
Special Article – Animal Brucellosis
J Bacteriol Mycol. 2017; 4(2): 1049.
Diversity of Phage-Host Specificity in Brucella Phage
Antadze I¹, Dadunashvili M¹, Burbutashvili T¹, Gunia S¹, Balarjishvili N¹, Tevdoradze E¹, Pataridze T¹, Obiso RJ², Hagius S³, Elzer P³ and Kutateladze M¹*
¹George Eliava Institute of Bacteriophages, Microbiology and Virology, Tbilisi, Georgia
²Avila Scientific, LLC, Christiansburg, Virginia, USA
³Louisiana Agricultural Experiment Station, Baton Rouge, Louisiana, USA
*Corresponding author: Mzia Kutateladze, George Eliava Institute of Bacteriophages, Microbiology and Virology, Tbilisi, Georgia
Received: March 24, 2017; Accepted: April 27, 2017; Published: May 04, 2017
Abstract
Bacteriophage typing of Brucella is accepted as an additional tool that can be used for the identification of bacterial species by the World Health Organization’s Expert Committee of Brucellosis. Phage typing is based on the host specificity of bacteriophages. Few phage for bacterial species are currently validated for typing purposes. Tbilisi (Tb) is a unique phage isolated in the 1950s at the Eliava Institute of Bacteriophages, Microbiology, and Virology in Tbilisi, Georgia. This paper describes some historical studies of Brucella phage performed at the Eliava Institute, as well as a comparative characterization of several novel Brucella bacteriophages from the Institute’s collection. Phage– host specificity was also examined using DNA restriction and is demonstrated by the efficient plating of bacteriophages grown on different Brucella hosts. The lytic reactions of the Brucella phage used in this study confirm the data that was also obtained by serological and molecular genotyping methods. Studies of the mechanisms of phage-host specificity support the use of phage typing schemes for the identification of bacterial strains of Brucella.
Keywords: Bacteriophage; Host-specificity; Efficiency of plating
Introduction
Brucellosis – a zoonotic disease and potential biological weapon - is caused by the bacterial genus of Brucella. The bacteria are transmitted from animals to humans by ingestion of infected food products, direct contact with an infected animal, or inhalation of aerosols. This disease continues to be a major public health concern worldwide and is a very common zoonotic infection throughout the world. Definitive diagnosis of this disease is based on culture (identification of the pathogen), serology, and molecular biological methods. Serological methods traditionally have been used to speciate Brucella isolates. Serotyping results should ideally be confirmed by molecular genotyping, a variety of which are available for this purpose: polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) [1]; cytoplasmic protein-specific gene probe analysis [2]; Multiple Locus Variable Number Tandem Repeat Analysis (MLVA) [3,4]; or typing with the rpoB gene coding the DNA-dependent RNA polymerase (RNAP) ß subunit [5,6]. In addition to these molecular methods, phage typing is used as an additional confirmatory tool for the identification of Brucella species.
Phage typing using species-specific bacteriophages has been used to differentiate Brucella species for many years [7]; the phage Tb, Iz, Wb, Bk, and S708 are those most often used for typing purposes [7]. Despite their utility, the biology of these bacterial viruses is not well understood, particularly with respect to their interactions with host bacterial cells. Brucella phage were first isolated at the Tbilisi Institute of Vaccines and Sera (the prior name of the Eliava Institute of Bacteriophages, Microbiology, and Virology) in Georgia by Nemsadze, Popkhadze, and Kilasonidze in 1952 [8] and used to evaluate the activity of phage filtrates on museum cultures and freshly isolated cultures of Brucella on solid media.
Improved methods for the isolation and target-specific reinforcement of phage, together with parallel research on the antigenic structure of Brucella, led to the isolation of a considerable number of phage from various sources from the 1950s through the 1970s [9-14]. Twenty-three Brucella bacteriophages, which were stable at high concentrations [titer 10-4 – 10 by the method developed by Appelmans [15]] were isolated between 1955 and 1962 at the Eliava Institute. Seven of these phage were isolated from the environment, 15 from the blood of human brucellosis patients, and one from a person vaccinated with a live Brucella vaccine [16,17]. All 23 phage were specific to B. abortus, despite the detection of B. melitensis in the blood of three of the brucellosis patients from whom the phage were isolated [16].
Brucella phage typing has a long history [18-23]. The stable Brucella phage named Tb, was first isolated from manure in 1955 at the Tbilisi Institute of Vaccines and Sera [23]. Subsequently, this phage was approved by the International Subcommittee on the Taxonomy of Brucella as a reference phage for the diagnostics and differentiation of Brucella strains [25]. Tb phage have been studied and used for typing purposes by many scientists [25-28]. One series of experiments showed that Tb phage are specific to B. abortus; plaques were not observed on lawns of B. suis, although phage at high concentrations (104 x routine test dilution or greater) did cause inhibition of growth that resembled lysis. High concentrations of Tb phage have also been shown to inhibit the growth of B. melitensis [29].
