
Special Article - Escherichia coli
J Bacteriol Mycol. 2018; 5(6): 1079.
Characterization and Utilization of Podoviridae Phages for Controlling Pathogenic Escherichia coli Serovar O168 from Ducklings in Egypt krusei
El-Daly RA1,3, Merwad AMA²*, Barakat AB¹, Hassan SE³ and Askora A4
¹Department of Microbiology, Faculty of Science, Ain Shams University, Cairo 11566, Egypt
²Department of Zoonoses, Faculty of Veterinary Medicine, Zagazig University, 44519, Zagazig, Egypt
³Animal Health Research Institute, Zagazig, Sharkia, Egypt
4Department of Microbiology and Botany, Faculty of Science, Zagazig University, 44519 Zagazig, Egypt
*Corresponding author: Abdallah MA Merwad, Elzerah Square, Zagazig, Faculty of Veterinary Medicine, Zagazig University, Egypt
Received: August 13, 2018; Accepted: September 12, 2018; Published: September 19, 2018
Abstract
This study was carried out to isolate and characterize lytic bacteriophages against the multidrug resistant and pathogenic Escherichia coli serotype O168 isolated from ducklings in Egypt; and also to study efficacy of single phage and cocktail phages on In vitro inactivation of E. coli O168. One hundred and fifteen samples from ducklings, including caecal contents, skin, minced breast, gizzard, swabs from mouth, cloacae and inner surface of liver, were collected, preenriched in peptone water and streaked MacConkey agar, and Eosin-Methylene Blue agar. The presumptive isolates of E. coli were subjected for biochemical and serological identifications. Antimicrobial susceptibility testing was performed by the disk diffusion method. Phages against E. coli serotype O168 were isolated from sewage samples using plaque assay test. An evaluation of phages efficacy was achieved separately and in cocktail to control the most multidrug resistant serotype E. coli O168. Three different single plaques with different plaque morphologies and diameters designated as ECa1, ECb1, and ECc1 were picked and chosen for further purification, amplification and characterization. The recovered phages were belonged to the family Podoviridae. The use of cocktail phages (ECa1/ECb1/ECc1) was significantly effective (reductions of 7.4 log CFU/ml 12hrs post treatment) than the use single phage suspensions. This study confirmed the higher efficacy of phage cocktails in controlling the infection of ducklings with multidrug resistant E. coli O168. These phages will reduce mortality in ducklings, and also protect human health from adverse side effects of antibiotic residues.
Keywords: Escherichia coli Serovar O168; Podoviridae Phages; Multidrug Resistance; Ducklings; One-Step Growth Curve; Phage Cocktails
Introduction
Duck meat is the most popular type of food in Egypt and various parts of the world. Ducks are excellent sources of animalderived high quality proteins that contain essential and nonessential amino acids. Pathogenic Escherichia coli infecting poultry is the causative agent of colibacillosis, one of the most important causes of economic losses in industry of poultry worldwide. In Egypt, five serotypes of E. coli (O86, O127, O114, O26 and O78) were identified from gizzard, heart, spleen and muscles of ducks [1]. Some bacterial diseases such as salmonellosis, colibacillosis, and pasteurelliosis infect a variety of organ systems with involving the alimentary tract. Escherichia coli is a food borne pathogen, and has a public health concern [2]. Recently, there has been attention that some multidrug resistant E. coli have represented a worldwide multiple food borne disease disorders related to the contaminated food consumption [3]. Also, the misuse of antimicrobial agents in poultry production for growth promotion and treatment purposes increases the major interest for the multidrug resistance that are frequently seen among serovars of E. coli and Salmonella [4]. Antimicrobial-resistant food borne pathogens is recognized as an essential public health in the developing countries and this resistance reduces the therapeutic options for treatment of human salmonellosis [5]. These aspects lead to a dangerous threat to public health and the searches for alternative strategies are crucial to overcome the spread of resistant E. coli and their evolution becomes an absolute necessity. Many studies have suggested that bacteriophages are considered as potential therapeutic agents for the biocontrol of multi- drug resistant bacteria in poultry [6,7]. Bacteriophages are viruses, which attack only bacterial cells and assume a dynamic part in the biology of indigenous habitats, affecting the dynamic of prokaryotic population [8]. The advantages of bacteriophages include the interaction ability with its target host cells, its lytic ability and its capability to multiply throughout the process of infection. Therefore, this study was aimed to isolate and characterize different bacteriophages against the multidrug resistant strains of pathogenic E. coli serovar O168 isolated from ducklings in Egypt as well as to evaluate the efficacy of phages separately and in cocktail on the In vitro inactivation for the growth of E. coli O168.
Materials and Methods
Ethical statement
The study was approved by Institutional Animal Care and Use Committee, Zagazig University (ZU-IACUC).
Collection and preparation of samples
One hundred and fifteen samples from ducklings with an age range (1-14 days) were collected from three commercial duckling farms and from Animal Health Research Institute (AHRI), Sharkia, Egypt. The samples included gizzard, liver and spleen and swabs from mouth and cloacae. All samples were collected from euthanized ducklings. Samples were directly transported to the Laboratory under aseptic conditions and kept in an insulated box with ice packs.
