
Research Article
J Bacteriol Mycol. 2018; 5(3): 1070.
Prevalence and Antimicrobial Susceptibility of Zoonotic Salmonella Species Isolated from Water Bodies in Bukombe District
Anatory YS1* and Machang’u RS2
¹Department of Microbiology and Parasitology, Mwalimu Julius K. Nyerere University of Agriculture and Technology, Tanzania
²Department of Microbiology and Parasitology, Sokoine University of Agriculture, Tanzania
*Corresponding author: Anatory YS, Department of Microbiology and Parasitology, Mwalimu Julius K. Nyerere University of Agriculture and Technology, Tanzania
Received: May 10, 2018; Accepted: June 18, 2018; Published: June 25, 2018
Abstract
A study on zoonotic Salmonella in water bodies was carried out between November 2016 and April 2017 in rural areas of Bukombe district, Tanzania. A total of 10 wards of Bukombe district were involved in the study. The main objective of the study was to estimate the burden of zoonotic Salmonella species (serovars Typhimurium and Enteritidis) as well as the antimicrobial resistance profile of isolates obtained from water bodies in Bukombe district. The specific objectives were to establish prevalence of zoonotic Salmonella spp from water, establish the antimicrobial resistance and to compare the prevalence of zoonotic Salmonella spp of multipurpose with single purpose water bodies in Bukombe district, Tanzania. A total of 240 water samples were examined for Salmonella contamination of which 50% were from multipurpose and the rest were from single purpose water bodies. Salmonella species were identified in 1.3% (3/240) of the 240 water samples. Multipurpose water bodies had a prevalence of 1.6% (2/120) while the single purpose water showed prevalence of 0.8% (1/120). The definitive confirmation by polymerase chain reaction (PCR) showed 1.3% isolates were positive for Salmonella. Based on biochemical test by lysine reaction 4.2% of the isolates were identified as Salmonella spp. The study observed that all isolated Salmonella spp. were resistant to penicillin (ampicillin) and cephalosporins (cephalexin), but were sensitive to floroquinolones (ciprofloxacin) and aminoglycosides (gentamicin).
Abbreviations and Symbols
Spp: Species; PCR: Polymerase Chain Reaction; Var: Serovar; Subsp: Subspecies; n: Number of samples; rpm: Rotation per minute; DNA: Deoxyribose Nucleic Acid; Bp: Base pair; Min: Minute; sec: Second; V: Voltage; BSL: Biosafety Level: MDR: Multidrug Resistant; UK: United Kingdom; DWAF: Department of Water Affairs and Forestry in South Africa; HIV: Human Immunodeficiency Virus; WHO: World Health Organization; TBE: Tris-Borate-EDTA
Introduction
Background information
Water bodies in Tanzania: Most of the water bodies in Tanzania are poorly sanitized and protected which makes them potential sources of infections such as zoonotic Salmonella. The scarcity of water has also led people and animals to share the water body which increases chances of salmonellosis transmission in humans and animals [1]. As of 2012 only 40.6% of the households have access to improved drinking water sources such as piped water, tube wells and protected springs. The remaining percentage uses non-improved drinking water sources [2].
Zoonotic salmonella: Salmonella species are gram-negative, rodshaped bacteria with broad host spectrum, including most animal species: mammals, birds and reptiles. Currently there are more than 2600 serotypes (serovars) of Salmonella classified into two species; Salmonella enterica and Salmonella bongori. S. enterica subsp. enterica I is divided into serotypes, for example serotypes enteriditis, typhimurium, typhi and choleraesuis [3]. Serotypes S. typhi and S. paratyphi A are referred to as typhoidal salmonella the rest are called non-typhoidal or zoonotic Salmonella species.
Epidemiology of zoonotic Salmonella: Salmonellosis ranks high in causing bacterial enteric illness in humans and animals. Zoonotic Salmonella spreads by direct or indirect means from infected animals through contaminated feed and water supplies. Water is among the main transmission routes for diseases such as salmonellosis in humans and animals [4]. It is among the common causes of illness and death among the poor populations in developing countries. Individuals infected with Salmonella spp. shed the organism in their faeces, which may contaminate drinking water sources through rain runoff [5]. Infection can still occur even if concentration of Salmonella in water is low, because the organism escapes the natural host defense mechanisms due to rapid passage of water through the stomach into the intestines without stimulating digestion.
Status of zoonotic Salmonella: The prevalence of salmonellas is varies slightly from one country to another. For example in Nepal 42 out of 300 drinking water samples were positive, with a total of 54 isolates identified to genus level by PCR detection of the virulence genes invA and spvC. The predominant serotypes were Salmonella typhimurium, followed by Salmonella typhi, Salmonella paratyphi A and Salmonella enteritidis [6,7].
Salmonellosis is an important but neglected disease in sub Saharan Africa, where food or fecal-oral associated transmissions are the primary causes of infections. The role of water-borne transmission is unclear, although significant prevalence has been demonstrated. For example, 6.5% of Salmonella spp was detected in dug wells in a rural area of Ghana [8]. A study by [9], showed the presence of Salmonella in water supplies to a slum and densely populated communities and a prevalence of 86 to 98.6% was reported in river Nile water in Egypt [10]. In Dar es salaam, Tanzania Salmonella spp. reported from different water bodies were; Salmonella ser. Paratyphi A (96.9%), and Salmonella choleraesuis serovar Choleraesuis (99.5%).
