
Special Article – Microbial Resistance
J Bacteriol Mycol. 2018; 5(5): 1078.
Profile of Microbial Resistance in a Clinical Laboratory of the Municipality of Itajaí krusei
Steps JVDM¹*, Valcarenghi D², Schmeling TB³ and Lamb F4
¹University of Vale do Itajaí, Rua Uruguay, Brazil
²Master’s Lecturer in Undergraduate Courses in Biomedicine and Pharmacy, University of Vale do Itajaí, Brazil
³Professor of the Physical Education course, University of Vale do Itajaí, Brazil
4Technical Head of the Laboratory of Clinical Analysis - LEAC, Brazil
*Corresponding author: João Victor de Mattia Passos, University of Vale do Itajaí, Rua Uruguay, Brazil
Received: August 01, 2018; Accepted: September 11, 2018; Published: September 18, 2018
Abstract
For some time, the presence of microorganisms with mechanisms of resistance were hospital exclusivity. Over the years, it has come into our daily lives, thanks to infections originating from collection points, health units and others, but also by the irrational use of antimicrobial. This work aims to trace the profile of microorganisms with isolated resistance mechanisms of patients attended by a clinical laboratory, the LEAC, located in the municipality of Itajaí, in the period between 2015 and 2016. Such mechanisms were identified in microbiological cultures of uroculture, blood culture, cerebrospinal fluid, coproculture, secretion in general and liquids such as ascitic and pleural. According to the results obtained in this study we can observe the prevalence of AmpC production, isolated, frequently, in cultures of secretion generally evidenced by the high number of strains of Pseudomonas aeruginosa from hospital samples and Escherichia coli isolated from ambulatory urocultures. Based on these data, we may suspect that Pseudomonas aeruginosa has a possible intrinsic resistance to AmpC expression. Microbial resistance is an emerging situation and it is necessary to implant care in these environments so that it does not occur, thus reducing financial costs and the number of deaths.
Keywords: Antibiotic; Bacterium; Outpatient; Hospital
Introduction
Bacteria are unicellular microorganisms, prokaryotes, contain both DNA and RNA, in their genetic material [1], have a short life span, are able to adapt rapidly to changes in the environment and are found in numerous anatomical sites and can be part of their microbiota or are pathogenic, thus causing infections [2].
Most infections are caused by the imbalance between the human microbiota and host defense mechanisms. This can occur due to the pathological mechanisms of the patient, invasive procedures and changes in the microbial population, which are often generated by the use of antibiotics [3]. Antibiotics are drugs used for the treatment of bacterial infections, produced in order to destroy or inhibit microorganism proliferation [4]. Its use has led to a reduction in the morbidity and mortality rates of these infections worldwide [5], but the use of these drugs causes the pathogen to develop antimicrobial defense mechanisms, making them resistant, causing continuous infections and ineffective treatments [4].
Health Care-Related Infections (IRAS) consist of adverse effects of invasive procedures performed in health facilities, such as Basic Health Units (BHU), collection points, emergency care units, hospital settings [6].
Microbial resistance is a worldwide problem, evidenced by the increase in the number of cases of IRAS caused by microorganisms resistant to the antibiotics used by the population [7].
Today, the indiscriminate use of antibiotics, that is, without medical prescription or without completing the treatment, provides the appearance of multiresistant microorganisms, provoking an outbreak of these biological agents in the population. Resistance cases, which were the reality of hospitals, are spreading to any health establishment, thus being routinely classified [8].
Bacterial resistance to antibiotics is one of the most significant public health problems today, due to the natural phenomenon of selective pressure exerted by its use [9].
When the bacteria present resistance to several classes of antibiotics, they are denominated as multiresistant bacteria, present mainly in Pseudomonas aeruginosa, in enterobacteria and Staphylococcus aureus. They can spread to patients through the hands of health professionals or contaminated equipment, because each Hospital Infection Control Commission (CCIH) has the responsibility to establish a policy of control of these microorganisms [10].
The microorganism acquires antimicrobial resistance through the acquisition of resistance genes, being divided into two types: intrinsic or innate resistance and acquired. The first is a characteristic of a species and dependent on its properties, we can mention the bacterium Escherichia coli that presents intrinsic resistance to vancomycin. Already acquired resistance occurs through mutation or gene transfer [11].
