
Special Article - Brucella
J Bacteriol Mycol. 2017; 4(3): 1054.
Molecular Typing of Brucella Species Strains from Georgia and Turkey
Sidamonidze K¹, Su W3, Zhgenti E¹, Buyuk F², Sahin M2, Trapaidze N4, Imnadze P1, Nikolich MP3,4, Obiso R5 and Kotorashvili A1*
¹National Center for Disease Control and Public Health, Tbilisi, Georgia
²Department of Microbiology, University of Kafkas, Turkey
²Walter Reed Army Institute of Research, USA
4US Army Medical Research Unit-Georgia, Georgia
5Avila Scientific, Virginia, USA
*Corresponding author: Kotorashvili A, National Center for Disease Control and Public Health, Tbilisi, Georgia
Received: July 28, 2017; Accepted: August 30, 2017; Published: September 06, 2017
Abstract
The genus Brucella consists of several species that infect different mammalian hosts including humans. Brucella infection can lead to abortion in many domestic animals, which causes significant economic losses. Human brucellosis is one of the most globally widespread and debilitating zoonotic diseases, and it remains endemic in both Georgia and Turkey. Precise genetic characterization of Brucella isolates by molecular typing methods can provide useful analysis for examining strain diversity; detection of clonal groups; determining sources of infection; and discrimination of naturally occurring outbreaks from a bioterrorist event. Recent studies have demonstrated that Multiple-Locus Variable-Number Tandem-Repeat Analysis (MLVA) is a high resolution genetic subtyping tool that can provide valuable information for epidemiological investigations on brucellosis. In this study, we assessed the genetic variability of 30 isolates of Brucella isolated in Turkey and 65 Brucella isolates from Georgia. All samples were identified as B. melitensis or B. abortus based on microbiological and Bruce-ladder PCR results. Discriminatory genotypes were determined using the MLVA approach using 15 mini-satellite markers. Relationships between Turkish and Georgian isolates were revealed and showed that strains are grouped by origin of country and species; however, there were two strains of B. melitensis from Turkish strains mixed within the Georgian group. Our results suggest that Brucella spp. is homogenous, however, two strains have been introduced from Turkey into Georgia and that cross-border migration is possible.
Keywords: Mammalian hosts; Bacterial zoonosis; Human brucella
Introduction
Brucellosis is globally the most common bacterial zoonosis, resulting in significant economic losses via the reduction of livestock productivity, and is a debilitating human disease. Although the picture of global brucellosis morbidity is not completely clear, brucellosis is found on almost every continent, with at least 500,000 people are infected annually [1]. Brucella melitensis is the most frequent cause of human brucellosis accounting for an estimated 96% of cases worldwide [2]. Human infection occurs from exposure to infected animal abortion products or contaminated dairy products, and it is accepted that control of the human disease can only occur through control and eradication of the disease in animals [3].
Brucellosis is currently under-diagnosed and under-reported in Turkey [4] and worldwide [5] due to the nonspecific clinical manifestations of the disease and the limitations of current diagnostic tests. Brucellosis infection in humans can be challenging to diagnose as its presentation is nondescript and mimics many other conditions [5]. In the early presentation (days to weeks after exposure), non-specific symptoms such as fever, fatigue/malaise, myalgia, and arthralgia predominate. Some patients proceed to develop focal complications, with osteoarticular complications being somewhat common and very debilitating [4]. Other patients may have chronic, mild symptoms for many months, making brucellosis difficult to recognize as the cause of their condition. These patients have traditionally been classified as having chronic brucellosis, but the utility of this classification is not established. Additionally, a certain percentage of patients relapse after treatment, usually with non-specific symptoms, even after an extensive course of initial therapy, however, the contribution of lack of treatment compliance and re-infection of these patients to relapse is not well known.
Human brucellosis is caused by five classical Brucella species (B. abortus, B. melitensis, B. suis, B. ovis, B. canis), of which B. melitensis is considered the most virulent for humans [2]. B. melitensis is highly infectious for humans with an ID50 of 10 cells per person, and it is virulent in other mammal hosts such as bovine cattle, though sheep and goats are its preferred host [3]. B. melitensis infections are especially problematic as the most commonly used attenuated B. abortus cattle vaccine (Strain 19) fails to protect from disease caused by B. melitensis [5]. Human cases of brucellosis are acquired through contact with infected animals and through consumption of contaminated dairy products. Adherence to traditional farming practices and the consumption of unpasteurized dairy products contribute to the high incidence of brucellosis in Turkey [4].