Problem statement and justification
Tanzania faces a serious lack of potable water which forces people to share surface water sources with animals (livestock and wild) on a day-to-day basis [11]. Water bodies are therefore areas where zoonotic diseases, including salmonellosis, may arise. Bukombe district possess a number of livestock, which at some point share same water bodies with humans. At these water bodies infected animals may shed the bacteria through faeces. Also, humans contaminate the water while carrying out different activities including: Bathing, swimming, fishing and irrigation. These situations justify establishing the magnitude of water-borne zoonotic Salmonella species in both multi-purpose water bodies (with high human-animal interaction) and singlepurpose water bodies (for humans use only) in communities such as Bukombe. So far limited studies have been conducted to address this concern in Bukombe district. This study aims at establishing the prevalence of zoonotic Salmonella spp. (serovars Typhimurium and Enteritidis) in the water bodies. Furthermore, the study determined the prevalence of antimicrobial resistance among Salmonella spp. isolated from the water bodies in Bukombe district.
Objectives
General objective: To estimate the burden of zoonotic Salmonella species (serovars Typhimurium and Enteritidis) in water bodies and establish antimicrobial resistance profile of the bacteria isolated from water bodies in Bukombe district.
Specific objectives:
1. To estimate prevalence of zoonotic Salmonella species from single and multi-purpose water bodies in Bukombe district.
2. To establish the antimicrobial resistance profile of zoonotic Salmonella species obtained from water bodies.
3. To compare the prevalence of zoonotic Salmonella spp of multipurpose versus single purpose water bodies in Bukombe district.
Research questions:
1. What is the prevalence of zoonotic Salmonella species in single and multi-purpose water bodies in Bukombe district?
2. What is the antimicrobial resistance profile of zoonotic Salmonella species obtained from the water bodies?
3. Is there any difference in the prevalence of zoonotic Salmonella species in multipurpose and single purpose water bodies?
Literature Review
Zoonotic Salmonella
Salmonella spp. comprise over 2600 serotypes and colonize range of animal hosts such as mammals, amphibians, reptiles, birds and insects [12]. The bacterial genus Salmonella causes a huge global burden of morbidity and mortality. With regard to human disease, salmonellae are divided into typhoidal serotypes (Salmonella enteric var Typhi) and Salmonella enteric var Paratyphi A) and thousands of non-typhoidal Salmonella serotypes [13]. Non-typhoidal Salmonella serotypes affect a wide range of hosts including human hence zoonotic, examples of these zoonotic Salmonella are S. Typhimurium and S. Enteritidis.
Epidemiology of zoonotic Salmonella
Water-borne diseases are a major public health concern worldwide, not only by the morbidity and mortality that they cause, but by the high cost associated with their prevention and treatment [14,15]. There has been increased number of epidemics in sub-saharan Africa, with the tendency towards more typhoid-like with regard to patterns of transmission and virulence [16]. Salmonella enterica var. Typhimurium, ST313 and Salmonella enteritidis are far more important in sub-Saharan Africa. HIV infections in adults, malaria and malnutrition in children have been identified as important risk factors [12].
A distinct genotype of Salmonella entericavar Typhimurium, ST313, has emerged as a new pathogenic clade in sub-Saharan Africa. This genotype might have adapted to cause invasive disease in humans [12]. According to [17], predominant serotypes have been shown to be Salmonella enterica serotype Typhimurium (54.9%) and Salmonella enterica serotype Enteritidis (64; 33.2%). It is estimated that 93.8 million cases of gastroenteritis due to Salmonella species occur globally each year, causing 155,000 deaths [18].
According to the WHO, the mortality of water associated diseases exceeds five million people per year with 50% of these deaths being microbial intestinal infections. Salmonella enterica subsp. Enterica serovar Enteritidis is the most frequently isolated serovar from humans all over the world. Humans can carry the bacteria in the gut without showing signs of disease for considerable periods of time [19]. Water-borne Salmonella spp is becoming a big problem. A study by [20], detected a relatively low contamination frequency of Salmonella spp in ground water around burial sites of culled animals in South Korea. Some studies have revealed high numbers of water-borne pathogens, including 6% Salmonella spp in harvested rainwater, which is gaining acceptance among many countries such as Australia, Germany, and South Africa [21]. Salmonella species loads in borehole water in South Africa were higher than the WHO threshold, which suggests that water from these sources may pose serious health risks if consumed without treatment [22]. It is the aim of this study therefore to estimate the magnitude of zoonotic Salmonella spp in single and multipurpose water bodies in Bukombe district.
Mode of infection and clinical signs
Most human Salmonella infections result in gastroenteritis, and are caused by Salmonella enteric serovar Typhimurium or Salmonella enteric serovar Enteritidis acquired from contaminated food and water. Apart from humans, other animals can also be infected with Salmonella Typhimurium. Some Salmonella spp are important pathogens that can be transmitted to people via food and water and can cause diseases characterized by mild to severe enteric and systemic illness [11]. Salmonella spp can arise from direct or indirect animal contact via an environment contaminated by clinically affected animals that exhibit a higher prevalence of shedding than apparently healthy animals, or transmission can occur through contaminated food and water to humans [23].
Diagnostic methods of Salmonella species
Several diagnostic methods currently in use for Salmonella species; bacterial culture by using selective culture media such as Xylose Lysine Deoxycholate (XLD) and Salmonella Shigella Agar (SSA). However, this method is time consuming and laborious [24].
PCR technology is widely used in detecting the presence of specific Salmonella species by amplification of a specific fragment of nucleic acid, genes such as invA, fliC are frequently targeted [25].
Serological methods are also used for the detection of Salmonella serotypes, one of them is ELISA, which may be indirect or double antibody-blocking assays using a range of antigens such as lipopolysaccharide, flagella and SEF14 fimbrial antigen [26].