Data from the World Health Organization (WHO) show that more than half of the world’s antibiotic prescriptions are inadequate. The inappropriate consumption of this type of drug is one of the causes that leads to the increase of microbial resistance [7].
The main mechanisms of resistance found today were divided by their morphotinory classification for Gram negative bacteria: through the production of enzymes such as AmpC (AmpC gene), Extended Spectrum Beta-lactamase (ESBL), carbapenemases (KPC) and resistance to polymyxin (MCR1 gene); and for Gram positive bacteria: Methicillin Resistant Staphylococcus aureus (MRSA), Vancomycin Resistant Enterococcus (VRE), Vancomycin Staphylococcus aureus (VRSA), and to terminate the MLSB Inducible Resistance (Macrolides, Lincosamine and Streptogramin B) [12].
AmpC-type beta-lactamase is mediated by plasmids expressing the AmpC gene, which originates from the transfer of the chromosome containing the gene, which has the ability to hydrolyze all beta-lactam antibiotics to 3rd generation cephalosporins. Identified mainly in Escherichia coli, Klebsiella spp, Proteus mirabilis [14].
One of the forms of mechanism of resistance to beta-lactam antibiotics is the production of enzymes, the most important is betalactamase [15].
Extended-spectrum beta-lactamase (ESBL) is mediated by non-inducible plasmid genes, which characterize TEM and SHV enzymes, capable of hydrolyzing the beta-lactam ring present in certain antimicrobials. Due to its spectrum of action can affect the broad spectrum beta-lactams, such as cephalosporins of up to 4th generation and monobacchaeam [16]. This mechanism of resistance is often found in enterobacteria, such as Escherichia coli, Klebsiella pneumoniae, Enterobacter sp [17].
Carbapenemase production (KPC) is a mechanism observed mainly in the bacterium Klebsiella pneumoniae, capable of hydrolyzing a broad spectrum of beta-lactamamines, including penicillins, cephalosporins, monobactams and carbapenems. It has high potential for dissemination because its location is in the plasmid, facilitating its dissemination [18].
Polymyxin is a widely used antibiotic for the treatment of infections by enterobacteria, such as Escherichia coli, Acinobacter baumanii, Pseudomonas aeruginosa and Klebsiella pneumoniae that present resistance mechanism. Over the years, these bacteria have acquired resistance to this antimicrobial, as it changes the posttranslational modification of lipopolysaccharide (LPS), altering the permeability of polymyxin at the target site. This situation is due to the acquisition of a plasmid containing the colistin resistance gene, mcr-1 [19].
One of the mechanisms expressed by Staphylococcus aureus is the coding of altered penicillin-binding protein (PBPs), termed PBP2a, consequently, no affinity to all beta-lactam antibiotics occurs. Such resistance is known as methicillin-resistant Staphylococcus aureus (MRSA) and the gene encoding this synthesis is mecA [20].
The mechanism of resistance involving the genus Enterococcus resistant to Vancomycin (VRE) occurs through the acquisition of the van gene (the types vanA and vanB are the prevalent) [21], through mutation or gene transfer, present in transposons [22]. There is a mutation in the terminal peptide, resulting in a D-alanyl-D-lactate residue, instead of D-alanyl-D-alanyl [23] which results in an altered peptideoglycan in the cell wall provoking little affinity to vancomycin, inhibition of its action on the blockade of cell wall synthesis [24].
Resistance to the MLSB group (Macrolide, Lincosamide, Streptogramina B) causes alteration in ribosomal antibiotic binding sites mediated by erm (erythromycin methylase ribosomal) gene [25]. This gene encodes enzymes that reduce the binding of MLSB class antibiotics to ribosomes [26]. Clindamycin is an antibiotic used for the treatment of Staphylococcus aureus infections because it acts on RNA and ribosomes. However, misuse of MLSB class antimicrobials led to resistance by several mechanisms, such as decreased permeability, altered ribosomal action site, enzymatic action and efflux pumps [27].
This work aims to trace the microbial resistance profile of patients attended by a clinical laboratory in the city of Itajaí to provide epidemiological data that help in the control of cases of bacterial resistance. Also, it can be used as a subsidy for protocol changes in the selection of antimicrobials in relation to the optimization of the laboratory routine.