The B. abortus RB51 vaccine has also been used successfully in selected countries to aid in the control of bovine brucellosis (caused by B. abortus). A complicating factor of vaccination is that classical serologic diagnostic protocols cannot distinguish animals vaccinated with Strain 19 from infected animals and diagnosis during latent infection can be particularly challenging. Only the use of the rough B. abortus vaccine RB51 in bovine cattle addresses this issue because it lacks the dominant antigen, LPS O-side chain, but this vaccine is yet to be accepted worldwide. Meanwhile the utility of vaccination of livestock with Strain 19 is significantly decreased by the inability to distinguish between vaccinated animals and those animals with active infections without sophisticated laboratory assays [5]. These issues underscore the need for improved detection and diagnostic capabilities in the primary livestock hosts of Brucella.
Genetic characterization of a bacterium is often used for molecular epidemiology purposes and can be useful to observe population structures. Owning to the high genetic identity among members of the Brucella genus [6-12], the physiological typing methodologies of the past showed little success [13]. However, in recent years, the application of Multiple-Locus Variable Number Tandem Repeats Analysis (MLVA) with different sets of repeat loci has gained momentum in Brucella typing. The MLVA typing assay consists of two complementary panels, panel 1 (8 markers) and panel 2 (7 markers); fifteen markers clearly presents a discriminative power in typing these microorganisms [13].
Previous studies to examine the genetic diversity and regional distribution of Brucella species in Georgia found high genetic diversity between isolates, and geographic distribution was dependent on the genotype. A study of 35 isolates of human and animal Brucella obtained in Georgia in 2009 and 2010 using MLVA-8 analysis illustrated the high diversity between isolates [6]. It was found that only 2-3 strains were located in one genotype cluster, with some genotypes showing restricted geographical distribution, and others were disseminated among different regions of Georgia. One genotype cluster contained animal and human isolates from the same region, revealing a potential source of human infection [6].
A study similar study was conducted in Turkey with 162 human Brucella isolates collected from 2001-2008, which were analyzed using MVLA-16 [7]. A total of 105 genotypes were recorded, with 73 being represented by a unique isolate, and 32 including 2-8 isolates. As was found in Georgia, several the isolates had restricted geographic distribution. In addition, spanning tree analysis of published B. melitensis isolates, using MLVA-11 data, indicted that Turkish isolates were most closely related to the neighboring countries’ isolates included in the East Mediterranean group [7].
The aim of this study was to genetically characterize Brucella spp. from strains isolated from both Georgia and Turkey by applying high-resolution genetic characterization techniques such as Bruce- Ladder PCR and Multiple Locus Variable-Number Tandem Repeat Analysis (MLVA) to determine the genetic relationships between pathogens and potential migration pattern across the borders. This study was designed to evaluate the molecular epidemiology of Brucella spp. from human and livestock of the Georgia and Turkey by applying these findings to future planning for disease control in the both countries.
Materials and Methods
Samples and study design
All Brucella samples used in the study were provided from the bacterial repository of Kafkas University (Kars, Turkey) and the repository of the National Center for Disease Control and Public Health of Georgia (NCDC&PH, Tbilisi, Georgia). It was the first time applying high resolution molecular approaches (Bruce-Ladder PCR and MLVA typing) for the selected samples. In total, 30 Turkish Brucella samples (Table 1) and 65 Georgian Brucella samples (Table 2) were used.DNA samples from Kafkas University were transported to NCDC using iced containers and kept at -20°C. General Directorate of Food and Control, Republic of Turkey Ministry of Food Agriculture and Livestock, Permission number 55016929-604.01-13587. Public of Health Agency of Turkey, Republic of Turkey Ministry of health, Permission number: 80962070.