Materials and Methods
Study area description
This study was conducted in Bukombe district which is located in the southern part of Geita region at the coordinates 03028’S 031054’E. The district has a total area of 8,055.59 km2. According to the 2012 census, the district has 75,668 cattle, 70,810 goats and 17,000 sheep and a human population of 224,542 individuals. The district is bordered to the East by Mbogwe District (Shinyanga), to the North by Chato District Geita), to the North-west by Biharamulo (Kagera) to the West by Kibondo District (Kigoma) and to the South by Urambo District (Tabora). The district eastern part constitutes Bugelenga, Bukombe, Butinzya, Ushirombo, and Lyamba mgongo wards whereas the western part consists of Namonge, Runzewe Mashariki, Runzewe Magharibi, Igulwa and Ng’anzo. The eastern part of Bukombe has a large number of livestock keeper’s as compared to the western part of the district.
As of 2012 only 40.6% of the households have access to improved drinking water sources such as piped water, tube wells and protected springs. The remaining percentage uses non-improved drinking water sources [2,26]. Prevalence of Salmonella spp and Escherichia coli in raw milk value chain in American Journal of Research Communication.
Study design and sampling techniques
cross-sectional study was done to estimate the prevalence of zoonotic Salmonella species in water bodies followed by the assessment of antimicrobial resistance of Salmonella isolates found in these water bodies in Bukombe district, Tanzania.
Sample size
The sample size was calculated based on the prevalence of the variables of interest with 0.05 sampling proportion assumed to have safe water with the least desired level of precision
n=z2 .p.qe2
Where: n= sample size, Z=standard normal deviation, set at 1.96 corresponding to 95% confidence level, p=proportion in the target population estimate; taken at 0.5
q=1.0-p =0.5, e=degree of precision desired, set at 0.05. Therefore the sample size calculated was as follows:
n=1.962x0.5x0.5/0.052=384 samples
Due to the small number of water bodies in the study area, only 240 water samples were examined, of which 120came from singlepurpose water bodies and the other 120 samples from multi-purpose water bodies.
Sampling and data collection methods
The study was undertaken in 10 wards of Bukombe district namely:Bugelenga, Bukombe, Butinzya, Igulwa, Lyambamgongo, Namonge, Ng’anzo, Runzewe Mashariki, Runzewe Magharibi, and Ushirombo. In each ward two villages were purposively selected based on the number of livestock, making a total of 20 villages. Four water bodies were selected from each village based on the purpose of each water body (single or multipurpose). Water bodies were checked for evidence of human activities and/or livestock depending on the purpose of the water body (Figure 1). Then three water samples were taken from three different water layers of each water body (at the bottom, middle, and surface).

Figure 1: Different water bodies in Bukombe district, single purpose uncovered water well in Namonge village (villagers fetching water and some laundering
clothes) (A) and multipurpose water pond in Bukombe village, (cattle are seen drinking water) (B).
Sampling procedure
For the surface water samples, the bottle cap was aseptically removed and the sterile collecting bottle was submerged to a depth of about 20cm with the opening facing slightly upward. The middle and bottom water samples were taken by tying the bottle on a weighted length of a rope by stone or piece of metal of about 500g and then was lowered into the water body to a depth of between one and two meters for the case of middle water sample and to the bottom for the case of bottom water sample. A total of 200ml of water samples were taken in each bottle. The bottles were capped and packed in cool boxes. The samples were temporary stored in refrigerator before being transported while in cool boxes to the Microbiology and Molecular Biology Laboratory, Department of Veterinary Microbiology and Biotechnology at Sokoine university of Agriculture in Morogoro for Laboratory analysis. The duration taken from sampling to laboratory analysis was 36 hours (Figure 1).
Laboratory analysis
Pre-enrichment of Salmonella spp: Volumes of 25ml of the water sample were mixed with 225ml of buffered peptone water and incubated for 18 hours at 37°C.
Enrichment of Salmonella spp: One ml of each peptone water culture was introduced separately into 10ml of Rapp port Vassiliadis broth using a sterile syringe and incubated at 42°C for 24h
Plating on selective medium: Each selective enrichment broth bottle was well shaken and a loopful from each was streaked onto plates of Xylose Lysine Deoxycholate (XLD) agar and the plates aerobically incubated at 37°C for 24h [26]. The characteristic Salmonella spp. colonies on XLD appeared red with black centres. All XLD cultures with characteristic colonies were subcultured on Mac Conkey agar so as to distinguish between lactose fermenters and non-lactose fermenters. The non-lactose fermenting colonies were subcultured on Blood agar to check for swarming property as a way of eliminating Proteus species which possess this feature.
Gram staining: The slide smears from suspect colonies were prepared and then heat fixed. The smears were flooded with crystal violet and left for 1 minute followed by rinsing with gentle stream of water. Then iodine was poured onto the slide for 1 minute followed by rinsing by alcohol acetone for 3 seconds. The slides were flooded with neutral red for a minute and gently washed with a stream of water. Lastly the prepared slides were observed under microscope at x100. Salmonella positive suspects showed gram negative reaction (red/ pinkcolour) with rod shaped organisms (Figure 2).
Figure 2: Gram stained smear showing gram negative rod shaped organisms
at x100.
Biochemical test: The suspected isolates were then inoculated on lysine slants and incubated at 37°C overnight; the positive samples had both the slant and the butt purple in colour. Samples positive for Salmonella species by the lysine test caused the medium colour to change into purple both at the slant and butt. The test helped to differentiate Salmonella species from other Enterobacteriaceae such as Citrobacter and Proteus spp. As shown in figure 3 below. Other tests such as Triple sugar Iron, Indole and Citrate were done to complement the results (Figure 3).

Figure 3: Colour change of lysine medium slants due to the presence of
Salmonella spp. after incubation overnight at 370C.