Materials and Methods
This was a cross-sectional descriptive research based on a documental analysis of data from patients attending the Laboratory of Clinical Analyzes (LEAC) at the University of Vale do Itajaí (UNIVALI), located in the city of Itajaí - SC, in the 2015 to 2016.
In this period, 9123 requests were made for microbiological cultures, such as uroculture, coproculture, liquor, blood culture, secretion in general (composed of ear secretion, abscess, osteomilite, wound, tracheal aspirate and catheter tip) and liquids such as ascites and pleural .
The samples were categorized as outpatient clinics: from the request of doctors to the Unified Health System (SUS), Family and Community Health Unit, dental clinic (both located at UNIVALI), private agreements; and hospital: from medical requests from Hospital University Little Angel (HUPA).
The laboratory provided the data regarding the date of collection, culture results and antibiograms of patients who were identified exclusively with the protocol number generated by their management system, TASY®, thus ensuring safety and confidentiality. These data were tabulated, using absolute and relative frequency, using Microsoft Excel®.
Ethics
The project was approved by the Research Ethics Committee (CEP) in humans of the University of Vale do Itajaí - UNIVALI, under the opinion number 2,267,570/2017.
Results and Discussion
In the years 2015 to 2016 a total of 9% (828/9123) microbiological cultures with bacterial growth of hospital and outpatient samples were performed by the Laboratory of Clinical Analysis (LEAC). Of the positive cultures, 15% (126/828) presented microorganism with resist Staphylococcus aureus ance mechanism, with a predominance of 62% (78/126) AmpC, 25% (31/126) ESBL, 6% (8/126) MRSA and Resistance to Macrolide, Lincosamide and Streptogramin B (RMLSB) group, 1% (1/126) KPC and 0% VRE, VRSA and Polymyxin Resistance. Only 1 (one) multiresistant microorganism characterized by AmpC and ESBL has been identified, this is Escherichia coli isolated from an outpatient uroculture.
In a study conducted in Iran by Ghotaslou et al. (2018) [28] analyzed samples from medical centers, obtaining 52.8% of microorganisms with resistance mechanism. Such a value is higher than the one found here, 15%. Still the prevalence was ESBL 42.7%, followed by 14% AmpC production and 4.9% carbapenemase.
Figure 1 shows the frequency of resistance mechanism of hospital samples from the years 2015 and 2016. A total of 66% (83/126) of positive microbiological cultures with resistance mechanism was identified. Of these, there was a predominance of AmpC, 60% (50/83), accompanied by 23% (19/83) ESBL, 10% (8/83) MRSA, 7% (6/83) RMLSB and finally 0% KPC and VRE (Figure 1).
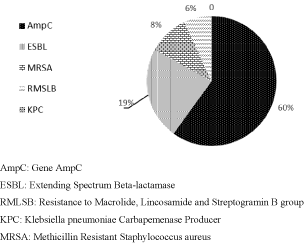
Figure 1: Resistance Mechanism Frequency in hospital samples in the years
2015 and 2016.
The presence of multiresistant microorganisms from the hospital environment is more prevalent, compared to community acquired infections due to the use of antimicrobials. Such microorganisms acquire resistance, often by the acquisition of plasmids containing resistance mediator-encoding genes [29].
Figure 2 shows the frequency of resistance mechanism in outpatient samples in the years 2015 and 2016. It is observed that 34% (43/126) of positive results for microorganisms with resistance mechanism. As in the outpatient samples, the prevalence of the same mechanism was found in the hospital samples, 65% (28/43) of AmpC, followed by 28% (12/43) ESBL, 5% (2) MLSB group resistance, 2% (1) KPC, and 0% MRSA (Figure 2).
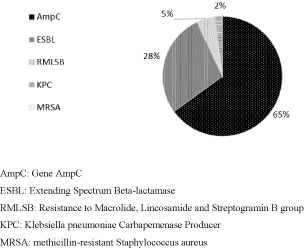
Figure 2: Resistance Mechanism Frequency in outpatient samples in the
years 2015 and 2016.