Phage typing has been used to confirm Brucella species. In two previous studies, 543 Brucella strains from different countries (former Soviet Union, United Kingdom, Poland, Germany, and South Africa) were identified [23,29]. Preliminary speciation was carried out using standard bacteriological and biochemical tests. The results of these studies indicated that 277 strains were B. melitensis, 62 strains were B. abortus, and 204 strains were B. suis. The results of the phage susceptibility testing demonstrated that 177 of the 277 B. melitensis strains in the Eliava Institute collection belonged to a single group according to the phage typing scheme described by Morgan [25]; a total of 99 non-Eliava strains also belonged to the same group. Seventy-three strains that were lysed by both dilutions of phage were identified as the B. abortus biotype V. All but five B. abortus strains were lysed by Tb phage; further investigation indicated that these five strains already contained temperate phage and were not lysed. Among the B. suis strains, 40 out of 204 were lysed by phage.
Currently, the Eliava Institute collection includes 44 Brucellaspecific bacteriophages, including the Tb phage; one phage isolated from B. canis; four phage isolated from B. ovis; 19 phage isolated from B. melitensis; 17 phage isolated from B. abortus; and two phage isolated from B. suis. All of the phage in the collection are specific; the phage isolated from B. abortus and B. melitensis cause lysis only in B. abortus and a limited number of B. melitensis strains. Phages isolated from B. ovis and B. canis fully lyse B. abortus and partially lyse B. melitensis but do not lyse B. ovis, B. suis, or B. canis. The current paper describes features of a set of 10 bacteriophages that have been selected for typing purposes for various Brucella species. The bacteriophages outlined in this paper were characterized, including an analysis of their reproduction parameters and lytic specificity against various species of Brucella.
Materials and Methods
Some of the bacterial strains of Brucella used in the study were from the Eliava Institute’s bacterial collection: B. abortus S19 vaccine strain; B. abortus 141, serotype I, originally isolated in Russia; B. abortus 544 serotype I; B. abortus 99 serotype V, was originally from the UK Weybridge collection; and B. abortus 64 serotype III, was isolated in Tbilisi. Among the B. melitensis strains in the Eliava museum, N7 was obtained from Saratov, Russia in 1963; and N16 was isolated in Moscow in 1962. The strain N110 was isolated from human synovial fluid in Tbilisi in 1942; N 237 and 238 were isolated from the blood of a brucellosis patient in Tbilisi in 1959. The strains N 71 m/z and 70 v/z were originally isolated from a patient in Bulgaria; 130 m/z was isolated from a patient in Germany; and 238 m/z and 254 m/z were originally isolated from a cow in England. Strain 63/9 was received from Almaty, Kazakhstan in 1979. Additional bacteriophages typing was conducted using bacterial strains that reside at the Louisiana State University (LSU) Ag Center and were as follows: B. inopinata BO1 from a breast cancer implant in a brucellosis patient [30]; B. inopinata BO2 from the lung biopsy of a patient with chronic pulmonary destructive pneumonia [31]; Brucella strain NF2653 from wild native rodents in Australia [32]; and SDRL an atypical B. abortus strain from a rat liver sample from San Diego, California [33]. The following Brucella phage were used for this study: Tb, originally isolated from manure; phage 1066 from B. canis; 281; 02 from B. ovis; 177; 110 from B. melitensis; V; 544; 141 from B. abortus; and 11sa from B. suis. All bacteriophages were propagated on two bacterial strains: B. abortus S19 and B. abortus 141 strains.
Phage spot test
The phage spot test is a common tool to determine phage host specificity. The host range of each phage (phage specificity) was determined by spotting 10 µL of a phage suspension (~109 PFU/mL) in nutrient broth onto freshly prepared bacterial lawns and counting the plaques that appeared after 24 and 48 hours of incubation at 37°C. Visual characteristics of the phage plaques (PFU/mL) were also evaluated. The Routine Test Dilution (RTD) is defined as the highest dilution of the phage stock that will produce confluent lysis of a lawn inoculum of the propagating strain. The results of the phage spot test were determined from three sets of experiments after each of the 24 hour and 48 hour time points.
Electron microscopy
The morphology of the phage particles was studied using an electron microscope JEM x100 (JEOL). Parlodion plates were overlaid with 1010 PFU/mL phage suspensions with uranyl acetate as a contrast agent.
Phage structural proteins
Phage suspensions (60 µL at 1010 to 1011 PFU/mL) were added to 13 µL of sample buffer containing 8% sodium dodecyl sulfate, 0.1% glycerol, and 0.5 % bromophenol blue; 5.0% ß-mercaptoethanol was added to the solution prior to use. Samples were boiled for ten minutes and then loaded onto 10% polyacrylamide gels [34]. Electrophoresis was carried out at 60 V, 9 mA for 18 hours in tris-glycine buffer.
Phage biology study
Biological properties, mainly phage-host interaction parameters including adsorption, latent period, lysis time, and average burst size, were calculated by standard methodology [35].
DNA isolation and restriction
Phage DNA was isolated by standard phenol/chloroform deproteinization [36] and with QIAamp DNA mini kits (Qiagen). Several restriction endonucleases were used according to manufacturer’s instructions (Biolab). Enzyme-restricted DNA fragments were subjected to electrophoresis on agarose gels. The gels were photographed with ultraviolet illumination.