Antimicrobial susceptibility testing: Antimicrobial sensitivity of the salmonella isolates were determined by the disc diffusion method on Muller Hinton agar. The bacteria were evenly spread on the medium by streaking the wire loop into all directions and planes. Four classes of antimicrobials namely: penicillins (ampicillin 10μg); cephalosporins (cephalexin 30μg); flouroquinolones (ciprofloxacin 5μg) and aminoglycosides (gentamicin 10μg) which were put at maximum distance from each other to avoid zones of inhibition from coalescing (Figure 4). Isolates were classified as sensitive, intermediate or resistant by measuring diameter of inhibition zone as provided by the Clinical and Laboratory Standards Institute as shown in table 1[27].
Drug
Sensitive
Intermediate
Resistant
Ampicilin
=17
14-16
=13
Cephalexin
=18
15-17
=14
Ciprofloxacin
=21
16-20
=15
Gentamycin
=15
13-14
=12
Table 1: Classification of Salmonella species based on diameter of zone of inhibition in mm.
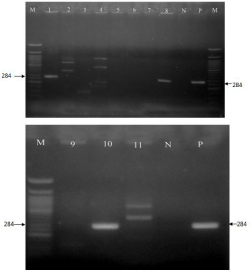
Figure 4: PCR products targeting inv Agene at 284bp, on 1.5% of agarose
gels for 1hr at 110V; ladder (M); positive for Salmonella spp. At 284bp (1, 8
and 10); negative control (N); positive control (P).
Molecular confirmation:
Primers: The following primers were used; 547-569 Forward-GTG ATC TGA AAT CCA GCT TCA AG, 1055-1078 Reverse-AAG TTT CGC ACT CTC GTT TTT GG (for S. Enteritidis), and 139 Forward- GTG AAA TTA TCG CCA CGT TCG GGC AA, 141 Reverse-TCA TCG CAC CGT CAA AGG AAC C (for S. Typhimurium) [28].
Sample preparation and DNA extraction: Samples of suspected Salmonella spp. were taken from individual colonies on the surface of nutrient agar plate, using a sterile plastic loop and resuspended in 4μl of DNA free water. Samples were heated at 100oC for 10min and centrifuged at 1500rpm for 5min to remove cell debris. The supernatant was then transferred into a new tube to be used as a source of DNA.
DNA amplification: The PCR reaction mixture contained 5μl of DNA template, 12.5μl of premix, 0.6μl of forward primer, 0.6μl of reverse primer and 6.3μl of DNA free water in the final volume of 25μl. For invA139 forward primer and invA 141 reverse primer (which targeted invA gene with a band size of 284bp), the reaction mixture was incubated at 950C for 2min, and then cycled 30 times through three steps: denaturing (95oC, 30s), annealing (50oC, 30s), primer extension (720C, 45s). This was followed by a final extension step at 72oC for 7min. For 547-569 forward primer and 1055-1078 reverse primer (which targeted fliC gene with a band size of 500bp), the PCR was carried out under the following conditions: One initial denaturation cycle was done at 95°C for 5min, followed by 30 cycles of 95°C for 40s, 58°C for 20s, and 72°C for 20s and one final extension cycle of 72°C for 7min.
Preparation of agarose gel: The 1.5% agarose gel was prepared by mixing 1.5g of agarose powder with 100ml of TBE (Tris-Borate- EDTA) buffer. This was followed by boiling the gel until became clear, and then it was left to cool. Lastly, Ethidium bromide was added into the gel and poured into the casting tray where the gel was left to solidify.
Loading of PCR products and Electropholesis: 8ul of 50bp DNA marker was loaded into the wells followed by a mixture of 8μl of DNA products and 2μlof blue loading dye. DNA fragments were separated by unidirectional electrophoresis at 110V for 1hr. During loading positive and negative controls were used.
Ethical considerations: The research was conducted according to the Sokoine University of Agriculture Code of Conduct for Research Ethics Section 2.0, Subsection 2.5. The research permit was granted from Faculty Research, Publication and Ethics Committee with reference number SUA/VET/016/25.
Biosafety measures: The study was carried out in compliance with Biosafety Level (BSL-2) necessary for all the Salmonella, except S. typhi where containment equipment and facilities were used during laboratory sample analysis.
Data management and analysis: Data entry and validation was carried out using MS-excel 2007 version. Epi Info TM 7 was used for data analysis. Descriptive statistics such as percentage was used to determine the prevalence/level of Salmonella spp. Chi-square test was used to compare the prevalence of the bacteria between single and multipurpose water bodies. The significance level for all tests was taken at 5% (a = 0.05).
Results
Prevalence of zoonotic Salmonella spp. from water bodies: Lysine test
Biochemical test using lysine reaction revealed that 10out of 240 water samples (4.2%) tested positive for Salmonella species (Table 2). Out of 120 water samples from multipurpose water bodies seven (5.8%) were positive for Salmonella spp. and three (2.5%) of the samples from single purpose water bodies were positive. Half of the positive samples (n=5) were from the eastern part of Bukombe district while the remaining half of the positive samples (n=5) came from the western part of the study area (Table 2).
Ward
Number of samples
positive
Prevalence (%)
Lyambamgongo
24
1
4.1
Iyogelo
24
4
16.7
Bukombe
24
1
4.1
Ushirombo
24
0
0
RunzeweMagharibi
24
1
4.1
RunzeweMashariki
24
1
4.1
Igulwa
24
0
0
Namonge
24
1
4.1
Ng’anzo
24
1
4.1
Bugelenga
24
0
0
Total
240
10
4.2
Table 2: Summary of the prevalence of zoonotic Salmonella isolates characterized by biochemical tests.