Multidrug-resistant microorganisms are usually associated with IRAS. But some of these can spread in the community, being one of the main causes of infections. This reality is a great risk to the population, because the community can acquire a bacterial resistance pattern that the antimicrobial used by the population becomes ineffective, thus necessitating the intervention of antibiotics that were just a hospital reality [30].
A study in Lebanon reported that the most frequent resistance mechanism was ESBL production, both in hospitalized patients (30.2%) and in the community (13.4%) [31]. These results are partially disagree according to the ones found here since the AmpC mechanism was more frequent in both environments.
It can be noticed that there was only 1 (one) microorganism that presented the mechanism of production of carbapenemase, from an outpatient sample. According to Alencar et al. (2016) [32] such mechanism is likely to occur in hospitalized or depressed patients with urinary tract infections and wounds, bacteremia, pneumonia, chronic atrophic rhinitis, arthritis, enteritis, meningitis in children, and sepsis . When related to the result found here, it is possible to suspect that the microorganism was isolated in one of the public assisted by the laboratory, which is a retirement home for the elderly. Such patients often go through the hospital environment and many of them are under the use of catheters and bladder catheters.
Based on the above results, we can understand that the hospital samples present a greater number of strains with resistance mechanism compared to outpatient ones. This situation is in agreement with the study of Matta et al. (2017) [31], who reports that the occurrence of these microorganisms with resistance mechanisms is prevalent in the hospital environment due to the situation that the patient is involved, such as immunosuppression, surgical procedures, age, sex, use of prolonged antibiotic therapy , neoplastic diseases and others.
Tables 1 and 2 demonstrate the relationship of the expression of resistance mechanisms to the frequency of the isolated microorganism. In the hospital samples the production of AmpC was identified more frequently in the Pseudomonas aeruginosa strains, 68% (34/50), as can be observed in Table 1. Also, in the same table we can identify that the production of ESBL was more frequent in Klebsiella pneumoniae, 37% (7/19). In the outpatient samples (Table 2), AmpC expression was more frequently identified in strains of Escherichia coli 53% (15/28). Khameneh et al. (2016) [11] is partially in agreement with this study, since it indicates that the microorganisms that present / display more frequency of mechanism of resistance are Pseudomonas aeruginosa, Acinobacter baunamanii, Escherichia coli and Klebsiella pneumonia.
Isolated Microorganism
Resistance Mechanism
AmpC
ESBL
MRSA
RMLSB
KPC
N
%
N
%
n
%
n
%
N
%
Pseudomonas aeruginosa
34
68
1
6
NA
-
0
-
0
-
Staphylococcus aureus
NA
-
NA
-
8
100
2
33
0
-
Klebsiella pneumoniae
2
4
7
37
NA
-
0
-
0
-
Enterobacter cloacae
4
8
2
10
NA
-
0
-
0
-
Enterobacter spp
1
2
4
21
NA
-
0
-
0
-
Escherichia coli
2
4
2
10
NA
-
0
-
0
-
Klebsiella spp
4
8
3
15
NA
-
0
-
0
-
Streptococcus spp
0
-
0
-
NA
-
4
67
0
-
Pseudomonas spp
3
6
0
-
NA
-
0
-
0
-
Total
50
60
19
23
8
10
6
7
0
-
Table 1: Relationship between isolated microorganisms with the production of resistance mechanism in hospital samples in the years 2015 and 2016.
Isolated Microorganism
Resistance Mechanism
AmpC
ESBL
MRSA
RMLSB
KPC
n
%
n
%
n
%
n
%
n
%
Escherichia coli
15
53
4
34
NA
-
0
-
0
-
Klebsiella pneumoniae
3
12
6
50
NA
-
0
-
1
100
Klebsiella spp
2
7
1
8
NA
-
0
-
0
-
Pseudomonas aeruginosa
2
7
0
-
NA
-
0
-
0
-
Enterobacter spp
3
12
0
-
NA
-
0
-
0
-
Proteus mirabilis
1
3
1
8
NA
-
0
-
0
-
Staphylococcus aureus
0
-
0
-
0
-
2
100
0
-
Serratia spp
1
3
0
-
NA
-
0
-
0
-
Morganella morganii
1
3
0
-
NA
-
0
-
0
-
Total
28
65
12
28
NA
-
2
5
1
0
NA: Not applicable
AmpC: Gene AmpC
ESBL: Extending Spectrum Beta-lactamase
MRSA: methicillin-resistant Staphylococcus aureus
RMLSB: Resistance to Macrolide, Lincosamide and Streptogramin B group
KPC: Klebsiella pneumoniae Carbapemenase Producer
Table 2: Relationship between isolated microorganisms with the production of resistance mechanism in outpatient samples in the years 2015 and 2016.