Isolation of Escherichia coli, Salmonella species and Pasteurella species
Isolation of E. coli from swab and tissue samples of ducklings was carried out according to the microbiological method listed in Bacteriological Analytical Manual with minor modifications [9]. The swabs from mouth and cloacae were pre-enriched in buffered peptone water; while 25g of each liver, spleen and gizzard was added to 225ml of buffered peptone water, then homogenized using a stomacher at 230rpm for 5min. The pre-enriched swabs and tissue mixtures were incubated at 37°C for 18hrs. One loopful of enriched culture was subjected to streaking on MacConkey agar plates, then followed by an incubation at 37°C for 18-24 hrs. Afterwards, reinoculation of lactose fermenting colonies was performed on the Eosin Methylene blue (EMB) agar plates, then plates were incubated at 37°C for 18-24 hrs. The presumptive E. coli isolates appearing as metallic green colonies on EMB agar plates were subjected for biochemical identification as previously described [10]. Serotyping of E. coli isolates was done using commercial antisera kits ((Difco, Detroit, MI, USA) at the Serology Unit, Animal Health Research Institute, Dokki, Giza, Egypt. Stock cultures of the isolates were stored in 50% glycerol at -80°C.
The isolation of Salmonella species from swab and tissue samples of ducklings was carried out according to the ISO-6579 method [11]. Briefly, 0.1ml of pre-enriched cultures was added to 10ml of Rappaport-Vassiliadis soy peptone (RVS, OXoid CM0669) broth then incubated at 41.5°C for 18-24 hrs. A loopul from enriched broth was streaked on the surface of Xylose Lysine Desoxycholate (XLD, Oxoid, CM0469) agar plate followed by incubation at 37°C for 24hrs. The isolates of salmonellae were biochemically identified. Serotyping of Salmonella isolates was carried out according to [12]. The preenriched swabs were streaked on 5% sheep blood agar plates for isolation of Pasteurella spp., and then incubated at 37°C for 24hrs. The identification of Pasteurella species was done as previously described [13].
Antimicrobial susceptibility testing
A total of 22 bacterial isolates including Escherichia coli serotype O168 (n=6), Salmonella Typhimurium (n=13) and Pasteurella anatipestifer (n=3) were tested for antimicrobial susceptibility against 10 antibiotics by the disk diffusion method [14]. The used antibiotics and their concentrations were including: Amoxicillin-clavulanic acid (30μg), gentamycin (10μg), chloramphenicol (30μg), doxycycline (30μg), imipenem (30μg), streptomycin (30μg), rifampin (10μg), ciprofloxacin (30μg), ampicillin (30μg) and sulfadimethoxineormetoprim (2μg. The test was performed by applying a bacterial inoculum of approximately 2×108 CFU/ml to the surface of Mueller- Hinton agar plate. The disks of antibiotics were distributed on the surface of inoculated agar plate, and incubated at 37°C for 18-24 hrs. The inhibition growth zone around each antibiotic disk is measured to the approximate millimeter. The diameters zone of each antibiotic was explained according to criteria documented by the European Committee on Antimicrobial Susceptibility Testing [15].
Isolation of lytic phages against Escherichia coli serotype O168
Duckling’s droppings from the same retail stores mentioned above and sewage water were collected from several stations in Zagazig, and 10th of Ramadan, then homogenized in 100ml of Luria-Bertani (LB) broth supplemented with 10ml mol/L CaCl2. Those homogenates were subjected to inoculation with overnight cultures (100ml) of 9 E. coli isolates as host cells, and then followed by incubation at 37°C for 24h. The inoculated samples (5ml) were harvested and centrifuged at 12,000x g for 5min. The supernatant was collected and filtrated through 0.45mm pore size filter (Millipore, France) to generate phage lysate. For screening of the existence of lytic phages, the lysate (10ml) was spotted onto the lawn of bacteria prepared from 9 bacterial hosts after overnight culture of on the double layer LB agar plates (Oxoid). These plates were dried at room temperature for 10 minutes, and then were exposed to overnight incubation at 37°C. Lysate showing clear lytic zones at the application point on the double layer LB plates were serially diluted in SM buffer (0.05mol/L Tris- HCl buffer, pH 7.5, containing 0.1mol/L NaCl, 0.008mol/L MgSO4, and 0.01% gelatin). The dilutions (100ml) were subjected to incubation with 100ml of corresponding host(s) at 37°C for 20min, mixed in 4ml molten agar, plated on LB and incubated at 37°C for 24hrs. The existence of plaques was checked in the plates. For phage isolation, we picked up the one clear and the most separated plaque that appeared on the double layer agar plates, then followed by serial dilution in SM buffer and incubated with bacterial host culture and plated on the double layer LB as previously stated. For the isolation of every phage, this procedure was repeated at least 3 times at least to prove the phage purity. The storage of final lysates was performed in SM buffer at 4°C until use.
Electron microscopy
A drop of three purified bacteriophage particles of a highly concentrated suspension (109PFU/ml) was spotted on the carboncoated copper grid, left to adsorb for 2min, and then followed by negative staining with 2% (w/v) uranyl acetate. Phage morphology was observed with electron microscope (Hitachi, H600A) at University of Mansoura, Egypt.
Phage adsorption
Exponential host bacterial cultures of E. coli isolate serotype (O168) were adjusted to a 0.8 O.D. at 600nm (corresponding to a cell density of 109CFU/ml). Phage suspensions (10μl) were added to 10ml of E. coli isolate to obtain a multiplicity of infection (MOI) of 0.001 and then incubated at 25°C [16]. Aliquots of mixture were gathered after zero, 5, 10, 15, 20, 25, 30, 40, 50, 60 and 70 minutes of incubation and chloroform (1% as a final volume) was added. The mixture was centrifuged at 12.000g for 5 minutes and supernatants were immediately filtered by using 0.2 (Millipore, France). Dilution and titration of unadsorbed phages were performed. Incubation of plates was carried out at 37°C followed by examination of plaques after 4-8 hrs. The adsorption was expressed as the percentage decrease of phage titer in the supernatant when compared to the zero time. The phage suspensions without any host cells were utilized as nonadsorption standard for calculations [16]. Three independent assays were performed.