Results and Discussion
For phage typing, ten bacteriophages (Tb, 141, 281, 544, 1066, 11sa, 02, 177, V, and 110) from the Eliava collection were selected based on the specificity of their lytic reaction on different Brucella species.
Morphology of phage
The size and shape of phage plaques varies considerably among Brucella phage [37-39]. In general, plaques range from 0.2 mm to 4 mm and are polymorphic on bacterial lawns; some of them are oval or perfectly round while others are more irregular. All Brucella phage from the Georgian collection, including the Tb phage, are morphologically identical and similar to other phage described to date [39-41]. They all have icosahedral heads (60-65 nm x 60-70 nm), short tails (14-20 nm; Figure 1), and belong to the Podoviridae family. Only minor differences are visible in the composition of structural proteins of Brucella phage
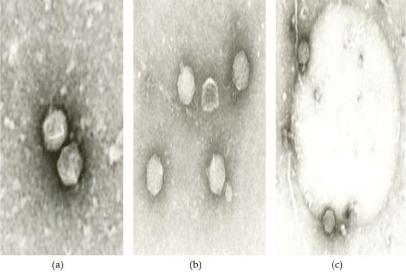
Figure 1: Electron micrographs of Brucella phage particles; (a) Phage 260
(magnification x 120,000); (b) phage X (magnification x 100,000), (c) Tb
phage adsorbed on Brucella cell (magnification x 96,000).
Phage reproduction parameters
The adsorption time of Brucella phages differ by host bacterial strain (B. abortus 141). Some phage (281, 544) adsorb on the host in less than two hours, while other phage require more time for adsorption (Table 1). Other parameters of reproduction on host bacterial cells for all the phage were prolonged too. The average burst size was comparatively low compared to other phage; the highest phage counts were recorded for phage 1066 (120-125 PFU/mL).
Phage
Isolation source
Time of adsorption (min)
% of adsorption
Latent period (min)
Lysis time (min)
Average burst size
(PFU/mL)
Tb
liquid manure
190
90
200
460
40-46
Tb*
liquid manure
180
81
210
460
50-53
110
B. melitensis
120
70
180-220
440
18-20
141
B. abortus
120
76
210-240
450
27-30
11sa
B. suis
120
53
180
410
28-30
1066
B. canis
150
98
180
425
110-120
02
B. ovis
120
82
240
400
36-40
544
B. abortus
90-100
82
270-300
480-490
30-35
281
B. ovis
90-100
70
250-280
450-460
60-65
177
B. melitensis
180
78
240
460
40-45
V
B. abortus
180
37
240
480
25-30
Table 1: Phage reproduction parameters on host bacteria.
Comparative DNA restriction
DNA restriction analysis of Brucella phage genomes show that they only differ slightly from each other, indicating that they are highly conserved. DNA restriction results observed in this study correlate well with the results presented by Rigby et al. [39]; the six Brucella phage (including Tb) targeted in this study could not be differentiated by restriction digestion profiles produced by BglI, EcoRI, HindIII, or PvuII. Nepean phage (Np) DNA differed from Tb phage with restriction by PvuII, and BglII but only by one band. The authors explained such a difference in possible self-ligating cos ends in submolar fragments of phage, although further procedures to prevent self-ligation did not demonstrate the existence of cos sequences. A comparison of the phage isolated from Georgia to the Np phage (isolated from an atypical B. abortus strain from a cow in Ontario, Canada [39] by DNA HindIII restriction showed that most of the phage from the Eliava Institute collection differ from both the Tb and the Np phage (Table 2). However, comparative restriction digestion of DNA from phage cultivated on various Brucella hosts indicated substantial differences. DNA from Tb phage cultivated on the vaccine strain B. abortus S19 was resistant to EcoRI digestion, while DNA from Tb phage grown on B. abortus 141 was susceptible to this enzyme. DNA restriction of phage 02 by EcoRV endonuclease also showed some differences: EcoRV digested DNA from phage 02 and from phage 1066 propagated on B. abortus S19, but EcoRV digestion did not cleave DNA extracted from phage 02 and from phage 1066 when they were cultivated on B. abortus 141. Further, KpnI did not digest 1066 phage DNA. The restriction digestion profiles by PvuII were identical for all phage. To examine these different DNA restriction patterns from phage propagated on different host bacteria, Tb phage that was initially cultivated on B. abortus 141 was propagated on B. abortus 19 and then B. abortus 141. DNA from all three phage preparations was then subjected to restriction analysis (Figure 2).
Similarity to Np phage
Bacteriophages
Similar DNA restriction
81, 2, 3, 5, 6, 27, 64, 43, 90, VII, VIII, XII, 12(B), 63, 110
Difference in restriction
Tb, 290, 100, BA, 248, 214, 19, 7sa, 141, 109, IV, OX, X, 273, 544, 281, 02, 1066, 177, VI, 11sa
Resistant to Hind III restriction
9, 147, 224, 239, 271
Table 2: Similarity in Hind III restriction of Georgian bacteriophages and Np bacteriophage.
