
Editorial
J Bacteriol Mycol. 2018; 5(7): 1083.
Gaseous Ligand Selectivity of Bacterial H-NOX Heme Sensor Proteins
Wu G* and Tsai A
Division of Hematology, Department of Internal Medicine, The University of Texas, Houston, USA
*Corresponding author: Wu G, Division of Hematology, Department of Internal Medicine, The University of Texas -McGovern Medical School, Houston, USA
Received: September 27, 2018; Accepted: October 11, 2018; Published: October 18, 2018
Editorial
For living organisms, diatomic gases Nitric Oxide (NO), Carbon Monoxide (CO), and O2 serve as messengers under various normal physiological and pathological conditions. To respond to the environmental changes of these gaseous messengers, living organisms have developed different types of messenger-sensing systems and heme sensor proteins are important components of these systems. Heme sensor proteins selectively bind NO, CO or O2 causing conformational changes which subsequently alter their interactions with the downstream effector proteins, leading to various responses. Six types of structural folds have been identified for heme sensor proteins and among them, Heme Nitric oxide/OXygen-binding proteins (H-NOXs) are found widely in bacteria [1-4]. Based on the significant sequence homology, the structural fold of heme domain in the β subunit of soluble guanylyl cyclase (sGC) is believed to be very similar to the bacterial H-NOXs. sGC is the only known authentic NO receptor in animals, and its enzymatic activity sGC that converts GTP to cGMP increases several hundred-fold above basal level upon binding to NO [5,6].
In facultative anaerobes, bacterial H-NOXs are encoded as standalone proteins but in the same operons with their effector proteins, putative histidine kinases or diguanylate cyclases; on the other hand, in obligate anaerobes, bacterial H-NOXs are encoded as domains of Methyl-accepting Chemotaxis Proteins (MCP) [2]. Recently the selectivity of several bacterial H-NOXs for gaseous ligands NO, CO, and O2 has been characterized in details through the determination of the kinetic parameters and affinities of their gaseous ligand bindings [7-10]. The bacterial H-NOXs include those from facultative anaerobes, Vibrio cholera (Vc H-NOX), Shewanella oneidensis (So H-NOX), and Nostoc sp. PCC7120 (Ns H-NOX), and the H-NOXs from obligate anaerobes, Clostridium botulinum (Cb H-NOX) and Caldanaerobacter subterraneus (Cs HNOX, previously known as Thermoanaerobacter tengcongensis H-NOX, Tt H-NOX). Each of these bacterial H-NOXs contains a 5-coordinate (5c) high-spin heme ligated to a neutral histidine residue and binds NO and CO with strong affinities [10] (Scheme 1 & Table 1). On the other hand, only Cb and Cs H-NOXs exhibit strong affinities for O2 and form stable oxyferrous complexes [8] (Scheme 1 & Table 1). Therefore, there is likely a correlation among a bacterium’s tolerance of O2, structure of genes, and its H-NOX’s capability to bind O2. In facultative anaerobes, their H-NOXs do not have any measureable affinity for O2 under atmospheric pressure, while on the other hand, H-NOXs from obligate anaerobes exhibit strong affinities for O2 (Scheme 1 & Table 1). Similar to the H-NOXs from facultative anaerobes, sGC totally excludes binding to O2, while binds NO with high affinity, although its affinities for NO and CO are all significantly weaker than those of bacterial H-NOXs (Table 1). The selectivity of these bacterial H-NOXs for gaseous ligands, NO, CO, and O2 can be clearly elucidated by the recently proposed “sliding scale rule” hypothesis [1,10,11]. The “sliding scale rule” is proposed based on extensive bioinformatics analysis of the bindings to NO, CO and O2 of more than a hundred hemeproteins with a 5c high-spin heme with a neutral proximal histidine ligand and model compounds [1,11]. The “sliding scale rule” reveals how the affinities of these hemeproteins for the gaseous ligands, KD(NO), KD(CO), and KD(O2), are modulated collectively by the proteinaceous environments and in some hemeproteins, either KD(NO) or KD(O2) is specifically decreased.
