
Special Article - Escherichia coli
J Bacteriol Mycol. 2019; 6(3): 1102.
Molecular Typing of β-Lactamase and Tetracycline Resistant Escherichia coli Strains Isolated from Imported Shrimp
Khan S¹, Campbell M², Alotaibi K³, Smani D4, Khan A4, Sung K4, Khan S4 and Nawaz M4*
¹University of Arkansas, USA
²Hendrix College, USA
³Department of Biology, Jazan University, Kingdom of Saudi Arabia
4Division of Microbiology, National Center for Toxicological Research, FDA, USA
*Corresponding author: Mohamed Nawaz, Division of Microbiology, National Center for Toxicological Research, Jefferson, AR 72079, USA
Received: February 14, 2019; Accepted: March 28, 2019; Published: April 04, 2019
Abstract
The misuse of antibiotics in commercial aquaculture may result in the selection of antibiotic-resistant bacteria. A study was undertaken to isolate and characterize the prevalence of β-lactam and tetracycline-resistant Escherichia coli from imported shrimp. All 55 strains of E. coli isolated from 207 shrimp samples were resistant to ampicillin, penicillin and tetracycline. These isolates were screened for 11 different β-lactam resistance genes by PCR. Oligonucleotide primers specific for the amplification of a 550-bp portion of the blaCTX-M gene amplified this gene from 31 of the 55 (56%) isolates. Oligonucleotide primers specific for the amplification of a 851-bp portion of the blaTEM gene amplified the genes from 9 (16%) of the isolates. Six (11%) were found to harbor both the 550-bp blaCTX-M and the 851-bp blaTEM genes. Template DNA were also screened for the presence of 6 different tetracycline resistance (tet) genes, PCR detected the presence tetB in a majority (39/55, 71.0%) of the isolates, followed by tetA (13.0%). Eleven percent contained both tetA and tetB genes. The PCR based Replicon Typing Method (PBRT) was used for typing plasmids from the 55 isolates by targeting the replicons of 15 major known plasmid families. Majority (44/55, 80.0%) of the isolates contained B/O plasmid replicon. Seven (12%) isolates also contained plasmids of the FIA family and 5 (9%) isolates contained both B/O and FIA plasmid families. The β-lactam and tetracycline resistant determinants were successfully transferred to E. coli J53 by conjugation. Our results indicate that imported shrimp may be a reservoir of the known β-lactam and tetracycline-resistance determinants.
Introduction
Asian nations, such as Thailand, Vietnam, Indonesia, China, India and Bangladesh, currently produce nearly 80% of the world’s farmed shrimp [1-3]. These Asian nations earn more than $5 billion by exporting 1.3 billion of farm-raised shrimp. Intense aquaculture demands the implementation of modern technologies to enhance production and profits [4,5]. However, aquaculture production is affected by several bacterial diseases. Thus, large amounts of antibiotics are broadly used in aquaculture farming to promote growth and to retard the incidence and effects of diseases caused by overcrowded aquaculture ponds [1,2]. The excess use of antibiotics in aquaculture ponds may select bacteria resistant to multiple antibiotics [1,5]. Several public health agencies in the United States limit the presence of antibiotic residues in food-producing animals to decrease the prevalence of antibiotic-resistant microflora in food-producing ecosystems [6].
β-Lactam antibiotics (including penicillin derivatives, cephalosporins, monobactams and carbapenems) and tetracyclines are widely used for the treatment of urinary tract, respiratory tract, the intestines, certain inflammatory disorders and numerous other life threatening infections [7]. However, the widespread use of the same types of drugs in aquaculture ponds to prevent the outbreak of diseases may select bacteria resistant to these life-saving drugs and reduce the efficacy of these drugs in clinical treatment of infectious diseases [1,5,6].
Several mechanisms of resistance to β-lactam and tetracycline antibiotics have been reported [8-10]. Additional mechanisms, such as altered cell membrane permeability and overexpression of multidrug efflux pumps, also contribute to high-level bacterial resistance to these antibiotics. However, limited information is available on the occurrence and prevalence of β-lactam and tetracycline-resistant determinants in E. coli in imported shrimp. There is also a paucity of information on the types of plasmid replicon prevalent in aquaculture ecosystem. Such information is indispensable for understanding the epidemiological dynamics and to devise interventional strategies to curtail the dissemination of specific plasmid replicons to other ecosystems. In this report, we describe the occurrence and prevalence of different β-lactam and tetracycline resistance determinants and plasmid replicons in E. coli isolated from imported shrimp samples.