Determination of Phage host range and analysis for Efficiency of Plating (EOP)
After phage isolation, the bacterial strains used in the present study were listed in (Table 3). The stocks of phages were prepared and the host range of isolated phages was detected by the spot testing as previously explained [17]. Each isolated phage was tested against 39 strains of pathogenic bacteria to determine the host range of each phage [18]. Briefly, phage stocks (10ml) were spotted onto each bacterial lawn, then it dried in a clean bench for 10 min and exposed to incubation at 37°C for 24hrs. The lytic zones of phages being visible at the application point were labeled as positive for lytic activities. Two categories of pathogenic bacteria were differentiated according to the clarity of spot including: Clear lysis zone (+), no lysis zone (-). Using the double-layer agar method, the EOP was determined for bacteria with positive spot tests (occurrence of a clear lysis zone) [17]. For each phage, three independent experiments were performed. The EOP was calculated by the following equation: Average PFU on target bacteria/average PFU on host bacteria with the standard deviation for the three measurements as previously explained [19].
Effect of temperature and pH on phages stability
For thermal stability test, the suspensions of phages were added to LB broth at different temperatures of 28, 37, 45 and 65°C, then exposed to incubation at the respective temperatures for one hour. For pH stability test, the suspension of phage was inoculated into the LB liquid medium with a pH range of 3-11 and the phage titer was determined after incubation at 37°C for one hour.
One single-step growth experiments
Adjustment of exponential host bacterial cultures of E. coli strain was done to a 0.8 O.D. at 600nm (corresponding to a cell density of 109 CFU/ml). The phage suspensions (10μl) were mixed with 10ml of the bacterial culture to have a MOI of 0.001 and followed by incubation at 28°C [20]. The mixture was centrifuged at 12.000g for 5 minutes, and then pellet was re-suspended in LB (10ml) at 28°C and then followed by perfect dilution and titration. During incubation at 37°C for 30-minutes, samples were removed intervals up to 5hrs and the phage titers were determined by the double-layer plaque assay. The first set of samples was subjected to dilution before titration. The second set of samples was treated with chloroform (1% V/v) for the release of intracellular phages to determine the eclipse period. Incubation of plates was performed at 37°C and examination of plates was done for presence of plaques after 4-8 hrs [20]. Three independent assays were done.
Challenge of E. coli O168 with three phages separately and in phage cocktail
The phages ECa1, ECb1 and ECc1 were separately tested then phage cocktails were tested (two or three phages mixed together at the same concentration). The two phage cocktails were: ECa1/ECb1, ECa1/ECc1, ECb1/ECc1 and ECa1/ ECb1/ECc1 phages. E. coli O168 was used as host at a MOI of 100 (based in preceding studies using different MOI: 1, 10, 100 and 1000, data not shown). To reach a MOI of 100, 2.5μl of E. coli serotype O168 culture (≈108 CFU.ml-1) and 20μl of phage suspension (109 PFU.ml-1) were added to LB medium (30ml) and then followed by incubation at 37°C without agitation. Two control samples were included in each assay: The bacterial control (BC) and the phage control (PC). The inoculation of BC was not done with the phages and the inoculation of PC was not performed with the phages but without the bacteria. One milliliter of test samples and BC and PC was withdrawn after 0, 2, 4, 6, 8, 10, 12 and 24 hrs of incubation. For all assays, determination of phage titer was carried out in duplicate using the double agar layer method after incubation at 37°C for 4-8 hrs. The reductions in bacterial counts were calculated by the differences in viable counts between the control and phage-treated samples.
Results
Bacterial isolation and identification
In this study, the overall presumptive isolates of E. coli, Salmonella spp. and Pasteurella spp. were detected in 140 of 150 (93.3%) samples of ducklings by plating onto selective media. The identification of all isolates showed that 101 isolates were confirmed as E. coli and 29 isolates were Salmonella spp. and 10 strains were identified into Pasteurella spp. (Table 1). The O-serogrouping of 27 E. coli strains showed that 15 out of 27 (55.6%) strains were O-serogroup untypeable (OUT); while the remaining12 out of 27 (44.4%) were typeable into 9 different groups; O1, O55, O78, O111, O114, O125, O127, O168, and O169 (Table 3). The serotyping of Salmonella isolates was ranging between serotype S. Typhimurium, S. Enteriditis. Also, Pasteurella anatipestifer was identified from ducklings.
Isolated pathogenic bacteria
Study location
Types of samples
E. coli
Salmonella
Pasteurella
Total No. of
spp.
spp.
isolates
Commercial duckling farms and
Cloacal swabs
70
8
2
80
Animal Health Research Institute
Liver
20
2
1
23
Gizzard
2
4
5
11
Mouth swabs
7
11
1
19
Spleen
2
4
1
7
Total isolates
101
29
10
140
Table 1: Isolation of pathogenic bacteria from different samples from ducklings.
Antimicrobial susceptibility testing
The antimicrobial resistance patterns of 22 bacterial isolates including E. coli (n=6), S. Typhimurium (n=13) and Pasteurella anatipestifer (n=3) were determined using 10 types of wide spectrum antibiotic by disc diffusion method (Table 2). Escherichia coli serotype O168 showed 100% resistance to each of the three tested antibiotics (amoxicillin-clavulanic acid, ciprofloxacin and imipenem) and also higher resistance (83.3%) to doxycycline when compared to the other antibiotics. In S. Typhimurium, the percentages of resistance to ampicillin (76.9%) and also for gentamycin (53.8%) were relatively higher than those detected in other tested antibiotics. Moreover, Pasteurella anatipestifer was resistant to sulfadimethoxineormetoprim with a percentage of 66.7%.