Turkish Brucella species
Strain ID
Sample Type
Origin
Year
Region
BRU-S-1
Sheep
Aborted Fetus
2005
Kars Center
BRU-S-2
Cattle
Aborted Fetus
2005
Kars Arpacay
BRU-S-3
Sheep
Aborted Fetus
2005
Kars Center
BRU-S-4
Sheep
Aborted Fetus
2006
Kars Center
BRU-S-5
Sheep
Aborted Fetus
2007
Kars Center
BRU-S-6
Cattle
Aborted Fetus
2007
Ardahan Prosof
BRU-S-7
Sheep
Aborted Fetus
2007
Kars Center
BRU-S-8
Sheep
Aborted Fetus
2008
Kars Center
BRU-S-9
Sheep
Aborted Fetus
2009
Kars Center
BRU-S-10
Sheep
Aborted Fetus
2009
Kars Center
BRU-S-11
Sheep
Aborted Fetus
2009
Kars Center
BRU-S-12
Cattle
Milk
2009
Kars Center
BRU-S-13
Cattle
Milk
2009
Kars Center
BRU-S-14
Cattle
Vaginal Secretion
2009
Kars Center
BRU-S-15
Cattle
Vaginal Secretion
2009
Kars Center
BRU-S-16
Cattle
Milk
2008
Kars Selim
BRU-S-17
Cattle
Vaginal Secretion
2008
Kars Center
BRU-S-18
Cattle
Milk
2010
Kars Selim
BRU-S-19
Cattle
Milk
2010
Kars Selim
BRU-S-20
Sheep
Aborted Fetus
2010
Kars Center
BRU-S-21
Cattle
Vaginal Secretion
2010
Kars Selim
BRU-S-22
Cattle
Vaginal Secretion
2010
Kars Selim
BRU-S-23
Sheep
Aborted Fetus
2011
Kars Center
BRU-S-24
Cattle
Aborted Fetus
2011
Kars Center
BRU-S-25
Sheep
Aborted Fetus
2011
Kars Akyaka
BRU-S-26
Sheep
Aborted Fetus
2012
Kars Center
BRU-S-27
Cattle
Aborted Fetus
2013
Kars Center
BRU-S-28
Sheep
Aborted Fetus
2013
Kars Center
BRU-S-29
Sheep
Aborted Fetus
2013
Kars Digor
BRU-S-30
Sheep
Aborted Fetus
2014
Kars Center
Table 1: Turkish Brucella spp. strains analyzed in this study and their phenotypic attributes.
Brucella spp. culturing and inactivation
Pure culture isolates of Brucella spp. from the Kafkas University were plated on Oxoid Brucella Agar plates (Oxoid, UK) for five days at 37°C. Several loops of culture were placed in 1.5mL micro centrifuge tubes and then samples were heat-inactivated for two hours at 80°C [8]. We determined sample sterility by pipetting 5% of the final volume (100μL) of the extracted DNA and incubating at 37°C in the same growth media used in bacterial culturing. At day +3, an aliquot of 5μL was streaked onto an agar plate (Oxoid, UK) and incubated at 37°C for 72 hours. At day +7, the streaking procedure was repeated. If no growth was observed at either time point, then the preparation was considered sterile. Primary and secondary containers were decontaminated for 30 minutes with 1% sodium hypochlorite, and stored at -20°C. After sterility confirmation and surface decontamination, samples required biosafety level-2 containment.
DNA extraction and PCR detection
Once sterility was confirmed, genomic DNA was extracted using a commercially available extraction kit (QIAamp DNA Mini Kit) according to the manufacturer’s instructions. Purified DNA was aliquoted in 95-100 μL volumes and stored at - 20°C. DNA concentrations were measured using Nano DROP 2000 spectrophotometer (Thermo Scientific). Standard PCR of these samples was conducted using one primer set (B4/5) as reported by Baily et al. [8]. The PCR reaction was composed of, 0.5μL dNTPs, 2.5μL buffer, 1.5μL of each primer, 1μL MgCl2, 0.5μL Taq polymerase, 15μLdH2O, and 2.5μL template DNA. The PCR cycling conditions were: Initial denaturation at 94°C for 3 minutes, followed by 35 cycles of 94°C for 60 seconds, 65°C for 60 seconds, and 72°C for 60 seconds, with a final elongation step of 72°C for 5 minutes. Control organisms B. abortus 544, B. melitensis 16M, and E. coli OP50 were also assessed. Amplicons were analyzed on a 1.5% agarose gel stained with SafeView nucleic acid stain (NBS Biologicals, Huntingdon, UK) using a horizontal gel electrophoresis system (BioRad). In summary, 7.5μL of PCR product was loaded with 1.5μL loading dye; 5μL of 100bp Hyper ladder Plus marker was used. The gel was run at 55 V-30mA for the first ten minutes followed by 80 V-30mA for the last 40 minutes;
Bruce-ladder PCR
Bruce-ladder PCR assay was performed as reported by García- Yoldi et al. [9]. PCR reaction were conducted in a 25μL volume and contained 2.5μL PCR buffer (x10), 2.5μL dNTP mix (2.5mM), 1.5μL MgCl2 (50mM), 2μL Bruce-ladder primer mix (10pmol) García- Yoldi et al. [9], 0.2μL Taq DNA polymerase (5u/μL), 15.3μL ddH2O, and 1μL template DNA (10ng/μL). Cycling conditions consisted of 35 seconds of initial denaturation at95°C, followed by 35 cycles of 35 secondsof denaturation at 95°C, 45 secondsof annealing at 63°C, and 180 seconds of extension at 72°C. PCR was completed with a final extension at 72°C for 3 minutes. Brucella reference strains (B. abortus biotype 1 (544) and B. melitensis biotype 1 (16M)) and a negative control (E. coli OP50) were used in the Bruce-ladder PCR as positive and negative controls, respectively. The amplified products were visualized by electrophoresis using a 1.5% agarose gel run at 80V-30mA for the 45 minutes followed by staining with ethidium bromide.