The prevalence of zoonotic Salmonella spp from water bodies by molecular method: PCR
All positive isolates (n=10) detected by biochemical test (Lysine reaction) were subjected to molecular characterization by PCR by targeting invA and fliC genes for S. typhimurium and S. enteritidis. Two sets of primers were used, each set targeting one gene. Only 1.3% (n=3) of 240 samples possessed the invA gene hence S. typhimurium. None of the isolates contained fliC gene therefore absence of S. enteritidis. Of the 120 samples from multipurpose water bodies, 1.6% (n=2) were S. typhimurium, where as 0.8% (1/120) sample from single purpose water bodies hadinvA gene (Figure 4).
The antimicrobial resistance profile of Salmonella spp. from water bodies
All three molecular confirmed Salmonella typhimurium isolates (100%) were resistant to penicillin (ampicillin) and cephalosporin (cephalexin), but were sensitive to aminoglycosides (gentamicin) and fluoroquinolones (ciprofloxacin) as shown in figure 5. Gentamicin and ciprofloxacin showed evident zones of inhibition that ranged from 21mm to 38mm (Table 3) (Figure 5).
Drugs
Isolate1
Isolate 2
Isolate 3
Gentamycin
22
24
21
Cephalexin
0
0
0
Ciprofloxacin
24
38
24
Ampicilin
0
0
0
Table 3: Diameters of zone of inhibition of the three S. typhimurium isolates in mm.
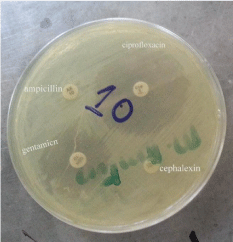
Figure 5: Antimicrobial discs showing zones of inhibition for ciprofloxacin and
gentamicin.
Variation of prevalence between the multipurpose and the single purpose water bodies
Both biochemical and molecular analyses revealed a varying prevalence between multipurpose (pond) and single purpose (well) water bodies. Biochemical tests showed that 5.8% (n = 7) of 120 water samples from multipurpose water bodies were positive for Salmonella spp. and 2.5% (n=3) of 120 samples from single purpose water bodies were positive. Molecular analyses confirmed that out of 120 samples from multipurpose water bodies, 1.6% (n=2) were positive for S. typhimurium and 0.8% (n=1) out of 120 samples from single purpose water bodies were positive for S. typhimurium. Statistical comparison between the two types of water bodies showed no significant difference in prevalence (in both biochemical and molecular analysis) (P>0.05) after data analysis by Epi info 7 software.
Discussion
The prevalence of zoonotic Salmonella isolates from water in Bukombe district
This study has established that the overall prevalence of zoonotic Salmonella species from water bodies was 1.3% suggesting that water borne salmonellosis might be a challenge in Bukombe district. This prevalence (1.3%) of zoonotic Salmonella observed from water in this study is much lower compared to study done by Mwang’onde, et al. which recorded a prevalence of 99.5% in Dares salaam in Tanzania. The latest study on water in Tanzania reported the prevalence of zoonotic Salmonella in northern Tanzania to be 26% [11]. Elsewhere, zoonotic Salmonella have been reported in water with the prevalence of 54.2% in Uganda [29], 49.4% in Cameroon [30]. 29.6% in Ghana [31]. In Burkina Faso (2% to 31%) [32], and Australia 3% [33,34]. Isolated in Morocco, three serotypes of S. enterica: Serotype Blockley (43.8 %), Kentucky (29.8%) and Senftenberg (26.3%).
Detection of Salmonellae in the corresponding water samples signifies microbiological contamination of open water sources. The presence of Salmonella spp. poses an instant risk to people who drink these waters if untreated [11] and Mwang’onde, et al. One of the sources of salmonellae in water from the rural areas of Bukombe may be fecal contamination from animals or humans. However in cities and towns contamination rises from percolation and infiltration of contaminants through the soil under waste disposal sites including pitting latrines [35]. A study done in Kondoa, Tanzania by [36], mentioned limited water supplies, coupled with the lack of hygienic and unreliable water sources, as factors that increase the chance of households using contaminated water during the dry seasons than wet seasons. Higher bacterial concentrations in the water sources are consistent with the scarcity of water due to continual fecal contamination mainly by livestock, wild animals and humans. The lower isolation rate of Salmonella spp. in water is probably related to lower incidence of human and animal salmonellosis in the district [37]. The findings of this study indicate that there is a risk for people living in the study area acquiring zoonotic Salmonella from the water sources used for domestic purpose. Water scarcity becomes prominent during the dry season which leads to sharing of water sources and consequent contamination of water sources and hence health risks to humans in Tanzania [38].
Although the prevalence (1.3%) of Salmonella found in this study appears low, the potential health risk to the public cannot be underrated. This is because carrier animals and humans do switch from one water body to another especially during dry seasons when water becomes scarce; which may lead to contamination of more water bodies. Salmonella has been considered a leading cause of water-borne disease outbreaks for the reason that it is commonly isolated from surface waters.
The antimicrobial resistance profile of Salmonella spp. from water bodies
The antimicrobial sensitivity test showed that all salmonella isolates (n=3) were sensitive to amino glycosides (gentamicin) and floroqunolones (ciprofloxacin). In contrast they were resistant to penicillin (ampicilin) and cephalosporins (cephalexin).The latest study done in water in northern Tanzania observed a prevalence of 88% of salmonella isolates being resistant to one or more antibiotics, with 26% of antibiotic resistant Salmonella spp. isolates having the capacity of transferring their resistance traits to Escherichia coli [11]. A part from drug resistant water-borne Salmonella isolates have been found in other samples; [39], observed resistance of salmonella isolates from beef to ampicillin (6.3%) and100% sensitivity to gentamicin. Elsewhere in Africa, drug resistant salmonella have been reported: In Morocco, (49.1%) and displayed resistance to ampicillin, nalidixic acid, sulfonamide compounds and tetracycline [34]. Multiple drug resistant (MDR) salmonella strains were also detected in surface water in developed countries [40]. Development of resistant Salmonella isolates in water from Bukombe district may have been attributed by improper usage of antibiotics in humans and animals such as over dosage and under dosage of these drugs [41]. Resistant strains eventually find their way into water bodies via human and animal faeces, or through in appropriate disposal of antibiotics into the environment [42,43], which ultimately reach water bodies through water runoff during rainy season [44]. Moreover, unregulated availability and low cost of these drugs are said to speedup rates of resistance [45]. Isolation of drug resistant Salmonella species from water is a public concern, taking into consideration the fact that these water bodies are shared between humans and animals during the dry seasons.