The “sliding scale rule” can be expressed in three statements: 1) model compounds with a 5c ferrous heme ligated to a neutral imidazolate binds NO, CO, and O2 with intrinsic ratios, KD(CO)/ KD(NO) ≈ KD(O2)/KD(CO) ≈ 103 -104; 2) the proteinaceous environments in hemeproteins with a 5c ferrous heme ligated to a neutral histidine simultaneously modulate the affinities of NO, CO, and O2 for heme but keep the ratios, KD(CO)/KD(NO) and KD(O2)/ KD(CO), unchanged as those in the model compounds; 3) specific structural elements in some hemeproteins greatly enhance the affinities for NO or O2, leading to KD(CO)/KD(NO) >> and KD(O2)/ KD(CO) << 103-104 [1,11]. In “sliding scale rule” plot, statements 1 and 2, which can be expressed as log KD(O2)-log KD(CO) ≈ log KD(CO)-log KD(NO), are demonstrated by the approximately parallel linear lines connecting each hemeprotein’s or model compound’s log KD(NO), log KD(CO), and log KD(O2) plotted versus the gaseous ligand type (Figure 1). The term “sliding scale” describes the simple vertical spread of these parallel lines (Figure 1). Statement 3 describes the deviations on either the KD(NO) or the KD(O2) end of such lines in some of the hemeproteins (Figure 1). Three governing factors have been identified for the vertical spread of the parallel log KD(NO)- log KD(CO)-log KD(O2) lines in different hemeproteins and the deviations exhibited in some hemeproteins can be ascribed to two specific factors.
First, the large binding selectivity of a heme among NO, CO, and O2, KD(CO)/KD(NO) ≈ KD(O2)/KD(CO) ≈ 103-104, is rendered by a proximal ligand with weak ligand field, a neutral histidine ligand in hemeproteins and a neutral imidazole ligand in model compounds such as PP(1-MeIm) (Figure 1). Heme ligated to a proximal ligand with significantly stronger ligand field, such as imidazolate, cysteine thiolate, or tyrosine phenolate anion, lose the selectivity for gaseous ligands [1,11]. Second, compared to those of a model compound(s), the KD(NO), KD(CO), and KD(O2) of a hemeprotein are simultaneously modulated through the distal steric hindrance to and the proximal strain of heme imposed by its proteinaceous environment. The steric hindrance describes the kinetic obstruction on the access of a gaseous ligand to heme from the distal side [1,11]. On the other hand, proximal stains applied to heme adjust the bond strength between the Fe atom and its proximal histidine ligand, and therefore modulate the bond formation between the Fe atom and a gaseous ligand from the distal side [1,11]. Distal steric hindrance or proximal strains do not affect the KD(CO)/KD(NO) and KD(O2)/KD(CO) ratios of a neutral histidine-ligated heme. In the “sliding scale rule” plot, the nearly constant KD(CO)/KD(NO) and KD(O2)/KD(CO) ratios in different hemeproteins determine that log KD(NO)-log KD(CO)-log KD(O2) lines of the hemeproteins remain approximately linear and parallel to each other (Figure 1) [1]. On the other hand, the modulations by distal steric hindrance and/or proximal strains introduce 108-109 fold variations in the KDs of hemeproteins for a particular gaseous ligand, leading to large vertical spread along the y-axis of the parallel log KD(NO)-log KD(CO)-log KD(O2) lines of different hemeproteins (Figure 1).
In the “sliding scale plot”, the log KD(NO)-log KD(CO) lines of bacterial Vc, So, Ns, Cb, and Cs H-NOXs are all parallel to but several orders above the corresponding line of model compound PP(1-MeIm) (Figure 1). Moreover, the log KD(NO)-log KD(CO) lines of bacterial H-NOXs cluster in a small range, consistent with the identical structural fold in these H-NOXs. The gaseous ligands bind bacterial H-NOXs essentially unhindered, indicating that modulation for gaseous ligand affinities are primarily due to the proximal strains [10]. Through the simultaneous decrease in the affinities for the gaseous ligands, Vc and So H-NOXs exclude binding to O2; their projected KD(O2)s based on the “sliding scale rule” are well above the atmospheric pressure (Figure 1). The proteinaceous environments in Vc, So, and Ns H-NOXs thus render these facultative anaerobes extraordinary capability to selectively bind NO while excluding the binding to O2 under the atmospheric pressure. Moreover, the affinities of Vc and So H-NOXs for NO are further enhanced, or significantly decreased KD, apparent (NO) (see below). On the other hand, Cb and Cs H-NOXs deviate from the “sliding scale rule”, showing strong affinities for O2. The factors causing these deviations are detailed below.