MLVA-15 analyses
Multiple Locus Variable-Number Tandem Repeat (VNTR) analysis (MLVA-15) was performed on Turkish and Georgian samples set in duplicate to examine the reproducibility of the analysis [10]. MLVA was performed as described previously [10]. The 15 primer pairs (Table 3) were divided into two groups: Panel 1 (MLVA- 8: eight loci including bruce03, bruce07, bruce14, bruce16, bruce20, bruce21, bruce25, and bruce28), panel 2 (MLVA-7: seven loci including bruce01, bruce02, bruce27, bruce29, bruce30, bruce31, and bruce33). DNA template stocks were diluted with sterile water to a final concentration of 10ng/μL. DNA concentrations were measured using NanoDROP 2000 spectrophotometer (Thermo Scientific). PCR amplifications were performed in 20μL reaction volumes. The PCR conditions were as follows: initial denaturation at 94°C for 3min, and then 35cycles of 94°C for 30s, 60°C for 30s, and 72°C for 50s, with a final extension of 72°C for 3min. PCR products for the 15 loci were denatured and resolved by capillary electrophoresis on an ABI Prism 3130xl automated fluorescent capillary DNA sequencer (Applied Biosystems). Fragments were sized following comparison with a ROX (carboxy-X-rhodamine)-labeled molecular ladder Liz 1200 (MapMaker 1000; Bioventures Inc., Murfreesboro, TN, USA) and Gene Mapper software version 4.0 (Applied Biosystems).
#
Primer Name
Primer Sequence (flanking VNTR locus)
1
Bruce1F-PET
ggcggacagagccgtcggtggttac
2
Bruce1R
cccgcgccggagattgtttttgattaatg
3
Bruce2F-VIC
gcggatcgacttcgagacattcacgctc
4
Bruce2R
gggtccgtaattgtcgggcgctcag
5
Bruce3F-VIC
tctcatcgacggcaagatcggcatcaagt
6
Bruce3R
cgcgaggacgaagagggcattgc
7
Bruce7F-FAM
gagcccgatatgcggccaacgat
8
Bruce7R
ggatattgacgatattcttgtgtcttccagcaaagtcac
9
Bruce14F-NED
ggcctggcgcatgccttggtg
10
Bruce14R
gcgatgtcctgcctgccccagttc
11
Bruce16F-VIC
ggctatgcgggcgtggagaacgaactc
12
Bruce16R
ctgcgcgctttgcaggatgctatgttagg
13
Bruce20F-NED
gctgcggccattaccacgc
14
Bruce20R
gccggcggttattcgtccggatcg
15
Bruce21F-FAM
ggaagcatgaaacaaacatcaataacgggaactg
16
Bruce21R
cacaacggccgccagaccgaatct
17
Bruce25F-FAM
gatgcgggtcgagggccttgagagtg
18
Bruce25R
gacaatggccgcaaaagcttccgaacc
19
Bruce27F-FAM
gcccgcgaccacgagcgtcaac
20
Bruce27R
gcccggcgaatctggctcgtcag
21
Bruce28F-FAM
gtgctgacgaagggaaggcaataaggcagtag
22
Bruce28R
gccaatggccgcaggaaag
23
Bruce29F-NED
gagcccgccattgcaatcgtgaacac
24
Bruce29R
caccgctgtccgcgcccacatc
25
Bruce30F-FAM
gccgaggcttgccattctgatcctttc
26
Bruce30R
gacgccagccttcaaatgttacctctctagcgc
27
Bruce31F-NED
cacgggcccctgcttttccattc
28
Bruce31R
ggcgctcgctgattggctgtgatatagg
29
Bruce33F-FAM
gatatcatgacacgcagcccgcgaac
30
Bruce33R
gaaattctggcgcccggctttttc
Table 3: List of the primers used for MLVA-15 analysis.