Variation in prevalence of Salmonella spp. isolates between the multipurpose versus the single purpose water bodies
For a while now, studies have labeled water bodies as a “one health” tool (where humans, livestock and wildlife animals interact) which plays an important role in disseminating zoonotic diseases such as salmonellosis. This is due to the fact that humans, livestock and wildlife converge at water sources and contaminate the water with zoonotic pathogens [46]. Water scarcity forces people and animals to use the same water sources for drinking and bathing and laundering, which leads to serious contamination of drinking water and increased risk of zoonotic diseases [47]. It is therefore worth saying that water bodies with high interaction (multipurpose water bodies) have high chances of being contaminated than those with no/limited interaction (single purpose water bodies). This study observed a prevalence of 1.6% of Salmonella species in multipurpose water bodies and 0.8% in single purpose water bodies. However, data analysis concluded that the difference in prevalence between the two types of water bodies is not statistically significant. Other studies have demonstrated high prevalence of Salmonella isolates in water wells. In Nigeria, three different water sources were contaminated with Salmonella enteric serovars with 35.0% being recorded in well water, while 71% of isolates were isolated from uncovered wells [48- 51]. The presence of Salmonella spp. From well water could have been attributed to contamination via unclean utensils used for fetching water and contaminated surface runoff during rainy season especially because the samples in this study were taken from uncovered wells.
Conclusions and Recommendations
Based on the results of this study in Bukombe district, the following are conclusions and recommendations to be drawn:
1. The observed prevalence of zoonotic Salmonella spp. In water was not as high as reported in other studies, but it is enough to raise public health concern. This calls for research to isolate and identify the zoonotic Salmonella species in livestock animals and humans in an effort to establish sources of the contaminant in water bodies. It is, therefore, important for hospitals to consider salmonellosis in differential diagnosis while dealing with diarrheal conditions.
2. The three molecular confirmed Salmonella typhimurium isolates showed drug resistance to ampicillin and cephalexin.
3. The study concluded that there was no difference in the prevalence of Salmonella species between multi- and single purpose water bodies.
4. There is a need for the authorities responsible for combating drug resistance in humans and animals to carry out educational campaigns to raise public awareness on causes of antimicrobial resistance and how to minimize it.
Acknowledgement
I am sincerely grateful to the Almighty Creator, God, who blessed my family and I with the gift of sound health throughout my studies. My heartfelt thanks go to my supervisors, Prof. Robert S. Machang’u and the late Dr. Huruma N. Tuntufye both of the Department of Microbiology, Parasitology and Immunology, College of Veterinary and Medical Sciences, Sokoine university of Agriculture, for their immense and close supervision, encouragement, discussion and constructive criticism during the development of this work. It was an honor to have the privilege to work under their supervision. I am also very thankful to Prof. Robinson H. Mdegela for having played a big role in refining the title of this work. His help is widely acknowledged.
This work would not have been accomplished without the assisting hand from my true friend, Dr. Salum Ahmed, who tirelessly supported me in the research from sample collection to laboratory work. I hereby give my thanks for his terrific assistance.
I am indebted to Mr. Jeremiah Mugusi and Mr. George Makingi for their enormous technical assistance during laboratory analysis of samples. Their time and skills did not go unnoticed, I sincerely thank them.
I also thank my loving classmates Mr. YusuphAron, Dr. Emil Mkemwa, and Dr. Glory Mkunde. I thank them for whatever resources they dedicated to make sure that this work brings fruitful results.
I thank all twenty villages in Bukombe as well as the district authority of Bukombe for allowing me to carry out this study in their areas. Their cooperation during the study is greatly noticed. Special thanks go to village executive officers, chairpersons and hamlet chairpersons for their valuable time while assisting me in locating water bodies in their respective areas. I also recognize the help rendered by the District Veterinary Officer of Bukombe, Dr. Sudi M. Kundyela who ensured that I got permission to conduct this work in his jurisdictional area. I thank him for such commitment.
This work was sponsored by One Health for Central and Eastern Africa (OHCEA) through the Sokoine University of Agriculture (SUA) and Higher Education Students Loan Board (HESLB), through the Mwl. Julius K. Nyerere University of Agriculture and Technology (MJNUAT). The invaluable financial backing is highly accredited.
Dedication
I dedicate this work to the beloved late Dr. Huruma Nelwike Tuntufye for his eagerness to render me memorable and treasured professional backup during the development stages of this work, his soul rest in eternity, AMEN.
References
- Mwang’onde BJ, Tibuhwa DD, Namkinga LA and Kweka EJ. Characterization of Salmonella species from water bodies in Dar-Es-Salaam city, Tanzania. J Health Biol Sci. 2013; 1: 16-20.
- Tanzanial National Bureau of Statistics. Basic demographic and socioeconomic profile Geita region. Dar es salaam, Government printer. 2016: 12-169.
- Radostits OM, Gay CC, Hinchcliff KW and Constable PD. Veterinary medicine: A textbook of the diseases of cattle, sheep, goats, pigs and horses. Edinburgh, Saunders elsevier. 10th ed. 1989: 160-167.
- Coquard D and Exeninger A. Routine detection of Salmonella Species in water: Comparative. Journal of Aoac International Aoac International. 1999; 82: 871-876.