Materials and Methods
Bacterial strains: All the strains of E. coli used in this study were isolated from shrimp (Penaeus monodon) imported to the US. The isolation, characterization and identification of these isolates were described earlier [11]. All isolates were stored in Luria Broth (LB) containing 20% glycerol at -70°C and were grown overnight at 37°C in LB or on Trypticase Soy Agar (TSA) plates supplemented with 5% sheep blood.
Determination of antibiotic susceptibility and the minimum inhibitory concentration (MIC) of the isolates: The antibiotic susceptibility of each isolate was determined by disk diffusion assay [12] and was interpreted as per the criteria specified by the Clinical and Laboratory Standards Institute (CLSI). The MICs for the antibiotics (tetracycline, ampicillin and penicillin) were determined by broth dilution using Mueller-Hinton broth [13].
Genomic DNA extraction
Genomic DNA was extracted from cells grown overnight at 37°C with the QIAamp DNA Mini Prep Kit (Qiagen, Valencia, CA).
Detection of β-lactamase genes from template DNA
The presence of various β-lactam resistance genes in the template DNA was determined by PCR [14]. The primers used for the amplification of these genes are listed in Table 1. PCR amplification of the β-lactam resistance genes was carried out in a reaction volume of 25 μl using a PCR Kit (Applied Biosystems, Foster City, CA). The thermal cycling conditions consisted of an initial denaturation of 94°C for 2 min followed by 35 cycles of amplification. Each cycle consisted of 94°C denaturation for 30 s, annealing for 1°C below the lowest Tm of a given primer pair, and 72°C extension for 1 min. The amplified PCR products were maintained at 4°C. A reagent blank contained all the components of the reaction mixture except template DNA, for which sterile distilled water was substituted. The PCR products were subjected to electrophoresis on 1.2% agarose gels in 1 x Tris-Borate- EDTA (TBE) buffer, visualized with UV, and photographed using an Eagle Eye II gel documentation system (Stratagene, La Jolla, CA). A 100-bp DNA ladder (Thermo Fisher Scientific, Grand Island, NY) was used as the size standard.
Primers
Nucleotide Sequence
Target gene
Size (bp)
blaOXAF
GCAGCGCCAGTGCATCAAC
OXA-1
198
blaOXAR
CCGCATCAAATGCCATAAGTG
blaPSEF
AGTAGGGCAGGCAATCACAC
PSE-1
421
blaPSER
GCGATCCGCAATGTTCCATC
blaSHVF
GCAAAACGCCGGGTTATTC
SHV-1
940
blaSHVR
GGTTAGCGTTGCCAGTGCT
blaTEMF
ATGAGTATTCAACATTTCCG
TEM
851
blaTEMR
TTAATCAGTGAGGCACCTAT
blaCTXMF
CGCTTTGCGATGTGCAG
CTX-M
550
blaCTXMR
ACCGCGATATCGTTGGT
blaCTXM9F
GTGACAAAGAGAGTGCAACGG
CTXM9
856
blaCTXM9R
ATGATTCTCGCCGCTGAAGCC
blaCMYF
TGGCCAGAACTGACAGGCAAA
CMY2
462
blaCMYR
TTTCTCCTGAACGTGGCTGGC
blaFOXF
AACATGGGGTATCAGGGAGATG
FOX
190
blaFOXR
CAAAGCGCGTAACCGGATTGG
blaIMPF
CATGGTTTGGTGGTTCTTGT
IMP
447
blaIMPR
ATAATTTGGCGGACTTTGGC
blaKPCF
CAGCTCATTCAAGGGCTTTC
KPC
533
blaKPCR
AGTCATTTGCCGTGCCATAC
blaVIMF
AGTGGTGAGTATCCGACAG
VIM
261
blaVIMR
ATGAAAGTGCGTGGAGAC
Table 1: Oligonucleotide primers used in the amplification of β-lactam resistance genes from E. coli strains isolated from imported shrimp.