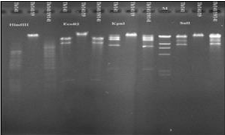
Figure 2: DNA restriction digestions of Tb phage cultivated on different host
bacteria.
Restriction endonuclease KpnI did not digest the DNA from Tb phage propagated on B. abortus 19; the restriction profile of KpnI digestion from Tb phage DNA propagated on B. abortus 141, and the profile of Tb phage DNA propagated on B. abortus 19 and then on B. abortus 141 were similar except for two extra high molecular weight bands. Slight differences were observed in structural proteins of Tb phage cultivated on different hosts (Figure 3).
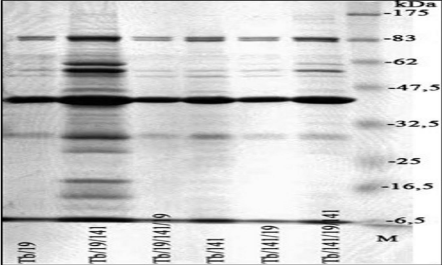
Figure 3: SDS-PAGE analysis of structural proteins of Tb phage propagated
on different host bacteria.
Efficiency of plating of phage propagated on different host bacteria
High Efficiency of Plating (EOP) of Tb phage propagated on various B. abortus strains has been demonstrated in several different studies [38-41]. In our investigations, the host specificity of Tb phage was evident in its EOP when grown on different host bacterial strains. A value more than 1 indicates a high infectivity of phage to the host bacteria. Phages were grown on one bacterial strain, then grown on a different bacterial strain, and then grown once more on the first bacterial strain. In general, the phages grown on different hosts revealed a high EOP on different bacterial strains, but B. abortus 141 might be considered the best host for the phages used in this study (Table 3).
Phages cultivated on
B. abortus strains
Bacterial strain of B. abortus
141
544
99
64
?b/B. abortus141
1
0.9
0.9
0.9
?b/ B.abortus141/B.a 99
1
0.9
1
0.7
?b/B.a.141/B.a.99/B.a 141
1
1
0.9
0.8
?b/B.a 141/B.a 64
1.2
0.9
1
1
?b/B.a. 141/B.a 64/B.a 141
1
1.1
0.9
0.6
?b/B.a. 99
1.1
0.9
1
1.3
?b/ B.a. 99/ B.a. 141
1
1.2
1.3
1.3
?b/ B.a. 99/ B.a. 141/ B.a. 99
0.9
1
1
0.9
?b/ B.a. 99/ B.a. 64
1.2
1.1
1
1
??/ B.a. 99/ B.a. 64/ B.a. 99
1
0.9
1
1
?b/ B.a. 64
1.3
1
1.4
1
?b/ B.a. 64/ B.a. 141
1
1.1
0.7
0.5
?b/ B.a. 64/ B.a. 141/ B.a. 64
1.5
1.3
1.3
1
??/ B.a. 64/ B.a. 99
1.2
1.3
1
1
??/ B.a. 64/ B.a. 99/ B.a. 64
1.1
1.1
1
1
?b/ B.a. 544
1.3
1
1.2
1
?b/ B.a. 544/ B.a. 99
1.1
1
1
1
?b / B.a. 544/ B.a. 99/ B.a. 544
0.8
1
1
1.1
?b/ B.a.544/ B.a. 64
2.9
2.3
2
1
?b/ B.a. 544/ B.a. 64/ B.a. 544
1.3
1
1.2
1
Table 3: Efficiency of plating of Tb phage on different host bacterial strains.
Lytic activity
Phage are used for typing at two concentrations, RTD and 104 x RTD, as recommended by the Subcommittee on the Taxonomy of Brucella. All smooth B. abortus strains are lysed by Tb phage at both concentrations. The set of ten phages used in this study were then used for typing 11 archival stains of B. melitensis. B. melitensis strains are generally resistant to Tb phage because these strains are smooth, although at high concentrations Tb phage may cause lysis. All smooth B. suis strains are resistant to Tb phage at RTD and are lysed at 104 x RTD. In this study, different bacteriophages at a high titer occasionally lysed the 11 archival strains of B. melitensis without infecting the organism (the phenomenon called “lysis from without” or soluble phage lysis effects) [42]. Semi-confluent lysis of strain N7 by phage Tb, 11sa, V, 177, 281, and 544 was observed; in addition the lysis of strain 16 by phage Tb, V, 110, 141, 177, 281, and 544. Bacterial strains 237, 238, 238 m/z, and 254 m/z were lysed at both dilutions and were therefore classified as B. abortus serotype V. Only one strain (63/9) was lysed by phage 1066 at both dilutions used in these experiments. Phage typing of the Brucella strains from the LSU AgCenter collection using ten phage cultivated on two hosts (B. abortus S19 and B. abortus 141) showed that B. canis and B. ovis are resistant to lysis by phage. The 10 phage used in this study also did not reveal any reaction against B. inopinata isolated from a patient with chronic lung infections; however, weak activity was observed against another strain of B. inopinata (Table 4). Several phages caused lysis of B. suis, though only in high concentrations that could be connected to lysis without infection, while B. neotomae was lysed by both concentrations of phage. In some cases, the susceptibility to a given phage varied depending on which host strains were used for cultivation. Phage 544 propagated on B. abortus 141 showed lytic reaction on strains B. abortus RB51 (with EOP 4x10-6) and non-typical B. abortus SDRL at both phage concentrations, while the same phage grown on B. abortus S19 did not lyse either of those strains. Phage 544 propagated on B. abortus 141 demonstrated low EOP on the phagesensitive B. abortus RB51 strain. The same phage grown on B. abortus S19 revealed a high efficiency on sensitive strain Brucella F2653 but low efficiency on B. neotomae (Table 5). Phage 544 cultivated on B. abortus 141 did not lyse the B. inopinata BO1 strain. The same phage grown on B. abortus S19 showed semi-confluent lysis at RTD and no reaction at higher dilutions, which would be indicative of lysis without infection.