- Waage AS, Vardund T, Lund V and Kapperud G. Detection of low numbers of Salmonella in environmental water, sewage and food samples by a nested polymerase chain reaction assay. Journal of Applied Microbiology. 1999; 87: 418-428.
- Bhatta DR, Bangtrakulnonth A, Tishyadhigama P, Saroj SD, Bandekar JR, Hendriksen RS. Serotyping, PCR, phage-typing and antibiotic sensitivity testing of Salmonella serovars isolated from urban drinking water supply systems of Nepal. Letters in Applied Microbiology. 2007; 44: 588-594.
- Bhasin S, Shukla AN, and Sharad S. Population dynamics of Salmonella in tropical river kshipra India with relation to water quality. European Journal of Pharmaceutical and Medical Research. 2015; 2: 461-481.
- Dekker DM, Krumkamp R, Sarpong N, Frickmann H, Boahen KG, Frimpong M. Drinking water from dug wells in rural Ghana-Salmonella contamination, environmental factors, and genotypes. International Journal of Environmental Research and Public Health. 2015; 12: 3535-3546.
- Haque A, Ahasan S and Rahman MM. Sanitary quality and public health significance of drinking water obtained from different communities. Bangladesh Research Publications Journal. 2010; 4: 226-234.
- El-Taweel GE, Moussa TAA, Samhan FA, El-Senousy WM and El-Lathy MA. Nested PCR and Conventional techniques for detection of Salmonella spp. in river Nile water, Egypt. Egypt Journal of Microbiology. 2010; 45: 63-76.
- Lyimo B, Buza J, Smith W, Subbiah M and Call DR. Surface waters in northern Tanzania harbor fecal coliform and antibiotic resistant Salmonella spp. capable of horizontal gene transfer. African Journal of Microbiology Research. 2016; 10: 348-356.
- Feasey NA, Dougan G, Kingsley RA, Heyderman RS and Gordon MA. Invasive non-typhoidal salmonella disease: An emerging and neglected tropical disease in Africa. The Lancet. 2012; 379: 2489-2499.
- Laupland KB, Schønheyder HC, Kennedy KJ, Lyytikäinen O, Valiquette L and Galbraith J. Salmonella enterica bacteraemia: A multi-national populationbased cohort study. BMC Infectious Diseases. 2010; 10: 1471-2334.
- Brenner FW, Villar RG, Angulo FJ, Tauxe R and Swaminathan B. Salmonella nomenclature. Journal of Clinical Microbiology. 2000; 38: 2465-2467.
- Ramírez-Castillo FY, Loera-Muro A, Jacques M, Garneau P, Avelar-Gonzáslez FJ, Harel J, et al. Waterborne pathogens: Detection methods and challenges. Pathogens (Basel, Switzerland). 2015; 4: 307-334.
- Singletary LA, Karlinsey JE, Libby SJ, Mooney JP, Lokken KL, Tsolis RM, et al. Loss of multicellular behavior in epidemic African Non-typhoidal Salmonella enterica serovar Typhimurium ST313 Strain D23580. mBio. 2016; 7: 1-11.
- Kariuki S, Revathi G, Kariuki N, Kiiru J, Mwituria J, Muyodi J, et al. Invasive multidrug-resistant non-typhoidal Salmonella infections in Africa: Zoonotic or anthroponotic transmission? Journal of Medical Microbiology. 2006; 55: 585- 591.
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ and Hoekstra RM. The global burden of nontyphoidal Salmonella Gastroenteritis. Clinical Infectious Diseases. 2010; 50: 882-889.
- Cabral JPS. Water microbiology. Bacterial pathogens and water. International Journal of Environmental Research and Public Health. 2010; 7: 3657-3703.
- Joung HK, Han SH, Park SJ, Jheong WH, Ahn TS, Lee JB, et al. Nationwide surveillance for pathogenic microorganisms in groundwater near carcass burials constructed in South Korea in 2010. International Journal of Environmental Research and Public Health. 2013; 10: 7126-7143.
- Dobrowsky PH, De Kwaadsteniet M, Cloete TE, and Khan W. Distribution of indigenous bacterial pathogens and potential pathogens associated with roof-harvested rainwater. Applied and Environmental Microbiology. 2014; 80: 2307-2316.
- Palamuleni L and Akoth M. Physico-chemical and microbial analysis of selected borehole water in Mahikeng, South Africa. International Journal of Environmental Research and Public Health. 2015; 12: 8619-8630.
- Hoelzer K, Switt AIM, and Wiedmann M. Animal contact as a source of human non-typhoidal salmonellosis. Veterinary Research. 2011; 42: 1-28
- Chiu CH and Ou JT. (1996). Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. Journal of Clinical Microbiology. 1996; 34: 2619-2622.
- Barrow PA. Serological diagnosis of Salmonella serotype enteritidis infections in poultry by ELISA and other tests. International Journal of Food Microbiology. 1994; 21: 55-68.
- Lubote R, Shahada F and Matemu A. Prevalence of Salmonella spp and Escherichia coli in raw milk value chain in Arusha, Tanzania. American Journal of Research Communication. 2014; 2: 1-13.
- Patel JB, Cockerill FR, Alder J, Bradford PA, Eliopoulos G and Hardy DJ. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational iupplement. CLSI. 2014; 34: 50-56.
- Herrera-leo S, Mcquiston JR, Usera MA, Fields PI, Garaizar J and Echeita MA. Multiplex PCR for distinguishing the most common phase-1 flagellar antigens of Salmonella spp. Journal of Clinical Microbiology. 2004; 42: 2581- 2586.
- Afema JA, Byarugaba DK, Shah DH and Atukwase E. Potential sources and transmission of Salmonella and antimicrobial resistance in Kampala, Uganda. PLOS One. 2016; 11: 1-21.
