaoui Z, Hosni H, et al. Escherichia coli colonizing healthy children in Tunisia: High prevalence of extra-intestinal pathovar and occurrence of non-extended-spectrum-β- lactamase-producing ST131 clone. Int J Antimicrob Agents. 2018; 52: 878- 885.
- Hu Y, Yang Y, Feng Y, Fang Q, Wang C, Zhao F, et al. Prevalence and clonal diversity of carbapenem-resistant Klebsiella pneumoniae causing neonatal infections: a systematic review of 128 articles across 30 countries. PLoS Med. 2023; 20: e1004233.
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci. 2015; 112: E3574-E3581.
- Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, et al. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015; 43: 6761-6771.
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nature com. 2018; 9: 5114.
- Azwai SM, Lawila AF, Eshamah HL, Sherif JA, Farag SA, Naas HT, et al. Antimicrobial susceptibility profile of Klebsiella pneumoniae isolated from some dairy products in Libya as a foodborne pathogen. Veterinary World. 2024: 17.
- Chaalal N, Touati A, Bakour S, Aissa MA, Sotto A, Lavigne JP, et al. Spread of OXA-48 and NDM-1-producing Klebsiella pneumoniae ST48 and ST101 in chicken meat in Western Algeria. Microb. Drug Resist. 2021; 27: 492-500.
- Rajput A, Chauhan SM, Mohite OS, Hyun JC, Ardalani O, Jahn LJ, et al. Pangenome analysis reveals the genetic basis for taxonomic classification of the Lactobacillaceae family. Food Microbiol. 2023; 115: 104334.