Detection of tetracycline resistance (tet) genes by PCR
The various tet genes were individually amplified by PCR [10] with oligonucleotide primers (Table 2). The universal PCR amplification included 30 thermal cycles of 30 s at 94°C, 60 s at 55°C, and 60 s at 72°C, with an additional extension in the last cycle for 300 s at 72°C. The amplified PCR products were maintained at 4°C. PCR products were subjected to electrophoresis on 1.2% agarose gels in 1 x TBE buffer, visualized with UV, and photographed using the Eagle Eye II gel documentation system.
Primers
Nucleotide Sequence
Target gene
Size (bp)
tetAF
GCTACATCCTGCTTGCCTTC
tet(A)
211
tetAR
GCATAGATCGCCGTGAAGAG
tetBF
TCATTGCCGATACCACCTCAG
tet(B)
391
tetBR
CCAACCATCATGCTATTCCATCC
tetCF
CTGCTCGCTTCGCTACTTG
tet(C)
897
tetCR
GCCTACAATCCATGCCAACC
tetDF
TGTGCTGTGGATGTTGTATCTC
tet(D)
844
tetDR
CAGTGCCGTGCCAATCAG
tetG_F
GCGCTNTATGCGTTGATGCA
tet(G)
803
tetG_R
ATGCCAACACCCCCGGCG
Table 2: Oligonucleotide primers for the amplification of tetracycline resistance (tet) genes from E. coli strains isolated from imported shrimp.
Isolation of plasmids
Plasmid DNA was isolated using a modified alkaline lysis method [14]. Samples were analyzed by electrophoresis in 1 X Tris acetate- EDTA buffer at 64 V for 2 h on 1.0% agarose gels. A supercoiled DNA ladder (Invitrogen, Carlsbad, CA) was used as a molecular weight marker.
Plasmid typing
Plasmids were typed by the PCR-Based Replicon Typing (PBRT) method [14,15] with primers listed in Table 3.
Replicon
Nucleotide Sequence
Target gene
Size (bp)
HI1
F-GGAGCGATGGATTACTTCAGTAC
parA-parB
471
R-TGCCGTTTCACCTCGTGAGTA
HI2
F-TTTCTCCTGAGTCACCTGTTAACAC
iterons
644
R-GGCTCACTACCGTTGTCATCCT
I1
F-CGAAAGCCGGACGGCAGAA
RNAI
139
R-TCGTCGTTCCGCCAAGTTCGT
X
F-AACCTTAGAGGCTATTTAAGTTGCTGAT
ori ?
376
R-TGAGAGTCAATTTTTATCTCATGTTTTAGC
L/M
F-GGATGAAAACTATCAGCATCTGAAG
rep A, B, C
785
R-CTGCAGGGGCGATTCTTTAGG
N
F-GTCTAACGAGCTTACCGAAG
rep A
559
R-GTTTCAACTCTGCCAAGTTC
FIA
F-CCATGCTGGTTCTAGAGAAGGTG
iterons
462
R-GTATATCCTTACTGGCTTCCGCAG
FIB
F-GGAGTTCTGACACACGATTTTCTG
rep A
702
R-CTCCCGTCGCTTCAGGGCATT
W
F-CCTAAGAACAACAAAGCCCCCG
rep A
242
R-GGTGCGCGGCATAGAACCGT
FIC
F-GTGAACTGGCAGATGAGGAAGG
rep A2
262
R-TTCTCCTCGTCGCCAAACTAGAT
A/C
F-GAGAACCAAAGACAAAGACCTGGA
rep A
465
R-ACGACAAACCTGAATTGCCTCCTT
T
F-TTGGCCTGTTTGTGCCTAAACCAT
rep A
750
R-CGTTGATTACACTTAGCTTTGGAC
FIIA (FIIS)
F-CTGTCGTAAGCTGATGGC
rep A
270
R-CTCTGCCACAAACTTCAGC
K/B
F-GCGGTCCGGAAAGCCAGAAAAC
RNAI
160
R-TCTTTCACGAGCCCGCCAAA
B/O
F-GCGGTCCGGAAAGCCAGAAAAC
RNAI
159
R-TCTGCGTTCCGCCAAGTTCGA
Table 3: Primers used in PCR for detection of plasmid replicon typing in E. coli strains isolated from imported shrimp.