Escherichia coli serotype(O168)
Salmonella typhimurium
Pasteurella anatipestifer
(n=6)
(n=13)
n=3)
Antimicrobials (Code, disc concentration)
R (%)*
I (%)
S (%)
R (%)
I (%)
S (%)
R (%)
I (%)
S (%)
Ampicillin (Amp, 30μg)
33.3
66.7
¥ND
76.9
15.3
7.7
ND
ND
100
Amoxicillin-clavulanic acid(AMC, 30μg)
100
ND
ND
38.4
46.1
15.3
33.3
66.7
ND
Gentamycin (CN, 10μg)
ND
83.3
16.7
53.8
46.1
ND
33.3
33.3
33.3
Chloramphenicol (CHL, 30μg)
50
33.3
16.7
46.1
30.7
23.07
ND
33.3
66.7
Doxycycline (DOX, 30μg)
83.3
50
20
ND
46.1
53.9
ND
66.7
33.3
Imipenem (IPM, 30μg)
100
ND
ND
23.07
15.3
61.5
33.3
33.3
33.3
Streptomycin(STR, 30μg)
66.7
33.3
ND
30.7
23.07
46.1
33.3
ND
66.7
Rifampin (RIF, 10μg)
50
83.3
33.3
46.15
30.7
23.07
ND
33.3
66.7
Ciprofloxacin(CIP, 30μg)
100
ND
ND
23.07
30.7
46.1
ND
ND
100
Sulfadimethoxine-ormetoprim (2μg)
66.6
33.3
ND
30.7
61.5
7.6
66.7
33.3
ND
*R: Resistant; I: Intermediate; S: Sensitive. ¥ND: Not detected any susceptibility of the bacterial isolate to the corresponding antimicrobial agent.
Table 2: Percentages of antibiotic resistance for the isolated Escherichia coli, Salmonella typhimurium and Pasteurella anatipestifer from ducklings.
Morphology and electron microscopy of isolated phages
Sewage samples of different stations were used to screen and isolate different bacteriophages active against the most drug resistant E. coli serotype O168 using the plaque assay test technique. Three different single plaques with different plaque morphologies and diameters designated as ECa1, ECb1, and ECc1 were picked and chosen for further purification, amplification and characterization. The isolated phages were assigned to the family Podoviridae on the basis of morphological features. The electron microscopic analysis revealed that phage ECb1 has an icosahedral capsid of approximately 57.1nm in diameter and with no tail. These characteristics suggest that phage ECb1 is a member of the Podoviridae family (Figure 1). Transmission electron microscopy (TEM) showed that ECa1 belonged to the C3 morphotype of the Podoviridae family according to their structure in electron micrograph (Figure 1). The phage had an elongated head of was 138.58x38.36 nm, and the tail was a little long, 16.12nm. While, the phage ECc1 had an icosahedra capsid of approximately 59×59 nm in diameter without tail.
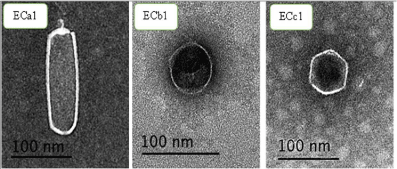
Figure 1: Electron micrographs of E. coli phages. (A) Phage ECa1; (B) Phage ECb1; (C) phage ECc1. The bars represent 100nm.
Host range of isolated phages and their susceptibility to Escherichia coli O168
The host range of the recovered bacteriophages ECa1, ECb1, ECc1 against different bacterial strains was determined by the spot test (Table 3). The results of the spot test indicated that phage ECa1 had the capacity to form completely cleared zones on 21 of the 39 strains, the phage ECb1 form cleared zones on 12 of the 39 strains, and the phage ECc1 form cleared zones on 19 of the 39 bacterial isolates (Table 3). However, EOP results indicated that the three Podoviridae phages formed phage lysis plates only in presence of their hosts. There was a degree of variability in the host range of each phage (Table 3). All tested E. coli strains were susceptible to most E. coli phages. In contrast, when examined individually, phages ECa1 only 53.8 tested isolates. Plaque assay results showed that bacterial strain E. coli O168 was the most sensitive strain to the three phages under investigation. These results explained why E. coli O168 was chosen for the following studies.
Bacterial strains
Spot test
Efficacy of plating (PFU mL-1)
ECa1 ECb1 ECc1
ECa1 ECb1 ECc1
Salmonella typhimurium ATCC 14,028
+
-
+
ND
*ND
0
Salmonella typhimurium ATCC13311
+
-
+
ND
ND
0
Salmonella enteriditis CVA
+
-
+
0
ND
0
S. enteriditis CVE
-
-
+
ND
ND
0
E. coli ATCC 25922
+
-
+
0
ND
0
E. coli ATCC 13706
-
-
+
ND
ND
0
E. coli BC30
+
-
+
0
ND
0
E. coli AE11
-
-
+
ND
ND
0
E. coli AF15
+
+
+
0
0
0
E. coli AD6
+
+
-
0
0
ND
E. coli O157
+
+
+
0
0
0
E. coli O55
+
-
-
ND
ND
ND
E. coli O26
+
-
+
ND
ND
0
E. coli O114
+
+
+
0
0
0
E. coli O111
+
-
-
ND
ND
ND
E. coli O168
+
-
-
9.21 × 10-6
1.55 × 10-1
1.00 × 10-4
E. coli O125
+
+
+
6.75 ×10-4
1.57 × 10-2
2.76 × 10-2
E. coli O78
+
+
+
ND
0
0
E. coli O1
+
+
+
5.8× 10-6
0
0
E. coli O127
+
+
+
0
3.14× 10-2
1.41× 10-2
Enterococcus faecalis
-
+
-
ND
0
ND
S. enteriditis CVB
+
+
-
8.13× 10-6
0
ND
S. enteriditis CVC
+
-
+
0
ND
0
S. enteriditis CVD
-
-
-
ND
ND
ND
Shigellaflexneri
-
-
-
ND
ND
ND
Citrobacterfreundii
-
-
-
ND
ND
ND
Providenciasp
-
-
-
ND
ND
ND
P. vermicola
-
-
-
ND
ND
ND
Proteus vulgaris
-
-
-
ND
ND
ND
Proteus mirabilis
-
-
-
ND
ND
ND
Klebsiellapenumoiea
+
-
-
0
ND
ND
Listeria innocua
-
-
-
ND
ND
ND
L. monocytogenes
-
-
-
ND
ND
ND
Vibrio parahaemolyticus
-
-
-
ND
ND
ND
V. fischeri
-
-
-
ND
ND
ND
Pseudomonas aeruginosa
+
+
+
9.21 × 10-6
1.55 × 10-1
1.00 × 10-4
P. fluorescens
-
-
-
ND
ND
ND
P. gingeri
-
-
-
ND
ND
ND
Shigella
_
+
+
ND
ND
0
*ND: Not detected efficacy of plating.