The two factors that specifically increase hemeproteins’ affinities for O2 and NO are distal hydrogen-bond donor(s) and multiple-step NO binding, respectively [1,11]. Hydrogen bond donor(s), existing on the distal side of heme in some hemeproteins form hydrogen-bond to O2 and stabilize the oxyferrous complex, significantly enhancing these hemeproteins’ affinities for O2. Both Cb and Cs H-NOXs have a distal hydrogen-bond donor, Y139 and Y140, respectively (Scheme 1), and therefore exhibit log KD(O2)s significantly lower than the predicted values extrapolated from their log KD(NO)–log KD(CO) lines, a deviation in “sliding scale rule” plot (Figure 1). A distal hydrogen–bond donor(s) preferentially enhances the binding of O2 but has little or no effect on CO or NO binding. On the other hand, in some hemeproteins, such as Cb, Vc, and So HNOXs, and sGC, 6c NO–heme–His complex is unstable and converts to 5c NO–heme complex due to the rupture of Fe–His bond (Scheme 1). Moreover, in the presence of excess NO, 6c NO heme His complex in these hemeproteins reacts readily with secondary NO to form 5c NO–heme complex (Scheme 1). The 5c NO–heme complexes have significantly slower NO dissociation constants compared to 6c NOheme– His complexes, koff, 5c(NO) << koff, 6c(NO) (Scheme 1). The subsequent chemical steps after the formation of 6c NO–heme in these hemeproteins lead to ultra–high apparent affinities for NO, KD, apparent (NO) << KD(NO) (Table 1). The KD(NO)s of the 6c NO-heme- His complexes in these hemeproteins do follow the “sliding scale rule”, but their much lower KD, apparent(NO)s show apparent deviations (Figure 1).
H-NOXsa
KD(O2), bM
KD(CO), cM
KD(NO), dM
KD, apparent(NO), eM
Cb H-NOX
5.3 × 10-5
6.7 × 10-7
8.0 × 10-11
7.3 × 10-12
Cs H-NOX
4.4 × 10-8
1.6 × 10-7
2.3 × 10-11
N/Ag
Ns H-NOX
1.3 × 10-2
1.4 × 10-6
1.7 × 10-10
N/Ah
So H-NOX
N/Af
8.5 × 10-7
1.5 × 10-10
8.6 × 10-13
Vc H-NOX
N/Af
7.7 × 10-7
2.7 × 10-10
9.1 × 10-13-10-12
sGC
N/Af
2.6 × 10-4
5.4 × 10-8
7.1 × 10-13-10-12
a: from ref. [1,7-10]; b: KD(O2)=koff(O2)/kon(O2); c: KD(CO)=koff(CO)/kon(CO); d: KD(NO) =koff,6c(NO)/kon(NO); e: KD, apparent(NO) = koff, 5c(NO)/kon(NO); f: unmeasurable; g: no 5c NO-heme complex formation is observed in Cs H-NOX; h: Ns H-NOX forms a secondary 6c NO complex with excess NO. The kinetic parameters kon and koff are available in the references.
Table 1: Affinities of H-NOXs for gaseous ligands NO, CO and O2.