According to Brooks, et al. (2014) [33], the bacterium Pseudomonas aeruginosa is a non-fermenting and opportunistic Gram negative bacillus. Commonly found in hospital settings, it causes wound infections, burns, patients undergoing chemotherapy, urinary and respiratory tract infections. It is considered one of the problems in hospitals, due to large cases of multiresistance, many of the cases arising from the production of beta-lactamases. According to the characteristics of the microorganism and the opportunism that the literature presents, corroborates with the results obtained in the isolation of the same biological sample processed in the cultures presented in this work.
Escherichia coli is a Gram-negative bacillus glucose fermenter often isolated in clinical laboratory and is associated with infectious diseases in several anatomical sites of man. It is considered one of the main etiological agents of sepsis, wounds, pneumonia in hospitalized patients immunosuppressed, meningitis in newborns and frequent in urinary tract infections [34].
The cultures with the most resistance mechanism were hemoculture in the hospital samples with 46% (38/83) and in the outpatient samples it was 100% uroculture. Blood culture is the gold standard for patients with suspected bacteremia. It also allows the isolation of the causative agent, consequently making changes in the antimicrobial therapy and generating a prognosis. It is the most used in the hospital environment [35]. In this study, blood culture was the most prevalent in hospital samples, reflecting that most microorganisms with resistance mechanism are isolated from the blood, which causes a worrying situation.
In this work, Escherichia coli with resistance mechanism was isolated in urocultures, and that the majority belongs to the female sex. According to Araújo et. al. (2012) [36], women, especially young women, are more likely to acquire compared to men, but the most predisposed population are children, pregnant women, elderly and immunocompromised patients, and the use of bladder catheters.
Following the protocol of BRASIL (2008) [37], sulfamethoxazole / trimethoprim can be administered for the treatment of outpatient Urinary Tract Infections (UTIs), and in the case of microorganisms with a resistance mechanism the quinolone class is used. Based on this work, the treatment of the patients treated by the LEAC would not suffer, because the mechanisms observed reach the beta-lactam antibiotics and not, the classes that are listed as first choice in the protocol of BRASIL (2008) [37].
However, when the expression of AmpC is related, it is the mechanism of resistance prevalent in hospital and outpatient samples. In outpatient samples, this mechanism was found in only 100% urocultures identified by the high number of strains of Escherichia coli corresponding to 53% (15/28). In table 03, it is observed that in the hospital environment the most frequent culture was in the secretions in general with 48% (24/50) was the prevalence of 59% (20/34) Pseudomonas aeruginosa (Table 3).
Type of Culture
Expression of AmpC
Hospital Cultures
n
%
Ascitic Fluid
1
2
General Secretion
24
48
urinalysis
8
16
Hemocultura
17
34
Total
50
64
Table 3: Relation of the isolation of the AmpC expression with the type of hospital cultures in the years of 2015 and 2016.
According to Santos (2014) [38], 63 samples of Pseudomonas aeruginosa were isolated from patients attending the Hospital Universitário de Santa Maria, since 68% (43) presented resistance mechanism mediated by AmpC production. The most prevalent cultures, with resistance mechanism, came from the respiratory tract 41% (23) (tracheal aspirate, sputum and bronchoalveolar lavage), followed by secretion in general 22% (14) (operative wound secretion, synovial fluid , peritoneal fluid, catheter tip). This study agrees with the mechanism of resistance prevalent, because both are AmpC. However, it contradicts itself regarding the clinical material, because in our study there was no case of isolated resistance of this sample.