For MLVA-15 analysis, 2μL of each PCR reaction was diluted 10- fold in 18μL of molecular biology grade water. A formamide/1200LIZ size standard mixture was prepared to serve as a denaturing solution and size standard for each sample. The solution was prepared by mixing 19μL of formamide and 0.5μL of 1200LIZ size standard.19.0μL of the formamide/1200LIZ size standard solution was combined with 1.0μL of the diluted multiplex samples in a 96-well plate (ABI platform compatible plate, e.g., MicroAmp™). Before loading plates on an ABI 3130xl platform, samples were denatured in a thermal cycler for five minutes at 95°C then placed on ice for three to five minutes. Fragment analysis was performed with GeneScan and GeneMapper software packages (Applied Biosystems, for 3130xl). To size fragment, samples were compared to a custom-made LIZ-dye-labeled size standard and custom macro programs were created in GeneMapper for automated scoring of VNTR alleles.
Fragments at each of 15 VNTR loci were normalized according to the expected size table for each isolate. The expected higher fragment size was taken if the differences between actual fragment size and the expected fragment size were greater than half a repeat in size; otherwise the expected lower fragment size was taken if the differences between actual fragment size and the expected fragment size were less than half a repeat in size. The resulting data were analyzed as a character dataset using the Bionumerics software package version 6.6 (Applied- Maths, Saint-Martens-Latem, Belgium). Clustering analysis was done using the categorical coefficient and Unweighted Paired Group Method Arithmetic Average (UPGMA) to generate dendrograms for our dataset. Diversity index (Hunt-Gaston) of each locus was also calculated using Bionumerics. To evaluate the discriminatory power of the selected loci, the Hunter and Gaston discrimination index was calculated for each of the 15 loci used in this study.
Results
Bruce-ladder analysis
The Bruce-ladder PCR analysis showed that there were 15 B. melitensis samples and 14 B. abortus samples present (Figure 1A, 1B). Out of 30 samples tested, only one sample failed (sample #28), which was repeated along with one B. abortus sample. The result of this repeated round of PCR is shown in Figure 1C; the repeated assay failed again. The 65 Georgian isolates were analyzed by the Bruce-ladder PCR to confirm their species identification (data are not shown).
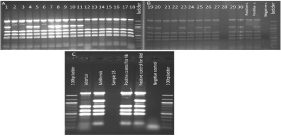
Figure 1: Agarose gel (1.5%) images of amplicons produced from the Bruce-ladder PCR assay performed on Turkish Brucella spp. samples. A) Samples 1-18, and
B) samples 19-30. Sample number 28 failed to be typed by Bruce-ladder PCR.C) Gel image of repeated analysis of sample 28, which failed to be typed.
MLVA typing
In this study, we analyze a total of 94 Brucella spp.: 29 Turkish strains (Table 1) and 65 Georgian strains (Table 2). Isolates were analyzed using MLVA-15 to differentiate closely related isolates within species. The pattern results were further analyzed and phylogenic trees were constructed using BioNumerics software; separate analysis was performed for Turkish strains to show genetic relationships between Turkish isolates (Figure 2). To evaluate the discriminatory power of the selected loci, the Hunter and Gaston discrimination index was calculated for each of the 15 loci used in this study. Among the 94 isolates analyzed, the diversity index value ranged from 0 to 0.96 (Figure 3). There were three loci (bruce 2, bruce 33 and bruce 1) that had no discriminatory power in this dataset.