Bacteriophages cultivated on different B. abortus strains
RTD
Bacterial strains
B. abortus 19
B. abortus 19
(Shreveport)
B. abortus RB51
B. inopinata BO1
Brucella species NF2653
B. neotomae
B. abortus SDRL
TB-19/TB-141
3 RTD
CL/CL
CL/CL
R/R
OL/OL
CL/CL
CL/CL
R/R
RTD
CL/CL
CL/CL
R/R
R/R
CL/CL
CL/CL
R/R
02-19/02-141
3RTD
CL/CL
CL/CL
R/R
R/R
CL/CL
CL/CL
R/R
RTD
SCL/CL
CL/CL
R/R
R/R
SCL/SCL
CL/CL
R/R
1066-19/
1066-141
3 RTD
CL/CL
CL/CL
R/R
R/R
CL/CL
CL/CL
R/R
RTD
CL/CL
CL/CL
R/R
R/R
SCL/SCL
CL/CL
R/R
281-19/281-141
3 RTD
CL/CL
CL/CL
R/R
R/R
CL/CL
CL/CL
R/R
RTD
CL/CL
CL/CL
R/R
R/R
CL/CL
CL/CL
R/R
177-19/177-141
3 RTD
CL/CL
CL/CL
R/R
OL/OL
CL/CL
CL/CL
R/R
RTD
CL/CL
CL/CL
R/R
R/R
SCL/SCL
SCL/CL
R/R
544-19/544-141
3 RTD
CL/CL
CL/CL
R/SCL
SCL/R
CL/CL
CL/CL
R/SCL
RTD
CL/CL
CL/CL
R/IPO4
R/R
CL/SCL
IPO100/SCL
R/IPO1
11sa-19/
11sa-141
3 RTD
CL/CL
CL/CL
R/R
OL/OL
CL/SCL
CL/CL
R/R
RTD
CL/CL
CL/CL
R/R
R/R
SCL/SCL
SCL/SCL
R/R
V-19/V-141
3 RTD
CL/CL
CL/CL
R/R
R/R
CL/CL
CL/CL
R/R
RTD
CL/CL
CL/CL
R/R
R/R
CL/SCL
R/R
R/R
141-19/141-141
3 RTD
CL/CL
CL/CL
R/R
OL/OL
CL/CL
CL/CL
R/R
RTD
CL/CL
CL/CL
R/R
R/R
CL/CL
SCL/IPO100
R/R
110-19/110-141
3 RTD
CL/CL
CL/CL
R/R
OL/OL
CL/CL
CL/CL
R/R
RTD
CL/CL
CL/CL
R/R
R/R
CL/CL
SCL/IPO500
R/R
Note: Bacteriophages were grown on two hosts: B. abortus S19 and B. abortus 141. 1* = phage at 1RTD; 2* = phage at 4 RTD; CL= Confluent Lysis; SCL = Semi-Confluent Lysis; OL = Opaque Lysis; IPO = Individual Plaques Opaque; R = Resistant.
Table 4: Lysis of LSU AgCenter Brucella strains by bacteriophages.
Bacterial strains
544 propagated on B. abortus 141
(titer 4.4x109 PFU/mL)
544 propagated on B. abortus S19
(titer 1.2x1010 PFU/mL)
Titer
EOP
Titer
EOP
B. abortus RB51
2x104
4x10-6
-
-
Brucella species NF2653
6.9x109
0.57
B. neotomae
2x10 7
1x10-4
B. inopinata BO1
complete lysis in initial dilution
-
Table 5: Efficiency of plating of phage 544 with different hosts on different bacterial strains.
Discussion
Research of Brucella bacteriophages has a long history, although the mechanisms of virus interaction with the host bacteria Brucella – a slow-growing organism - remains unknown. Further, there is a wealth of Brucella phage work in older scientific publications from the former Soviet Union that is still not available to the scientific society. In this paper, we tried to present some limited information about the Brucella phage studies from the old Soviet periodicals. Brucella bacteriophages are used to type various Brucella species; and when taken together with other identification methods, phage typing is a useful tool for such purposes. A set of typing phages and phage-typing schemes has existed for many years, but this study is an attempt to include more phages into typing schemes, providing a better understanding of phage biology and bacterial phage-host interactions.