Table 3: Lytic spectra of three E. coli phages isolates determined on 39 bacterial strains.
One-step growth curve of isolated phages
Phage adsorption assays with phage ECc1, ECb1showed that approximately 38% of the phage particles adsorbed to the host cell after 40min and 97% adsorbed after 70min (Figure 2). For phage ECa146% of particles adsorbed to E. coli O168within 20min and 98% after 50min. One-step growth curve experiment was performed to estimate the latent time period and burst size of the phage, which are the two most important characteristics of phage infection process. The one-step growth curve for the isolated phages ECa1, ECb1, and ECc1 was determined (Figure 2). From the triphasic curves obtained, an eclipse period of 30min, a latent period of 20min and a burst size of 17±2 PFU/host cell were calculated for phage ECa1. The phages ECb1, ECc1are characterized by an eclipse period of 50min, the latent time was at 40min and each infected bacterium produced 117±11 PFU/ host cell.
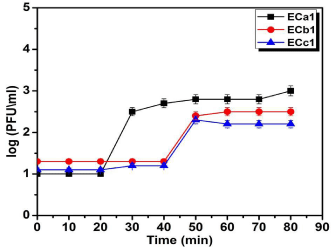
Figure 2: One-step growth curves of ECa1, ECb1 and ECc1 phages in
the presence of E. coli O168 as host. Values represent the mean of three
experiments. Adsorption of bacteriophages ECa1, ECb1 and ECc1 o E. coli
O168. Percentage of unadsorbed phage is the ratio of PFU in the supernatant
to the initial PFU and was determined by tittering an equivalent dilution of
the phage in the absence of host cells. Values represent the mean of three
experiments.
Effects of pH on phage viability
The isolates appeared to be stable at the pH range 5-9 but inactivation is evident at the very low pH of 3 and very high pH level of 11 (Figure 3).
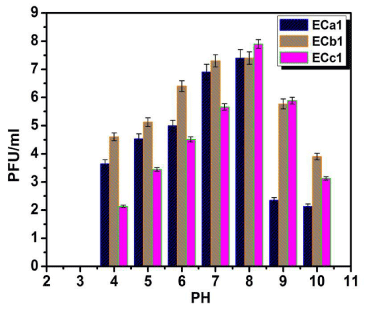
Figure 3: Effect of pH on the stability of phages ECa1, ECb1 and ECc1.
Assays were performed in triplicate and phage titers were expressed as the
mean ± standard deviation.
Challenge of E. coli O168 with three phages separately and phage cocktail
The mixture of E. coli O168 and three phages separately and in a cocktail form was incubated at 37°C up to 24hrs in LB broth to examine the lytic potential as therapeutic agents efficacy. In Figure 4A and B we noticed that the addition of the phages results in differences of bacterial counts to treatment and non-treatment cultures. The bacterial inactivation with the ECa1 phage was 4 log after 4hrs of incubation and after 12hrs was still significantly higher (4.2 log) and the host cells were undetectable at 24hrs incubation (<10CFU/ml) (Figure 4A). The phage concentration was significantly increased after 24hrs the when this phage was subjected to incubation in the presence of the host, relatively to phage control (1.6 log) (Figure 4B). When the ECb1 phage inactivate bacteria, we observed that after 4hrs (3.8 log) and after 12hrs the rate of inactivation was still significantly high (4.2 log) relatively to the bacterial control, the host cells were undetectable at 24hrs incubation (<10 CFU/ml) (Figure 4A). A significant increase of 1.7 log in phage concentration was observed after 24hrs relatively to phage control (Figure 4B). The bacterium inactivation by the ECc1 phage was 2.6 log, 3 log after 4, 6 hrs respectively. After 12hrs, the bacterium inactivation was still high (2.5 log) relatively to the bacterial control (Figure 4A). The phage control remained constant since the beginning of the treatment, but the phage concentration increased considerably in the presence of the host, by 1.8 log, (Figure 4B). The inactivation of the bacteria by the phage ECc1 was less efficient than phages ECa1 and ECb1 until 4hrs of treatment as they were more efficient after 6hrs (Figure 4A).