Georgian Brucella species
Strain ID
Sample Type
Source
Year
Region
G-Bru-1
Human
Blood
2009
Kvemo Kartli,Marneuli
G-Bru-2
Human
Blood
2009
Imereti, Samtredia
G-Bru-3
Human
Blood
2009
Samegrelo, Senaki
G-Bru-4
Human
Blood
2009
Samegrelo, Senaki
G-Bru-6
Bovine
Fetus
2009
Imereti, Kutaisi
G-Bru-7
Bovine
Blood
2009
Kakheti, Gurjaani
G-Bru-10
Bovine
Blood
2009
Kvemo Kartli, Marneuli
G-Bru-11
Bovine
Blood
2009
Kakheti, Signagi
G-Bru-12
Bovine
Milk
2009
Imereti, Kutaisi
G-Bru-14
Bovine
Milk
2009
Kakheti, Gurjaani
G-Bru-13
Bovine
Milk
2009
Imereti, Kutaisi
G-Bru-18
Bovine
Milk
2009
Kakheti, Gurjaani
G-Bru-19
Bovine
Milk
2009
Tbilisi, Vake
G-Bru-20
Bovine
Milk
2009
Kvemo Kartli, Marneuli
G-Bru-21
Sheep
Milk
2009
Kvemo Kartli, Marneuli
G-Bru-22
Sheep
Blood
2009
Kvemo Kartli, Marneuli
G-Bru-23
Bovine
Milk
2009
Kvemo Kartli, Marneuli
G-Bru-24
Bovine
Milk
2009
Mtskheta-Mtianeti, Mtskheta
G-Bru-61
Bovine
Blood
2009
Kvemo Kartli, Marneuli
G-Bru-44
Human
Blood
2009
Kakheti, Kvareli
G-Bru-54
Human
Blood
2009
Kvemo Kartli, Dmanisi
G-Bru-30
Human
Blood
2010
Kvemo Kartli, Marneuli
G-Bru-53
Human
Blood
2010
Kakheti, Gurjaani
G-Bru-40
Human
Blood
2010
Kakheti, Gurjaani
G-Bru-29
Human
Blood
2010
Kvemo Kartli, Marneuli
G-Bru-72
Human
Blood
2010
Kvemo Kartli, Dmanisi
G-Bru-34
Human
Blood
2010
Kakheti, Dedoplistkaro
G-Bru-55
Human
Blood
2010
Mtskheta-Mtianeti, Kazbegi
G-Bru-62
Human
Blood
2010
Kakheti, Sagarejo
G-Bru-67
Human
Blood
2010
Tbilisi
G-Bru-50
Human
Blood
2010
Tbilisi
G-Bru-28
Human
Blood
2010
Shida Qartli, Kaspi
G-Bru-42
Human
Blood
2010
Kvemo Kartli, Bolnisi
G-Bru-51
Human
Blood
2010
Kvemo KartliQ, Dmanisi
G-Bru-58
Human
Blood
2010
Kvemo Kartli,Marneuli
G-Bru-71
Human
Blood
2010
Kakheti, Sagarejo
G-Bru-63
Human
Blood
2010
Kvemo Kartli, Bolnisi
G-Bru-64
Human
Blood
2010
Kakheti, Akhmeta
G-Bru-41
Human
Blood
2010
Kakheti, Akhmeta
G-Bru-49
Human
Blood
2010
Kakheti, Sagarejo
G-Bru-68
Human
Blood
2010
Kvemo Kartli, Gardabani
G-Bru-33
Human
Blood
2010
Kvemo Kartli,Marneuli
G-Bru-36
Human
Blood
2010
Kakheti, Sagarejo
G-Bru-32
Human
Blood
2011
Kakheti, Sagarejo
G-Bru-46
Human
Blood
2011
Kakheti, Lagodekhi
G-Bru-35
Human
Blood
2011
Kvemo Kartli, Bolnisi
G-Bru-45
Human
Blood
2011
Mtskheta-Mtianeti, Kazbegi
G-Bru-37
Human
Blood
2011
Kakheti, Sagarejo
G-Bru-26
Human
Blood
2011
Kvemo Kartli,Marneuli
G-Bru-65
Human
Blood
2011
Kvemo Kartli, Dmanisi
G-Bru-27
Human
Blood
2011
Samtskhe Javakheti, Ninotsminda
G-Bru-56
Human
Blood
2011
Kakheti, Akhmeta
G-Bru-25
Human
Blood
2011
Kakheti, Akhmeta
G-Bru-38
Human
Blood
2011
Kvemo Kartli, Bolnisi
G-Bru-43
Human
Blood
2011
Kvemo Kartli, Gardabani
G-Bru-59
Human
Blood
2011
Kvemo Kartli, Gardabani
G-Bru-69
Human
Blood
2011
Kakheti, Telavi
G-Bru-66
Human
Blood
2011
Kvemo Kartli, Dmanisi
G-Bru-31
Human
Blood
2011
Kvemo Kartli,Marneuli
G-Bru-60
Human
Blood
2011
Kakheti, Sagarejo
G-Bru-39
Bovine
Milk
2011
Kvemo Kartli, Bolnisi
G-Bru-48
Bovine
Milk
2011
Kvemo Kartli, Gardabani
G-Bru-57
Sheep
Milk
2011
Kvemo Kartli,Marneuli
G-Bru-8
Human
Blood
2011
Kvemo Kartli, Gardabani
G-Bru-70
Sheep
Milk
2011
Kvemo Kartli,Marneuli
Table 2: Georgian Brucella spp. strains used in this study and their phenotypic attributes.