Based on the historic scientific literature, phages against Brucella species are relatively homogenic. In this study, ten phages were selected for typing purposes. These phages exhibited differences in phage plaques on solid media, which was generally a characteristic feature for Brucella phages. All the phage particles have an icosahedral head and short tail. The reproduction cycle of these phages of these slowly growing bacteria is also very slow. Reproduction parameters for Tb phage were previously studied by Antadze and Popkhadze [38]. Each of the phage analyzed in this study demonstrated a slow growth cycle on host bacterial cells.
Phage host bacteria cell interaction mechanisms are not well determined for Brucella phages. Efficiency of plating, which is derived from the ratio of the phage titer on the target strains to the phage titer on the host strains, was examined for phages cultivated on different hosts. Phages were grown on one strain, then grown on a different strain, and then grown once more on the first strain. An EOP value greater than 1 indicates a high efficiency of cultivation. The efficiency of cultivation of Tb grown on different hosts is generally high, but Tb cultivated on B. abortus strain 64 and then B. abortus strain 544 showed higher EOP than that on other hosts. The EOP of bacteriophages cultivated on B. abortus strain S19 followed by B. abortus strain 141 was greater than 1, indicating that phage have high efficiency on B. abortus 141.
total of 11 archival bacterial strains of B. melitensis from the Eliava Institute were typed by ten bacteriophages in this study. The data show that four bacterial strains out of 11 were lysed by both dilutions (RTD and 4TD) of phages, therefore they were identified as B. abortus serotype V. Lytic reactions against B. melitensis were caused by the phenomenon characteristic of some temperate phages.
Brucella species are very similar [46]; the DNA restriction maps of different Brucella strains show that a 640-kb insertion was found in the small chromosome of B. abortus 544 biovar 1 [47]. Division of the genus into species and biovars is based solely upon a small number of metabolic traits, two lipopolysaccharide epitopes, and sensitivity to a set of phage that are host-range variants of the same ancestor [48]. Although phage susceptibility to different species is evident, it indicates the significance of phage typing as a tool for Brucella species differentiation. The sequence of the omp2 locus in different Brucella species supports the existence of species-specific lineages. A close relationship has been found between B. melitensis and B. abortus, while an extreme divergence of B. ovis and B. neotomae from the other species was observed. This divergence may be the result of a gene conversion for this locus in these two species [48]. In this study, we found that B. neotomae strains are only partially susceptible to Tb phage, producing a few isolated plaques at RTD. They are completely lysed by the phage at 104 x RTD. B. ovis and B. canis strains are invariably resistant to the phage at both dilutions.
It has been shown that cultivating bacteriophages through several rounds on different hosts will increase phage lytic activity; this is known as a way to adapt phage on various hosts. Even after adaptation, phages preserve their specificity against various species of Brucella. The phages in the study do not show any activity against B. canis and B. ovis; however, several phages caused lysis of B. suis, though only in high concentrations, which may be connected to the phenomenon of lysis without infection, while B. neotomae was lysed by both concentrations of phage.
Host specificity plays an important role in the biology of Brucella phage. DNA restriction of different bacteriophages against Brucella shows homogeneity of these bacterial viruses. Evident differences are shown in susceptibility to the restriction enzymes of phages grown on different hosts. Differences in DNA restriction patterns of various phage propagated on different hosts might be explained by the temperate nature of Brucella phage, particularly by possible horizontal gene transfer between host bacteria facilitated by bacteriophages. Since most Brucella bacteriophages are isolated from bacterial strains, there is a high probability of detecting phage genome fragments in the host. However, this hypothesis does not align with the results from previous experiments by Rigby et al. Southern blot analysis of 32P-labelled Tb DNA did not hybridize to any fragment of HindIIIor EcoRI-digested chromosomal DNA of B. abortus S19 or any other Brucella species [39].
Brucella phage described in different scientific publications are members of the same family Podoviridae. These phages have the same morphology (icosahedral head and short tail) and have similar DNA restriction profiles with only some minor differences (the extra bands visible in some patterns may be due to repetitions or shared base sequences that affect the locations of endonuclease recognition sites). Phage Tb produces divergent PCR profiles with various arbitrary primers compared to host strain B. abortus S19 fragments. The complete genome of this unique phage is already sequenced [44]; genome analysis will explain all functional peculiarities connected to phage-host interaction. The most notable differences among the Brucella phage reside in their host ranges and phage-specific lytic reactions to different bacterial strains [43]. In a previous study by Tevdoradze [49], the full genomic sequences of several of the bacteriophages used in this study were reported, revealing pronounced sequence homogeneity. Interestingly, finescale genetic variability of these phages grown on multiple hosts within a single Brucella species remains unknown; and the genomic changes, as a result of passaging, were observed in similar genes and predominantly occurred at identical sites in separate phages. This 2015 study also showed that there were multiple instances of ‘withinsample’ genetic heterogeneity observed often at shared genomics positions across phages. Our study supports the previous work and shows that bacteriophages propagated on two distinct host bacterial strains illustrates multiple common sequence variations which frequently display within-sample genetic heterogeneity [49].