Recent studies have shown that many facultative anaerobes, such So, Sw (Shewanella woodyi), and Vc, respond to low level of NO to elicit physiological responses such as biofilm formation, quorum sensing, and symbiosis [12,13]. Although not fully elucidated, these signaling pathways are likely starting from the binding of NO to their H-NOXs which modulates the activities of downstream histidine kinase/ diguanylate cyclase [13]. To selectively bind NO in the presence of often much higher level of O2, the proteinaceous environments of their H-NOX sensors simultaneously down-modulate the intrinsic affinities of heme for each gaseous ligand to exclude the binding of O2 under atmospheric pressure while substantially enhancing the affinities of their H-NOXs for NO through the multiple-step NO binding (Figure 1). The same mechanism also renders sGC dramatic selectivity for NO; its extremely strong apparent affinity for NO but complete exclusion of O2 binding enables it to selectively bind low level of NO in the presence of much higher level of O2 [1]. On the other hand, in obligate anaerobes Cb and Cs, the distal hydrogen-bond donating tyrosine residues in their H-NOXs render them capable of sensing the low level of O2. The capability for sensing low O2 is critical for obligate anaerobes since living environments with extremely low [O2] are crucial for the survival of these organisms. However, studies on physiological responses due to the binding of O2 to their H-NOXs in obligate anaerobes are still lacking. Whether Cb or Cs H-NOX is involved in such signal pathways is currently unclear.
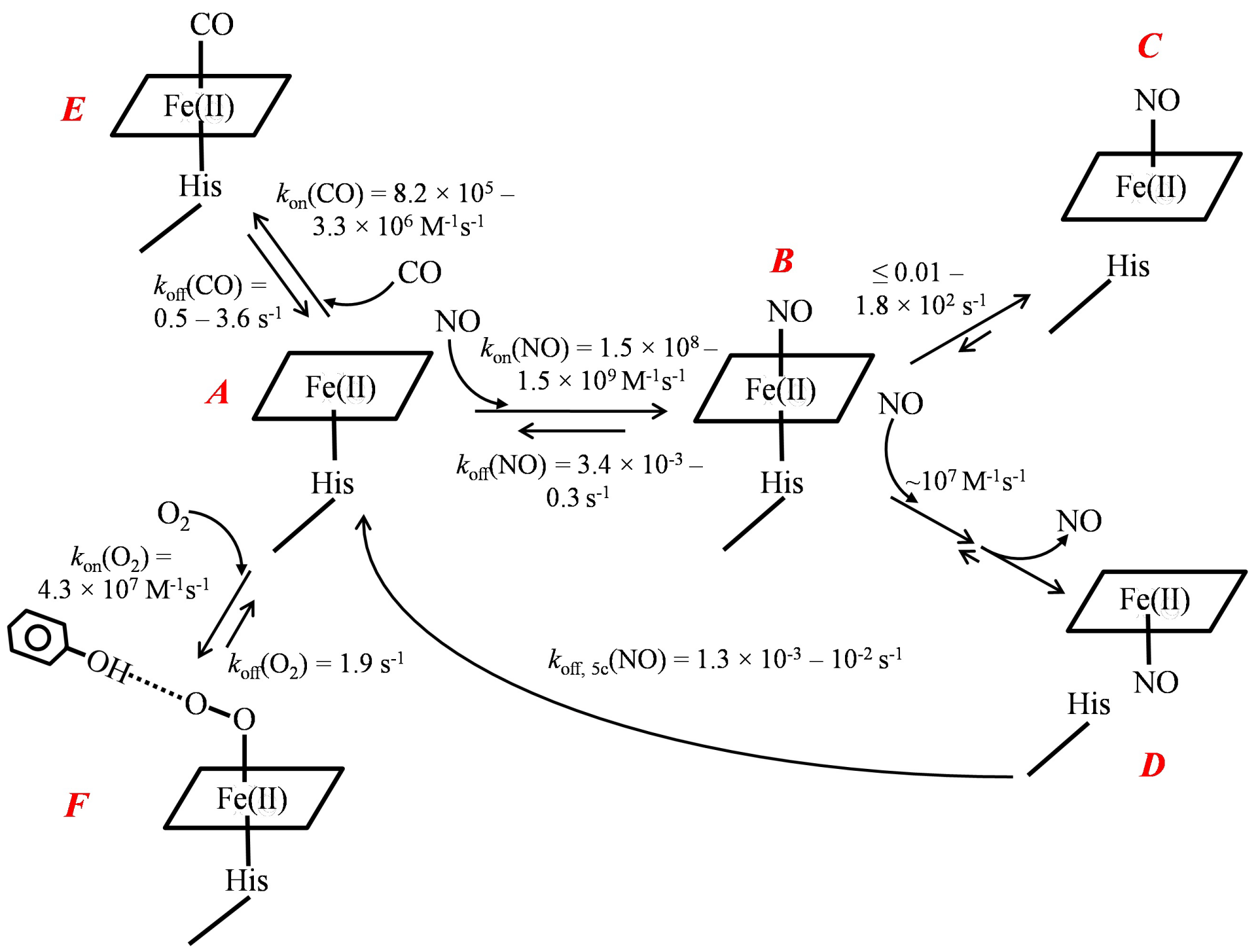
Figure 1: “Sliding scale rule” plot of H-NOXs.