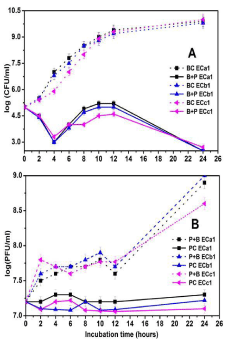
Figure 4: In vitro inactivation of E. coli O168 using the three phages (ECa1,
ECb1 and ECc1) separately in LB broth at a MOI of 100 during 24hrs.
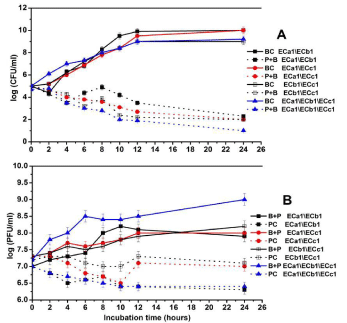
Figure 5: In vitro inactivation of E. coli serotype O168 using a cocktail of three
phages (ECa1, ECb1 and ECc1) in LB broth at a MOI of 100 during 24hrs.
A. Bacterial concentration: BC: Bacteria control; B + P: Bacteria plus phage.
Phage concentration: PC: Phage Control; B + P: Bacteria Plus Phage. Values
represent the mean of three independent experiments; error bars represent
the standard deviation.
When the ECa1, ECb1 phage cocktail was used, the utmost of bacterial inactivation was 2.8 log after 4hrs of incubation, after 12hrs the inactivation was 6.4 log. These results are significantly higher from the values obtained for the phage ECb1 alone (Figure 5A). There was an increasing in phage concentration in the presence of the host during the study time by1.6 log (Figure 5B). Then the phage cocktail ECa1\ECc1 was tested and the maximum of inactivation was 2 and 6.2 log, respectively, after 4hrs and 12hrs of bacterial incubation. The inactivation was significantly higher from the value obtained when phage ECc1 was used alone. However, when compared to ECa1 inactivation the results were not significantly different (Figure 5A). The phage concentration was constant during study period in the absence of the host and in the presence of its host the phage concentration increased significantly by 1 log. There was an increasing in phage concentration in the presence of the host during the study time of 1 log (Figure 5B). Phage cocktail ECb1\ECc1recorded a maximum inactivation after 4 and 12 hrs of bacterial incubation by 1.9, 6.8 respectively. These results were differing from assays with both phages separately. There was an increasing in phage concentration in the presence of the host during the study time of 1.1 log (Figure 5B). The phage cocktail ECa1\ECb1\ ECc1 recorded a bacterial inactivation by 2.5 log achieved after 4hrs of incubation and of 7.4 log after 12hrs. Results are significantly different from the results obtained using ECa1 and ECc1 alone but are comparable with the results obtained using ECb1 phage. The phage titer was constant during the study period and an increase of 1 log in phage concentration in the presence of bacterial host was observed (Figure 5B). The inactivation of the bacteria by the phage cocktails ECa1/ECc1, ECb1/ECc1 was similar, reductions of 2-1.9 log after 4hrs of phage addition, but the phage cocktail ECa1/ECb1 was more effective (reductions of 2.8 log after 4hrs of phage addition) than the to inactivate the bacteria (Figure 5A). However, the inactivation of the bacteria by the phage cocktails ECa1/ECb1\ECc1 was the most effective one as recorded reductions by 3.5 log after 4hrs of phage addition.
Discussion
The present study was conducted to isolate and characterize phages against multi drug resistant isolates of E. coli from ducklings in Egypt and also to investigate the efficacy of isolated phages on the In vitro inactivation of pathogenic E. coli. The O-serogroup typing of 27 E. coli isolates from different sources (swabs from cloaca, mouth, liver, gizzard & spleen) of ducklings showed that 12 out of 27(44.4%) isolates were typeable O-serogroups including O1, O55, O78, O111, O114, O125, O127, O168, and O169. Moreover, S. Typhimurium, S. Enteriditis and Pasteurella atipestifer were identified from different samples of ducklings. The antimicrobial resistance patterns of 22 bacterial strains including E. coli (n=6), S. Typhimurium (n=13) and Pasteurella anatipestifer (n=3) were estimated by 10 types of wide spectrum antibiotic using the disc diffusion method. Escherchia coli serotype O168 was found as the most multidrug resistant strain to the amoxicillin-clavulanic acid, imipenem and ciprofloxacin with a resistance percentage of 100%; while E. coli O168 had a resistance percentage of 83.3% for doxycycline.
Therefore, the use of bacteriophages is considered as a possible alternative tool to antimicrobials against multi drug pathogens [21]. In this study, three bacteriophages of different plaques morphology and size targeting these multidrug resistant E. coli O168 had been isolated from different sewages samples. The isolated phages were named as ECa1, ECb1, and ECc1 and recovered phages showed variations in their abilities to infect and induce lysis for the target pathogen. The morphology of phages revealed different structural features and dimensions by TEM. The isolated bacteriophages were assigned to the family Podoviridae based on their morphological features. The phage ECa1 was belonged to of the C3 morphotype and this was compatible with the results suggested by the E. coli phage phiEco32 [22]. Other analyzed C3 phages growing on Gram negative enterobacteria include the Serratia marcescens phage KSP100 isolated in Japan [23], The Salmonella entrica, Newport phage 7-11 [24,25]. While the ECb1 phage had an icosahedral capsid of approximately 57.1nm in diameter and with no tail and the Phage ECc1 had an icosahedral capsid of approximately 59×59 nm in diameter without tail.