According to David and David (2016) [30] bacterial infections originating from the community caused by the bacterium Pseudomonas aeruginosa with resistance mechanism are considered uncommon, in agreement with our study. In a cohort of 60 patients with community-acquired bloodstream infections, all strains of this microorganism were susceptible to meropenem, piperacillin / tazobactam and ceftazidime. This study corroborates the work in question, since there were few samples with isolation of Pseudomonas aeruginosa with resistance mechanism in outpatient samples.
A study carried out by Klein and Goulart (2008) [39] from a laboratory in Uruguaiana analyzed 48 laboratory reports with isolation of Staphylococcus aureus from a hospital environment. Of these samples, 46% had a mechanism of resistance to methicillin, often resulting from secretion in general (59%), followed by urine (9%), catheter and sputum (4%). These results are in disagreement with those found here in this work, it is possible to relate the amount of Staphylococcus aureus strains with only 8 cases resistant to methicillin isolated from blood culture and secretion in general.
In Lago e Fuentefria and Fuentefria, (2010) [17] analyzed positive microbiological cultures with ESBL resistance mechanism. 1546 (31.6%) positive cultures from the total of 4888 were identified, as 54.2% were enterobacteria, of these 46% were ESBL-producing Escherichia coli, followed by Enterobacter sp. ESBL isolates (n=208) were identified mainly from urine samples (n=79), followed by tracheal secretion (n=23). The results of this study are also different, such as the frequency of Escherichia coli (in our study were only 19% (23/126), and the predominant mechanism was AmpC, however it is similar to the type of culture, such as uroculture.
Different results were found in Donoso (2009) [40]. About 370 microbiological cultures were positive, most of them coming from bronchial aspirates. Klebsiella pneumoniae was isolated in 80% of cultures, different from the study presented here, which was the Pseudomonas aeruginosa producing AmpC. For Gram-positive bacteria, microorganisms such as Staphylococcus aureus and Staphylococcus epidermidis, of the MRSA type, were prevalent. This is discouraging to the work, since there were few microorganisms with such a mechanism of resistance.
Infection by these microorganisms is a serious situation, as it results in increased morbidity and mortality, length of stay, reduction or loss of protection for patients undergoing various procedures, difficulty in choosing the appropriate antimicrobial for treatment . Consequently, it generates excessive expenditure, overloading the services of the public health system [8].
In the world, microbial resistance has been increasing over the years, a major challenge for physicians. Although excessive use of these drugs has begun to exert selective pressure, resistance was unknown in the past, the extent of new antibiotics [41].
Conclusion
In the study period, there was a prevalence of the production of the AmpC mechanism in both samples. In the hospital, the high number of Pseudomonas aeruginosa isolated from cultures of secretion in general was observed, suggesting the expression of an intrinsic resistance. In outpatient clinics, it was Escherichia coli in urocultures, corroborating with the literature and fomenting epidemiological data.
It is concluded that microbial resistance is a frequent situation in hospital environments, due mainly to the indiscriminate use of antibiotics, since it is necessary that there is control to decrease its spread, thus reducing financial costs and cases of death. LEAC microbial resistance numbers are lower than other national and global laboratories.
The information contained in this study is important for health professionals with the intention of promoting protocol changes in the selection of antibiotics for the treatment of infections and patient care, in order to reduce the number of cases of microbial resistance.
References
- Jr Winn WC, Allen SD, Janda WM, Koneman EW, Procop GW, Schreckenberger PC, et al. Microbiological Diagnosis, Text and Atlas Colotrido. Guanabara Kooban. 6th Koneman. 2014.
- Santos NQ. The Bacterial Resistance in the Context of Hospital Infection. Text Context Nursing. Federal University of Santa Catarina. 2014: 64-70.
- Pereira MS, Souza ACS, Tipple AFV, Prado MA. The Infection Hospital and its Implications for Nursing Care. Texto contexto Nursing. 2005; 14: 250-257.
- Costa APM, Pires Ekds, Souza GLFD, Fonseca L, Silva NCDS. Antimicrobial Resistance and the Implementation of RDC 20/2011. 2011.
- Costa ALP, Silva Junior ACS. Bacterial Resistance to Antibiotics and Public Health: A Brief Literature Review. Scientific station. UNIFAP. Macapá. 2017; 7: 45-57.
- National Health Surveillance Agency (Brazil). Measures to Prevent Infections Related to Health Care. Module 4. 2013.