The logarithms of the measured KDs of NO/CO/O2 to the ferrous bacterial
H-NOXs and sGC are plotted versus the ligand type. Parameters measured
for H61L leghemoglobin (Lb) and heme model Fe(II) PP(1-MeIm) are also
plotted for comparisons and demonstrating the large dynamic ranges of
KD which are modulated by the proteinaceous environments. sGC, black
triangle up; Cb H-NOX, blue diamond; Cs H-NOX, green square; Ns H-NOX,
red triangle down; So H-NOX, yellow triangle up; Vc H-NOX, pink square;
Fe(II) PP(1-MeIm), purple circle, and H61L Lb, black circle. The dashed
lines represent the predicted KD(O2)s for sGC, So, and Vc HNOXs based on
the “sliding scale rule”. The KD(O2) of Ns H-NOXs is measured using high
pressure cell and the straight log KD(NO) - log KD(CO) - log KD(O2) line of
Ns H-NOXprovides one of the best demonstrations for the “sliding scale
rule”; however, the high pressure cell method does not work for So and Vc
H-NOXs due to much faster autoxidation in these proteins [7,8,10]. The [O2]
in aqueous buffers under 1 atm of pure O2 gas and air are marked with two
blue dashed lines. The dash-dot-dot lines connect log KD(CO) of Cb, So, and
Vc HNOXs and sGC to corresponding log KD, apparent (NO), which are also
indicated by arrows.
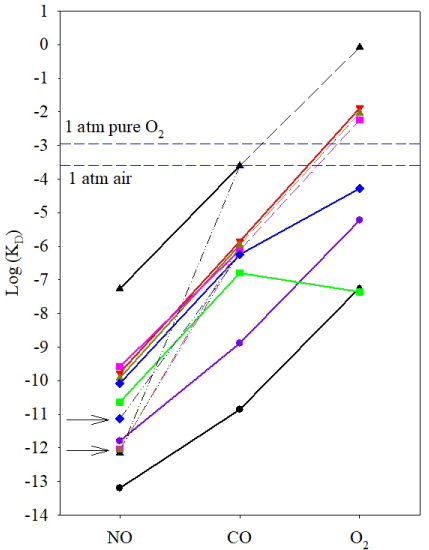
Figure 2: Binding of gaseous ligands NO, CO, and O2 to bacterial H-NOXs.
The kinetic parameters are experimentally determined for Cb, Cs, Ns, So, and
Vc H-NOXs [7-10]. A, ferrous heme with a neutral histidine proximal ligand;
B, 6c NO-heme complex; C and D, 5c NO-heme complexes; E, 6c CO-heme
complex; F, oxyferrous complex. The essentially irreversible conversion of 6c
NO-heme complex to 5c NO-heme complex is observed in Cb, So, and Vc
H-NOXs. The 6c NO-heme complexes in Cb, So, and Vc H-NOXs also react
with secondary NO in the presence of excess NO, generating 5c NO-heme
complex, most likely through a transient quaternary intermediate (not shown)
and loss of NO from the distal side. The 5c NO complexes with NO on either
distal (C) or proximal side (D) are not distinguished in the text. Only Cb and
Cs H-NOXs form stable oxyferrous complexes and exhibit strong affinities
for O2 due to distal hydrogen-bond donor, Y139 and Y140, respectively. The
kon(O2) and koff(O2) are those of Cs H-NOX; the oxyferrous complex in Cb
H-NOX forms extremely fast, cannot be measured by stopped-flow method.