The one step growth curve for these phages was determined and the results of burst sizes and latent periods were estimated for ECa1, ECb1, ECc1 phages. The ECa1 phage showed higher merit for the inactivation of E. coli O168 than the ECb1 and ECc1 phages and had a higher burst size and a shorter latent period than the other two phages. This finding indicated that the use of high burst sizes and short lytic cycles plainly improved the efficiency of phage therapy. The ECa1 phage having the highest burst size (more than 3 times of those of ECb1 and ECc1 phages) and the shortest lytic cycle (half of those of ECb1 and ECc1 phages), was more efficient to inactivate E. coli O168 than ECb1 and ECc1 phages (more 2 log of inactivation, with the maximum of inactivation occurring 2-3 hrs before than those of ECb1 and ECc1 phages). Our findings agreed with previous study which elucidated that burst size upsurge might participate to higher burst size and larger plaques [26].
The survival and stability of phages are influenced by physical and chemical factors such as pH, temperature and storage [27]. Our results showed that the three Podoviridae phages against E. coli O168 could survive at pH range of 4 to 10. However, at extreme low pH (3.0), phage viability was greatly impaired. This was consistent with findings of previous studies, where the incubation at pH value of 3.5, but not pH value of 10 significantly affected the phage survival compared to the more neutral ph value of 7.4 [28,29].
The three phages ECa1, ECb1 and ECc1 infect a semi-similar host range, which can be confirmed by the fact that all of these bacteriophages were isolated using the same strain of E. coli O168 as a target host. The ECa1, ECb1 and ECc1 phages could inactivate different pathogenic bacteria with high efficiency. These results were supported with the previous studies that some phages might infect more than one related species of bacteria or even genus [30,31]. In our study, the isolated Podoviridae phages had a broad host range. The phage ECa1 had the capacity to form completely cleared zones of lysis on 21 of the 39 bacterial strains and the phage ECb1 formed cleared zones of lysis on 12 of the 39 strains and the phage ECc1 formed cleared zones of lysis on 19 of the 39 strains. Furthermore, these phages showed lytic effect on different bacterial strains tested indicating that these bacteriophages were polyvalent. This finding was coincided with previous studies, where the isolated phages had the ability to infect E. coli strains and Salmonella serovars [32,33]. For phage therapy, a wide host range phage that kills multiple species of bacteria would be the same of a broad-spectrum antimicrobial agent.
Thus, the use of phage therapy for inactivation of pathogenic bacteria from infected ducklings could offer a fast and relatively inexpensive technology. Therefore, we tested the phages (ECa1, ECb1 and ECc1) against the most multidrug resistant strain E. coli O168 either used individually or in cocktails. The inactivation of E. coli O168 by the phage ECc1 was less efficient than phages ECa1 and ECb1 until 4hrs of treatment as they were more efficient after 6hrs. Of interest, the phage cocktail ECa1\ECb1\ECc1 recorded a bacterial inactivation by 2.5 log achieved after 4hrs of incubation and of 7.4 log after 12hrs. Results obtained by cocktail of three phages (ECa1\ ECb1\ ECc1) were significantly different than the findings obtained by using ECa1 and ECc1 alone, but were comparable with the results obtained using ECb1 phage. The phage titer was constant during the study period and an increase of 1 log in phage concentration in the presence of bacterial host was noticed. The inactivation of the E. coli O168 by the phage cocktails ECa1/ECc1, ECb1/ECc1 was similar, reductions of 2-1.9 log after 4hrs of phage addition, but the phage cocktail ECa1/ECb1 was more effective (reductions of 2.8 log after 4hrs of phage addition) than the ECa1/ECc1, ECb1/ECc1 to inactivate the bacteria. However, the inactivation of E. coli O168 by the phage cocktails ECa1, ECb1, ECc1 was the most effective one as recorded reductions by 3.5 log after 4hrs of phage addition. Thereby, the level of bacterial reduction was gradually increased when phage cocktail introduced the re-growth of the host cells in the treatment was not observed. This was proved in a study, where phage cocktails were prepared to inactivate pathogenic bacteria, it was important to consider the receptors utilized by the phage to infect the host; and to improve the bacterial inactivation effectiveness [34].
Conclusion
This study showed that ECa1 phage had higher merit for the inactivation of multi drug resistant E. coli O168 than the ECb1 and ECc1 phages. The higher burst size and a shorter latent period of ECa1 compared to the other two phages improved the efficiency of phage therapy. Also, the inactivation of E. coli O168 by the Podoviridae phage cocktails (ECa1/ECb1/ECc1) was the most effective one due to the reductions by 3.5 log after 4hrs of phage treatment. Therefore, Podoviridae phage cocktails could be used as an alternative tool for antibiotics and a biological method to treat mutltidrug resistant E. coli O168 infecting ducklings.
Acknowledgment
The authors would like to acknowledge the members of Serology Unit, Animal Health Research Unit, Doki, Giza, Egypt for their support in serotyping of E. coli isolates.
References
- Darwish WS, Saad Eldin WF, Eldesoky KI. Prevalence, molecular characterization and antibiotic susceptibility of Escherichia coli isolated from duck meat and giblets. J. Food Saf. 2015; 35: 410-415.
- Ahmed AM, Shimamoto T. Isolation and molecular characterization of Salmonella enterica, Escherichia coli O157: H7 and Shigella from meat and dairy products in Egypt. Int. J. Food Microbiol. 2014; 168-169: 57-62.
- Yamasaki E, Watahiki M, Isobe J, Sata T, Nair, GB, Kurazono H. Quantitative detection of Shiga toxins directly from stool specimens of patients associated with an outbreak of enterohemorrhagic Escherichia coli in Japan-quantitative shiga toxin detection from stool during EHEC outbreak. Toxins (Basel). 2015; 7: 4381- 4389.
- Duong VN, Paulsen P, Suriyasathaporn W, Smulders FJ, Kyule MN, Baumann MP. Preliminary analysis of tetracycline residues in marketed pork in Hanoi, Vietnam. Annals New York Acad. Sci. 2016; 1081: 534-542.