- National Health Surveillance Agency (Brazil). Know the Anvisa Actions to Combat Microbial Resistance.
- National Health Surveillance Agency (Brazil). National Plan for the Prevention and Control of Microbial Resistance in Health Services Brasília. 2017.
- Loureiro RJ, Roque F, Rodrigues AT, Herdeiro MT, Ramalheira E. The Use of Antibiotics and Bacterial Resistances: Brief Notes on Its Evolution. Revista Portuguesa de Saude Publica. 2016; 34: 77-84.
- National Health Surveillance Agency (Brazil). Interventions and Measures of Prevention and Control of Microbial Resistance. Module 5.
- Khameneh B, Diab R, Ghazvini K, Bazzaz Bsf. Breakthroughs in Bacterial Resistance Mechanisms and the Pontecial. Microbial Pathogenesis. 2016; 95: 32-42.
- National Health Surveillance Agency (Brazil). Microbial Resistance - Mechanisms and Clinical Impact, Module 3.
- Gude Mj, Seral C, Sáenz Y, Cebollada R, González-Dominguez M, Torres C, et al. Molecular Epidemiology, Resistance Profiles and Clinical Features in Clinical Plasmid-mediated AmpC- producing Enterobacteriaceae. International Journal of Medical Microbiology. 2013; 303: 553-557.
- Zhang Q, Zhang W, Li Z, Bai C, Li D, Zheng S, et al. Bacteremia due to AmpC-lactamase-producing Escherichia coli in Hospitalized Cancer Patients: Risk Factors, Antibiotic Therapy, and Outcomes. Diagnostic Microbiology and Infectious Disease. 2017; 88: 247-251.
- Rodrigues FCB, Mesquita ARCD. Amplified Spectrum Beta-lactamase Enterobacteria (ESBL) in Kidney Transplanted Urocultures: Frequency and Resistance Profile. Brazilian Journal of Clinical Analysis. 2016; 48: 129-132.
- Gram-Negative Bacilli: Beta-lactamases. In: Rossi, F, Andreazzi, D, Bacterial Resistance-Interpreting the antimicrobial. Publisher Atheneu. Sao Paulo. 2005: 65.
- Lake A, Fuentefria SR, Fuentefria DB. Enterobacteria producing ESBL in Passo Fundo, State of Rio Grande do Sul, Brazil. Journal of the Brazilian Society of Tropical Medicine. Deep step. 2010; 43: 430-434.
- Rechenchoski Dz, Dambrozio AML, Vivan ACP, Schuroff PA, Burgos TDN, Pelisson M, et al. Antimicrobial Activity Evaluation and Comparison of Methods of Susceptibility for Klebsiella pneumoniae carbapenemase (KPC) - producing Enterobacter spp. Isolates. Brazilian Journal of Microbiology. 2017; 48: 509-514.
- Jeannot K, Bolard A, Plésiat P. Resistance to Polymyxins in Gram-negative Organisms. International Journal of Antimicrobial Agenes. 2017; 49: 526-535.
- Lima MFP, Borges MA, Sr Parent, JR Victoria RC, Oliveira ME. Staphylococcus aureus and Hospital Infections - Literature Review. Revista Uningá Review. 2015; 21: 32-39.
- Holzknecht BJ, Hansen DS, Nielsen L, Kailow A, Jarlev JO. Screening for Vancomycin-resistant Enterococci with Xpert® vanA/vanB: Diagnostic Accuracy and Impact on Infection Control Decision Making. New Microbes and New Infections. 2017; 16: 54-59.
- Vargas AVS, Cordero RB, García, WB, Santamaría FG, Valverde EB. Prevalence and Genotypic Identification of Resistant Vancomycin Enterococci in Patients in a Hospital Setting. Acta Médica Costarricense. 2004; 16-26.
- Silveira GP, Name F, Gesser JC, SÁ MM, Terenzi H. Strategies Used in Combating Bacterial Resistance. New Chemistry. 2006; 29: 844-855.
- Furtado GHC, Martins ST, Coutinho AP, Soares GMM, Wey SB, Medeiros EAS. Incidence of Vancomycin Resistant Enterococcus in a University Hospital in Brazil. Journal of Public Health. 2005; 39: 41-46.