The phage-host bacterial interaction study clearly demonstrated in Brucella-specific bacteriophages is an interesting issue of phage biology. Over the course of their evolution, bacteriophages have developed unique responses to their hosts; although the biological features of temperate phage are generally dependent on host bacteria. Using phage typing as an additional tool for the differentiation of Brucella species is still a reliable methodology and has been validated by scientists and medical workers worldwide. Identification of Brucella species based on phage-specific lytic reactions can be used to confirm the results obtained by serological and molecular genotyping methods. The mechanisms of host specificity of phage, however, are not completely clear; and further investigation of phage-bacterial genomics is needed to elucidate Brucella phage biology and explain the unique interactions that have been observed between these bacteria and their viruses.
Acknowledgments
This research was made possible by financial support provided by the U.S. Department of Defense (DoD), Defense Threat Reduction Agency (DTRA), through the Cooperative Biological Engagement Program’s (CBEP) Cooperative Biological Research Project (CBR) GG-17, in Georgia. The findings, opinions and views expressed herein belong to the authors and do not reflect an official position of any organization listed. The authors are grateful to the US Centers for Disease Control and Prevention for supplying the atypical Brucella strains to the LSU Ag Center. The authors also thank Attimo Research and Development, and Sonya Narodny for editorial assistance.
Author Contributions
R.O. and M.K. conceived and designed the experiments; I.A., M.D., T.B., S.G., N.B., E.T., T.P., and M.K. performed the experiments; I.A., M.D., T.B., S.G., N.B., E.T., T.P., R.O., S.H., P.E., and M.K. analyzed the data; S.H., P.E., and M.K. contributed reagents/materials/analysis tools; R.O., S.H., P.E., and M.K. wrote the paper.”
Conflicts of Interest
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Al Dahouk S, Tomaso H, Prenger-Berninghoff E, Splettstoesser WD, Scholz HC, Neubauer H. Identification of Brucella species and biotypes using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP). Critical reviews in microbiology. 2005; 31: 191-196.
- Verger JM, Grayon M, Tibor A, Wansard V, Letesson JJ, Cloeckaert A. Differentiation of Brucella melitensis, B. ovis and B. suis biovar 2 strains by use of membrane protein- or cytoplasmic protein-specific gene probes. Research in microbiology. 1998; 149: 509-517.
- Bricker BJ, Ewalt DR, Halling SM. Brucella 'HOOF-Prints': strain typing by multi-locus analysis of Variable Number Tandem Repeats (VNTRs). BMC microbiology. 2003; 3: 15.
- Le Fleche P, Jacques I, Grayon M, Al Dahouk S, Bouchon P, Denoeud F, et al. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC microbiology. 2006; 6: 9.
- Marianelli C, Ciuchini F, Tarantino M, Pasquali P, Adone R. Molecular characterization of the rpoB gene in Brucella species: new potential molecular markers for genotyping. Microbes and infection / Institut Pasteur. 2006; 8: 860-865.
- Sayan M, Yumuk Z, Bilenoglu O, Erdenlig S, Willke A. Genotyping of Brucella melitensis by rpoB gene analysis and re-evaluation of conventional serotyping method. Japanese journal of infectious diseases. 2009; 62: 160-163.
- Gwatkin H. J Infect Dis. 1931; 48: 104.
- Antadze VS, Popkhadze MZ, Abashidze TG. The correlation between bacteriophages isolation and Brucella type. Summaries of papers from Second Scientific Conference on Bacteriophages. Tbilisi, Geaorgia. 1952; 43.
- Dimitriu O, Cerbu A, Vasilesco T. La sensibilitk de quelques souches de Brucella if l'6gard du phage anti Brucella. Arch Pathol Exp Microbiol. 1959; 18: 475-480.
- Drozevkina MS. Brucella bacteriophage and its use for the identification of cultures. Proceedings of a Conference of scientific and practical workers on Brucellosis control; Rostov-on-Don, Russia. 1952; 78.
- Drozevkina MS. Brucella phage and prospects for its use. Collected papers of an inter-institute scientific conference on bacteriophagy. Tbilisi, Georgia. 1955; 5: 355.
- Drozevkina MS. The present position in Brucella phage research. Bull World Health Organ. 1963; 29: 43-57.
- Parnas J, Bulikowski W. Arch Ins Pasteur Tunis. 1973; 35.
- Pickett MJ, Nelson EL. Observations on the problem of Brucella blood cultures. Journal of bacteriology. 1951: 61: 229-237.
- Appelmans R. Le dosage du bacte´riophage. CR Soc Biol. 1921; 85: 1098-1099.
- Antadze VS, Popkhadze MZ, Abashidze TG. Correlation between bacteriophage isolation and Brucella type. Book of articles, Scientific-research Institute of Vaccines and Sera. Tbilisi, Georgia. 1961; 5.
- Antadze VS, Popkhadze MZ, Abashidze TG, Karichasvili L. Analyses of dynamics of isolation of Brucella bacteriophage from various stages of infection and immunization with a live Brucella vaccine. Book of articles, Scientific-research Institute of Vaccines and Sera. Tbilisi, Georgia. 1967; 4.
- Drozevkina MS. Bacteriological diagnosis of Brucella, taking into account their variability. Live Vaccines. Moskva, Russia. 1956; 231.