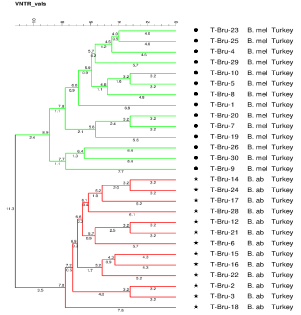
Figure 2: MLVA-15 based phylogeny of all Turkish (Kafkas University,
Turkey) strains. Green color refers to Brucella melitensis, read color refers
to Brucella abortus.
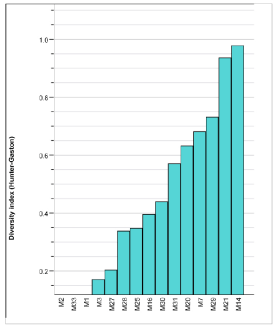
Figure 3: The Hunter and Gaston discrimination index was calculated for
each of the 15 loci used in this study. Among the isolates analyzed, the
diversity index value ranged from 0 to 0.96). There were three loci (bruce 2,
bruce 33 and bruce 1) that had no discriminatory power in this dataset.
Turkish Brucella spp. isolates analyzed in this project were divided into two clusters according to their relationships based on MLVA-15 genotyping. Based on the results of the Bruce-ladder PCR (Figure 1A, 1B) one MLVA cluster was confirmed as B. abortus, and the second cluster was confirmed as B. melitensis. The B. abortus cluster displays more diversity than does the B. melitensis cluster. The B. melitensis cluster comprised isolates from four animals from the same geographic location with the same genetic subtype, which suggests a common origin of infection for this group of isolates. The Bruceladder assay as well as MLVA-15 failed to type sample 28 (Figure 1B, 1C). Using new technology such as whole genome sequencing may be appropriate to gain further data on this sample, which may have been misidentified as Brucella.
We noted that analyzed strains, in general, are grouped by origin of country and species, but interestingly, comparison of Georgian and Turkish strains showed two Turkish B. melitensisstrains mixed in the Georgian group. This finding might suggest that these two B. melitensis strains in Georgiamay have come from Turkey (Figure 4).
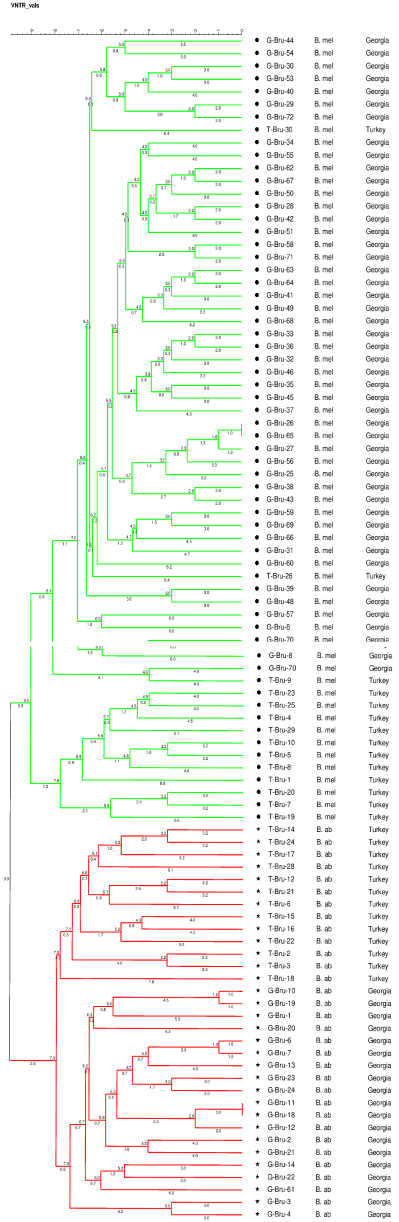
Figure 4: MLVA-15 based comparative phylogeny of Turkish (repository of
Kafkas University) and Georgian (repository of National Center for Disease
Control and Public Health) strains. Green color refers to Georgian and
Turkish B. melitensis, red color refers to Georgian and Turkish B. abortus.
Discussion
Research in Brucella has a long history, although the mechanisms for disease and disease transmission are somewhat unknown. There is a wealth of information in the literature that describes incidence of disease and epidemiology of disease in many countries around the world. Based on the historic scientific literature, Brucella species are relatively homogeneous.