- Moore ZS, Jerris RC, Hilinski JA. High prevalence of inducible clindamycin resistance among Staphylococcus aureus isolates from patients with cystic fibrosis. Journal of Cystic Fibrosis. 2008; 7: 206-209.
- Eksi F, Gayyurhan ED, Bayram A, Karsligil T. Determination of Antimicrobial Susceptibility Patterns and Inducible Clindamycin Resistance in Staphylococcus aureus Strains Recovered from Southeastern Turkey. Journal of Microbiology, Immunology and Infection. 2011; 44: 57-62.
- Silva ACO, Silva RCG, Oliveira SR. Clindamycin microbial resistance in clinical isolates of Staphylococcus derived from blood cultures of hospitalized patients. Jornal Brasileiro de Patologia e Medicina Laboratorial. 2016; 52: 165-170.
- Ghotaslou R, Sadeghi MR, Akhi MT, Asgharzadeh M. Prevalence and Antimicrobial Susceptibility Patterns of ESBL, AmpC and Carbapenemase-producing Enterobactericeae Isolated from Hospitalized Patients in Azerbaijan, Iran. Irian Journal of Pharmaceutical Research. Azerbaijan. 2018; 17: 79-88.
- Levinson W. Fármacos Antimicrobianos: Resistência. In: Levinson W. Medical Microbiology and Immunology. Porto Alegre, ARTMED ed. 10th 2010; 94.
- David VD, David LP. Multidrug-Resistant Bacteria in the Community. 2016; 30: 377-390.
- Matta R, Hallit S, Hallit R, Bawab W, Rougues A, Salameh P. Epidemiology and Microbiological Profile Comparison Between Community and Hospital Acquired Infecctions: A Multicenter Retrospective Study in Lebanon. Journal of Infection and Public Health. 2017: 1-7.
- Alencar MPID, Silva JMD, Vidal ME, Vandesmet LCS. Klebsiella pneumoniae: A Bibliographic Review. Scientific Show in Biomedicine. 2016.
- Carrol KC. Pseudomonas, Acinobacter and Gram-negative Bacteria Uncommon. In: Brooks GF, Carroll KC, Buttel JS, Morse SA, Mietzner TA. Medical Microbiology of Jawetz, Melnick and Adelberg. Porto Alegre, AMGH ed., 26th 2014; 245-248.
- As Enterobacteriaceae. In: Júnior Winn WC, Allen SD, Janda WM, Koneman EW, Procop GW, Schrenckenberger PC, Woods GL, Koneman. Microbiological Diagnosis, Text and Colotid Atlas. Guanabara Koogan: 6th 2012: 208-299.
- Pardinas-Llergo MJ, Alarcón-Sotelo A, Ramírez-Angulo C, Rodríguez-Weber F, Díaz-Greene EJ. Probability of Success in Obtaining a Positive Hemoculture. Internal Medicine of Mexico. 2017; 33: 28-40.
- Araújo K, Queiroz AC. Analysis of the Profile of Agents Causing Urinary Tract Infection and Carrier Patients, Served at Hospital and Maternidade Metropolitano - SP. Journal of Science. 2012; 30; 7-12.
- National Agency of Sanitary Surveillance (Brasil). Treatment of Major Community and Health-Related Infections and Antimicrobial Prophylaxis in Surgery, Module 3.
- Santos SO. Study of Clinical Isolates of Pseudomonas aeruginosa and Acinobacter spp. Multiresistants of the University Hospital of Santa Maria. (Master of Science in Pharmaceutical Sciences). UFSM. 2014.
- Klein G, Goulart LS. Prevalence of Multiresistant Staphylococcus aureus in Biological Samples of Osvaldo Cruz Laboratory, Uruguaiana-RS. Brazilian Journal of Pharmacy. 2008; 89: 121-124.
- Donoso NAA. Bacterial Resistance in the Adult Intensive Care Unit of the Medilase Neiva Clinic - Colombia, between January and December 2008. Revista Facultad de Salud. 2009: 31-37.
- Theuretzabacter U. Global Antibacterial Resistance: The Never-ending Story. Journal of Global Antimicrobial Resistance. 2013; 1: 63-69.