- Jones LM, Merz GS, Wilson JB. Phage typing reactions on Brucella species. Applied microbiology. 1968; 16: 1179-1190.
- Lazuga R, Renoux G. Arch Inst Pasteur Tunis. 1960; 37.
- Organization FaA, Organization WH. Joint FAO/WHO Expert Committee on Brucellosis. 1953.
- Parnas J, Kotlinska E. J Vet Med. 1961; 2: 41.
- Popkhadze MZ, Abashidze TG. Characteristics of Brucella bacteriophages isolated at the Tbilisi scientific-research Institute of vaccines and sera. Bacteriophagy. 1957; 5: 7.
- International bulletin of bacteriological nomenclature and taxonomy. 1963; 13: 145-158.
- Brinley-Morgan WJ. The examination of Brucella cultures for lysis by phage. 1962.
- Brinley-Morgan WJ, Kay D, Bradley DE. Brucella bacteriophage. Nature. 1960; 188: 74-75.
- Jones LM. Comparison of phage typing with standard methods of species differentiation in Brucella. Bulletin of the World Health Organization. 1960; 23: 130-133.
- Meyer ME, Cameron HS. Metabolic characterization of the genus Brucella. II. Oxidative metabolic patterns of the described biotypes. Journal of bacteriology. 1961; 82: 396-400.
- Popkhadze MZ. [Use of brucellosis Tb bacteriophage for differentiation of Brucella]. Zhurnal mikrobiologii, epidemiologii, immunobiologii. 1968; 45: 119-124.
- Scholz HC, Nockler K, Gollner C, Bahn P, Vergnaud G, Tomaso H, et al. Brucella inopinata sp. nov., isolated from a breast implant infection. International journal of systematic and evolutionary microbiology. 2010; 60: 801-808.
- Tiller RV, Gee JE, Lonsway DR, Gribble S, Bell SC, Jennison AV, et al. Identification of an unusual Brucella strain (BO2) from a lung biopsy in a 52 year-old patient with chronic destructive pneumonia. BMC microbiology. 2010; 10: 23.
- Tiller RV, Gee JE, Frace MA, Taylor TK, Setubal JC, Hoffmaster AR, et al. Characterization of novel Brucella strains originating from wild native rodent species in North Queensland, Australia. Applied and environmental microbiology. 2010; 76: 5837-5845.
- Cook I, Campbell RW, Barrow G. Brucellosis in North Queensland rodents. Australian veterinary journal. 1966; 42: 5-8.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227: 680-685.
- Adams MH. Bacteriophages. Moscow, Russia. 1961.
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed: Cold Spring Harbor Laboratory Press 1989.
- Morris JA, Corbel MJ. Properties of a new phage lytic for Brucella suis. The Journal of general virology. 1973; 21: 539-544.
- Popkhadze MZ, Antadze I. Comparative studies of phages isolated from the different species of Brucella. Bacteriophages. Moscow, Russia. 1973.
- Rigby CE, Cerqueira-Campos ML, Kelly HA, Surujballi OP. Properties and partial genetic characterization of Nepean phage and other lytic phages of Brucella species. Canadian journal of veterinary research. Revue canadienne de recherche veterinaire. 1989; 53: 319-325.
- Corbel MJ, Thomas EL. The Brucella phages: their properties, characterization and applications. Ministry of Agriculture, Fisheries, and Food. 1973.
- Thomas EL, Corbel MJ. Isolation of a phage lytic for several Brucella species following propagation of Tbilisi phage in the presence of mitomycin C. Archives of Virology. 1977; 54: 259-261.
- Goldfarb DM Bacteriophages. Chapter VII. Moscow. 1961; 109-114.
- Zhu CZ, Xiong HY, Han J, Cui BY, Piao DR, Li YF, et al. Molecular characterization of Tb, a new approach for an ancient Brucellaphage. International journal of molecular sciences. 2009; 10: 2999-3011.
- Flores V, Lopez-Merino A, Mendoza-Hernandez G, Guarneros G. Comparative genomic analysis of two Brucellaphages of distant origins. Genomics. 2012; 99: 233-240.
- International committee on systemic bacteriology subcommittee on the taxonomy of Brucella. 1988; 38.
- Corbel MJ, Brinley-Morgan WJ. Proposal for minimal standards for description of new species and biotypes of the genus Brucella. Int J Syst Bacteriol. 1975; 25: 83-89.
- Michaux-Charachon S, Bourg G, Jumas-Bilak E, Guigue-Talet P, Allardet-Servent A, O'Callaghan D, et al. Genome structure and phylogeny in the genus Brucella. Journal of bacteriology. 1997; 179: 3244-3249.
- Halling SM, Zehr ES. Polymorphism in Brucella spp. due to highly repeated DNA. Journal of bacteriology. 1990; 172: 6637-6640.
- Tevdoradze E, Farlow J, Kotorashvili A, Skhirtladze N, Antadze I, Gunia S, et al. Kutateladze Whole genome sequence comparison of ten diagnostic bacteriophages propagated on two Brucella abortus hosts. Virology Journal. 2015; 12: 66.