- Foley SL, Nayak R, Hanning IB, Johnson TJ, Han J, Ricke SC. Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl. Environ. Microbiol. 2011; 77: 4273-4279.
- Mahony J, McAuliffe O, Ross RP, van Sinderen D. Bacteriophages as biocontrol agents of food pathogens. Curr. Opin. Biotechnol. 2011; 22: 157-163.
- Jassim SA, Limoges RG. Natural solution to antibiotic resistance: Bacteriophages "The Living Drugs". World J. Microbiol. Biotechnol. 2014; 30: 2153-2170.
- Parsley LC, Consuegra EJ, Thomas SJ, Bhavsar J, Land AM, Bhuiyan NN, et al. Census of the viral metagenome within an activated sludge microbial assemblage. Appl. Environ. Microbiol. 2010; 76: 2673-2677.
- S. Food and Drug Administration. Bacteriological Analytical Manual, 8th ed. AOAC International, Gaithersburg MD. 1998.
- Cloud SS, Rosenberger JK, Fries PA, Wilson RA, Odor EM. In vitro and in vivo characterization of avian Escherichia coli. I. Serotypes, metabolic activity, and antibiotic sensitivity. Avian Dis. 1985; 29: 1084-1093.
- International organization for standardization 6579: 2002(e): Microbiology of food and animal feeding stuffs-horizontal method for detection of Salmonella spp. 2002.
- Kauffmann F, Das-Kauffmann W. Antigenic formulas of the Salmonella serovars 8th WHO co-operating Centre for Reference and Research on Salmonella. 2001.
- Bisgaard M, Mutters R Taxon. Characterization of some previously unclassified Pasteurella obtained from the oral cavity of dogs and cats and description of a new species tentatively classified with the family Pasteurellaceae Pohl 1981 and provisionally called. Taxon 16. Acta Pathol. Microbiol. Immunol. Scand. Series B: Microbiology Banner. 1986; 94: 177-184.
- Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966; 45: 493-496.
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, version 4.0. 2014.
- Stuer-Lauridsen B, Janzen T, Schnabl J, Johansen E. Identification of the host determinant of two prolate-headed phages infecting Lactococcus lactis. Virology. 2003; 309: 10-17.
- Adams MH. Bacteriophages. T4-Related Bacteriophage LIMEstone Isolates for the Control of Soft Rot on Potato Caused by ‘Dickeya solani’. Interscience Publishers. New York. 1959: 14-15.
- Carvalho C, Susano M, Fernandes E, Santos S, Gannon B, Nicolau A, et al. Method for bacteriophage isolation against target Campylobacter strains. Lett. Appl. Microbiolol. 2010; 50: 192-197.
- Kutter E. Phage host range and efficiency of plating. In: Bacteriophages. Methods Mol. Biol. 2009; 501: 141-149.
- Mateus L, Costa L, Silva YJ, Pereira C, Cunha A, Almeida A. Efficiency of phage cocktails in the inactivation of Vibrio in aquaculture. Aquaculture. 2014; 424-425: 167-173.
- Akhtar M, Viazis S, Diez-Gonzalez F. Isolation, identification and characterization of lytic, wide host range bacteriophages from waste effluents against Salmonella enterica Food Control. 2014; 38: 67-74.
- Savalia D, Westblade LF, Goel M, Florens L, Kemp P, Akulenko N, et al. Genomic and proteomic analysis of phiEco32, a novel Escherichia coli J. Mol. Biol. 2008; 377: 774-789.
- Matsushita K, Uchiyama J, Kato S, Ujihara T, Hoshiba H, Sugihara S, et al. Morphological and genetic analysis of three bacteriophages of Serratia marcescens isolated from environmental water. FEMS Microbiol. Lett. 2009; 291: 201-208.
- Ackermann HW, Petrow S, Kasatiya SS. Unusual bacteriophages in Salmonella J. Virol. 1974; 13: 706-711.
- Kropinski AM, Lingohr EJ, Ackermann HW. The genome sequence of enterobacterial phage 7-11, which possesses an unusually elongated head. Arch. Virol. 2011; 156: 149-151.
- Abedon ST, Culler RR. Optimizing bacteriophage plaque fecundity. J. Theor. Biol. 2007; 249: 582-592.
- Jonczyk E, Klak M, Miedzybrodzki R, Górski A. The influence of external factors on bacteriophages-review. Folia. Microbiol. 2011; 56: 191-200.
- Nakai T, Sugimoto R, Park KH, Matsuoka S, Mori K, Nishioka T. Maruyama K. Protective effects of bacteriophage on experimental Lactococcus garvieae infection in yellow tail. Dis. Aquat. Organ. 1999; 37: 33-41.
- Nakai T, Park SC. Bacteriophage therapy of infectious diseases in aquaculture. Res. Microbiol. 2002; 153: 13-18.
- Carlton RM. Phage therapy: Past history and future prospects. Arch. Immunol. Ther. Exp. (Warsz). 1999; 47: 267-274.
- Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol and Oceanogr. 2000; 45: 1320-328.
- Bielke LR, Higgins SE, Donoghue AM, Donoghue DJ, Hargis, BM, Tellez G. Use of wide-host- range bacteriophages to reduce Salmonella on poultry products. Int. J. Poult. Sci. 2007; 6: 754-757.
- Parra B, Robeson J. Selection of polyvalent bacteriophages infecting Salmonella enterica serovar Cholerae suis. Elect. J. Biotechnol. 2016; 21: 72-76.
- Chan BK, Abedon ST, Loc-Carrillo C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013; 8: 769-783.