This study represents the first attempt to describe local patterns of brucellosis between Turkey and Georgia. We used analytical methods to analyze genetic differences in Brucella in these countries. Our analyses confirm that brucellosis persists throughout much of these countries. These analyses affirm the notion that brucellosis is endemically established since the first documented reporting in the 1920s. This study also represents the first time that the molecular characterization of Brucella isolates originated from the neighboring countries of Georgia and Turkey. As brucellosis is a noncommunicable disease in humans, clusters of cases are most likely a result of shared food sources, animal processing, more intensive agricultural production zones, or similar socio-cultural practices. Clusters of disease in this case are also most likely indicative of a larger underlying prevalence of the disease in the local livestock population. These results contribute to better understanding geographical transmission patterns of Brucella in human and animal and would be important to implement specific control measures in the future.
Conclusions
In conclusion, our finding suggests that etiological agent of human and animal brucellosis (B. abortus and B. melitensis) circulating in the regions of Georgia and Turkey are closely related and mostly grouped by country origin. Interestingly two B. melitensis strains were mixed, which might suggest that strain migration across the borders occurs. Our results highlight the usefulness of Bruce-ladder multiplex PCR and MLVA as epidemiological tools for investigations of Brucella infections. MLVA 15 segregated Brucella strains into groups corresponding to the recognized phylogenetic lineages in the genus Brucella. MLVA-15 displayed good discriminatory power for the typing of Brucella isolates from this region of endemicity and may be used as epidemiological tools for the resolution of strains.
Acknowledgments
The work was made possible by support provided by the US Defense Threat Reduction Agency (TAP-10 project) through the Cooperative Biological Engagement Program in Georgia. The findings, opinions and views expressed herein belong to the authors and do not reflect an official position of the Department of the Army, Department of Defense or the US Government, or any other organization listed.
References
- Seleem MN, Boyle SM, Sriranganathan N. Brucellosis: a re-emerging zoonosis. Vet. Microbiol. 2010; 140: 392-398.
- Corbel MJ. Brucellosis in humans and animals. WHO Press, Geneva, Switzerland. 2006.
- He, Y. Analyses of Brucella pathogenesis, host immunity, and vaccine targets using systems biology and bioinformatics. Front. Cell. Infect. 2012.
- Yumuk Z, O’Callaghan D. Brucellosis in Turkey -- an overview. Int. J. Infect. Dis. Ijid Off. Publ. Int. Soc. Infect. Dis. 2012; 16: 228-235.
- Barrett ADT, Stanberry LD. Vaccines for biodefense and emerging and neglected diseases. Academic Press. 2009.
- Zhgenti E, Sidamonidze K, Zakalashvili M, Malania L, Ramishvili M, Grdzelidze M, et al. Genetic diversity of Brucella isolates in Georgia. IMED conference, Vienna Austria. 2013.
- Kiliç S, Ivanov IN, Durmaz R, Bayraktar MR, Ayaslioglu E, Uyanik MH, Aliskan H, et al. Multiple-locus variable-number repeat analysis of Brucella species. 2011; 49: 3276-3283.
- Bailey GG, Krahn JB, Drasar BS, Stoker NG. Detection of Brucella melitensis and Brucella abortus by DNA amplification. The Journal of tropical medicine and hygiene. 1992; 95: 271-275.
- García-Yoldi D, Marín CM, de Miguel MJ, Muñoz PM, Vizmanos JL, López I. Multiplex PCR Assay for the identification and differentiation of all Brucellaspecies and the vaccine strains Brucella abortus S19 and RB51 and Brucella melitensis Rev1. Clin Chem. 2006; 54: 779-781.
- Huynh LY, Van Ert MN, Hadfield T, Probert WS, Bellaire BH, M Dobson, et al. Multiple Locus Variable Number Tandem Repeat (VNTR) Analysis (MLVA) of Brucella spp. identifies species specific markers and insights into phylogenetic relationships. Georgiev V, Western KA, McGowan JJ, editors. In: National Institute of Allergy and Infectious Disease, NIH, vol. 1. Frontiers in research. Humana Press, Totowa, NJ. 2008.
- Sidamonidze K, Hang J, Yang Y, Dzavashvili G, Zghenti E, Trapaidze N, et al. Genome sequences of human and livestock isolates of Brucella melitensis and Brucella abortus from the country of Georgia. Genome Announc. 2017; 5: 1516-1518.
- Verger JM, Grimont F, Grimont PA, Grayon M. Taxonomy of the genus Brucella. Ann. Inst. Pasteur Microbiolo. 1987; 138: 235-238.
- Le Fleche P, Jacques I, Grayon M, Dahouk AL, Bouchon P, Denoeud F, et al. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 2006; 9: 9.