The “sliding scale rule” reveals why heme sensor proteins possess a 5c heme ligated to a neutral histidine and how the proteinaceous structures modify the intrinsic gas binding properties of such a heme to achieve specific gaseous ligand selectivity. The generality of “sliding scale rule” has been confirmed by the data of each hemeprotein with a 5c neutral histidine-ligated heme [1,11]. A most recent example is the selective binding of NO, CO, and O2 to dehaloperoxidase, a globin with many types of enzymatic activities found in terebellid Polychaeta Amphitrite ornate [14].
References
- Tsai A-L, Berka V, Martin E, Olson JS. A "sliding scale rule" for selectivity among NO, CO and O2 by heme protein sensors. Biochemistry. 2012; 51: 172-186.
- Boon EM, Marletta MA. Ligand specificity of H-NOX domains: From sGC to bacterial NO sensors. J Inorg Biochem. 2005; 99: 892-902.
- Shimizu T, Huang D, Yan F, Stranava M, Bartosova M, Fojtikova V, et al. Gaseous O2, NO and CO in signal transduction: Structure and function relationships of heme-based gas sensors and heme-redox sensors. Chem Rev. 2015; 115: 6491-6533.
- Jain R, Chan MK. Mechanisms of ligand discrimination by heme proteins. J Biol Inorg Chem. 2003; 8: 1-11.
- Derbyshire ER, Marletta MA. Biochemistry of soluble guanylate cyclase. Handb Exp Pharmacol. Springer. 2009: 17-31.
- Boon EM, Marletta MA. Ligand discrimination in soluble guanylate cyclase and the H-NOX family of heme sensor proteins. Curr Opin Chem Biol. 2005; 9: 441-446.
- Wu G, Liu W, Berka V, Tsai A-L. The Selectivity of Vibrio cholerae H-NOX for Gaseous Ligands follows the "Sliding Scale Rule" Hypothesis. Ligand Interactions with both Ferrous and Ferric Vc H-NOX. Biochemistry. 2013; 52: 9432-9446.
- Wu G, Liu W, Berka V, Tsai A-L. H-NOX from Clostridium botulinum, like H-NOX from Thermoanaerobacter tengcongensis, Binds Oxygen but with a Less Stable Oxyferrous Heme Intermediate. Biochemistry. 2015; 54: 7098-7109.
- Tsai A-L, Berka V, Martin F, Ma X, Van den Akker F, Fabian M, et al. Is Nostoc HNOX a NO sensor or redox switch? Biochemistry. 2010; 49: 6587-6599.
- Wu G, Liu W, Berka V, Tsai A-L. Gaseous ligand selectivity of the H-NOX sensor protein from Shewanella oneidensis and comparison to those of other bacterial H-NOXs and soluble guanylyl cyclase. Biochimie. 2017; 140: 82-92.
- Tsai A-L, Martin E, Berka V, Olson JS. How do heme-protein sensors exclude oxygen? Lessons learned from cytochrome c', Nostoc puntiforme heme nitric oxide/oxygen binding domain, and soluble guanylyl cyclase. Antioxid Redox Signal. 2012; 17: 1246-1263.
- Hossain S, Nisbett LM, Boon EM. Discovery of Two Bacterial Nitric Oxide-Responsive Proteins and Their Roles in Bacterial Biofilm Regulation. Acc Chem Res. 2017; 50: 1633-1639.
- Nisbett LM, Boon EM. Nitric Oxide Regulation of H-NOX Signaling Pathways in Bacteria. Biochemistry. 2016; 55: 4873-4884.
- Wu G, Zhao J, Franzen S, Tsai A-L. Bindings of NO, CO, and O2 to multifunctional globin type dehaloperoxidase follow the 'sliding scale rule'. Biochem J. 2017; 474: 3485-3498.