- Henriette AB, Ebiane NM, Olive NE, Kweyang T, Elvis TR, Jean-christel M and Thomas N. Occurrence of Salmonella spp in surface waters of Yaoundé, Cameroon. Journal of Environmental Science and Water Resources. 2012; 1: 243-250.
- Feglo PK and Dakorah MP. Contribution of dug-out wells to Salmonella dissemination in Kwaebibirem District of Ghana. European Journal of Scientific Research. 2017; 13: 124-134.
- Traoré O, Nyholm O, Siitonen A, Bonkoungou IJO, Traoré AS and Barro N. Prevalence and diversity of Salmonella enterica in water, fish and lettuce in Ouagadougou, Burkina Faso. BMC Microbiology. 2015; 15: 1-7.
- Ahmed W, Sawant S, Huygens F, Goonetilleke A and Gardner T. Prevalence and occurrence of zoonotic bacterial pathogens in surface saters determined by quantitative PCR. Elsevier. 2009; 43: 4918-4928.
- Setti I, Rodriguez-castro A, Pata MP, Cadarso-suarez C, Yacoubi B, Bensmael L, et al. Characteristics and dynamics of Salmonella contamination along the Coast of Agadir , Morocco. Applied and Environmental Microbiology. 2009; 75: 7700-7709.
- Abdulkadir RS, Mahmoud AM, Adnan A, Shamsuddeen U, Adamu RT and Yunusa I. Effect of pit latrine leaks on shallow well water. International Journal of Microbiology and Application. 2015; 1: 46-51.
- Aller DM, Lwiza KMM, Pizer ME and Aller JY. Water source quality in northern and central Tanzania: Implications for rural communities. Journal of Environmental Protection. 2013; 4: 389-404.
- Arvanitidou M, Papa A, Constantinidis TC, Danielides V and Katsouyannopoulos V. The occurrence of Listeria spp. and Salmonella spp. in surface waters. Microbiological Research. 1997; 152: 395-397.
- Kusiluka LJM, Karimuribo ED, Mdegela RH, Luoga EJ, Munishi PKT, Mlozi MRS, et al. Prevalence and impact of water-borne zoonotic pathogens in water, cattle and humans in selected villages in Dodoma Rural and Bagamoyo districts, Tanzania. Physics and Chemistry of the Earth. 2005; 30: 818-825.
- Murutu R. Prevalence of antibiotic resistant Salmonella isolated from beef in Arusha. American Journal of Research Communication. 2016; 4: 1-16.
- Levantesi C, Bonadonna L, Briancesco R, Grohmann E, Toze S, and Tandoi V. Salmonella in surface and drinking water: Occurrence and water-mediated transmission. Food Research International. 2012; 45: 587-602.
- Mshana SE, Matee M and Rweyemamu M. Antimicrobial resistance in human and animal pathogens in Zambia, Democratic Republic of Congo, Mozambique and Tanzania: An urgent need of a sustainable surveillance system. Annals of Clinical Microbiology and Antimicrobials. 2013; 12: 28.
- Fick J, Sodersttrom H, Lindberg RH, Phan Ch, Tysklind M and Laesson DGJ. Contamination of surface, ground, and drinking water from pharmaceutical production. Environment Toxicology and Chemistry. 2009; 28: 2522-2527.
- Larsson DGJ and Fick J. Transparency throughout the production chain-a way to reduce pollution from the manufacturing of pharmaceuticals? Regulatory Toxicology and Pharmacology. 2009; 53: 161-163.
- Kistemann T, Claßen T, Koch C, Dangendorf F, Fischeder R, Gebel J and Exner M. Microbial Load of drinking water reservoir tributaries during extreme rainfall and runoff. Applied and Environmental Microbiology. 2002; 68: 2188- 2197.
- Van den Boogaard J, Semvua HH, Boeree MJ, Aarnoutse RE and Kibiki GS. Assessment of antibacterial sale by using the Anatomic Therapeutic Chemical classification and Defined Daily Dose methodology in Moshi Municipality, northern Tanzania. Tanzania Journal of Health Research. 2010; 12: 208-212.
- Jofre J, Blanch AR and Lucena F. Water-borne infectious disease outbreaks associated with water scarcity and rainfall events. In S. Sabater and D. Barceló (Eds.), Water scarcity in the Mediterranean: Perspectives under global change Berlin: Springer Verlag Berlin Heidelberg. 2010: 147-159.
- Mazet JAK, Clifford DL, Coppolillo PB, Deolalikar AB, Erickson JD and Kazwala RR. A “One Health” approach to address emerging zoonoses: The HALI project in Tanzania. PLOS Medicine. 2009; 6: 1-6.
- Akinyemi KO, Iwalokun BA, Foli F, Oshodi K and Coker AO. Prevalence of multiple drug resistance and screening of enterotoxin (stn) gene in Salmonella enterica serovars from water sources in Lagos, Nigeria. Public Health. 2011; 125: 65-71.
- Momba MNB, Malakate VK and Theron J. Abundance of pathogenic Escherichia coli, Salmonella typhimurium and Vibrio cholerae in Nkonkobe drinking water sources. Journal of Water and Health. 2006; 4: 289-296.
- Bahwere P, Levy J, Hennart P, Donnen P, Lomoyo W, Dramaix-Wilmet M and De Mol P. Community-acquired bacteremia among hospitalized children in rural central Africa. International Journal of Infectious Diseases. 2001; 5: 180-188.
- Stone GG, Oberst RD, Hays MP, McVey S and Chengappa MM. Detection of Salmonella serovars from clinical samples by enrichment broth cultivation- PCR procedure. Journal of Clinical Microbiology. 1994; 32: 1742-1749.