Conjugation
Four tetracycline, ampicillin resistant E.coli strains isolated from shrimp aquaculture samples were selected as donors. A tetracyclinesensitive E. coli J53 strain was used as a recipient for conjugation experiments to determine transferability of β-lactamase and tetracycline resistant genes. The broth mating conjugation method was performed as previously described [16]. LB agar plates containing tetracycline (256 μg/mL) and sodium azide (800 μg/mL) were used to select transconjugants harboring genes conferring β-lactam and tetracycline resistance.
Results
Determination of the Minimum Inhibitory Concentrations (MICs) of the isolates for ampicillin, penicillin and tetracycline
Wide ranges of MICs were observed for all three antibiotics tested. Thirty-two of the 55 strains (58%) were resistant to the highest concentration of ampicillin (256 μg/mL) tested; and 42 (76%) of the strains were resistant to the highest concentration of penicillin (256 μg/mL) tested; and 14 (25%) were resistant to the highest concentration of tetracycline (256 μg/mL) tested. Thirteen of the 55 isolates (24%) were resistant to the highest concentrations (256 μg/ mL) for all three antibiotics tested and 15 of the 55 isolates that had MICs of 256 μg/mL for the two β-lactam antibiotics and variable MICs of 32-192 μg/mL for tetracycline (data not shown).
PCR amplification of β-lactam and tetracycline (tet) resistance genes from the template DNA by PCR
Template DNAs from the 55 β-lactam and tetracycline-resistant E. coli strains from the imported shrimp were screened for the presence of 11 different β-lactam resistance genes. Oligonucleotide primers specific for 550-bp portion of the blaCTX-M gene amplified the gene from the template DNA of 31 of the 55 (56%) of the isolates (Figure 1A). Oligonucleotide primers specific for a 851-bp portion of the blaTEM gene amplified the gene from the template DNA of 9 (16%) of the isolates (Figure 1B). Six (11%) of the isolates harbored both the 550-bp blaCTX-M and the 851-bp blaTEM genes in the template DNA (Figure 1C). PCR failed to amplify the other nine β-lactam resistance genes from the template DNA of any of the 55 isolates. The template DNA was also screened for the 5 tet genes (tetA, tetB, tetC, tetD and tetG). Oligonucleotide primers specific for a 957-bp region of tetA and for a 436-bp region of tetB successfully amplified the PCR amplicons from the template DNA of 7 (13%, Figure 2A & 2C) and 39 (71%, Figure 2B & 2C) of the isolates respectively. Additionally, 5 (9%) of the strains harbored both tetA and tetB genes (Figure 2C). PCR primers specific for the amplification of tetC, tetD, and tetG failed to amplify these genes from the template DNA of any of the isolates.
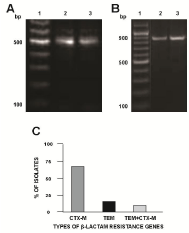
Figure 1: Detection and quantification of β-lactamase genes in the template
DNA of the E.coli isolated from imported shrimp. (A) lane 1, 100-bp molecular
weight marker; lanes 2-3, 550-bp blaCTX-M amplified from the template DNA.
(B) lane 1, 100-bp molecular weight marker; lanes 2-3, 851-bp blaTEM
amplified from the template DNA of the isolates. (C) Quantification of the
occurrence of blaCTX-M, blaTEM and a combination of the genes in the template
DNA of the isolates.
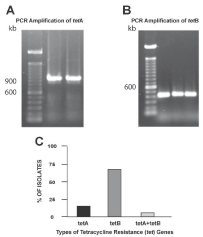
Figure 2: Detection and quantification of tetracycline resistant (tet) genes
in the template DNA of the E. coli isolated from imported shrimp. (A) lane 1,
100-bp molecular weight marker; lanes 2-3, 851-bp tetA amplified from the
template DNA (B) lane 1, 100-bp molecular weight marker; lanes 2-4, 436-
bp region of the tetB (C) Quantification of the occurrence of tetA, tetB and a
combination of these genes in the template DNA of the isolates.
Plasmid identification and typing by PCR
Primers specific for a 159-bp portion of the B/O plasmid [15] were amplified from the template DNA of 44 (80%) isolates (Figure 3A-C). Additionally, oligonucleotides specific for the FIA plasmid successfully amplified a 462-bp region of the replicon from the template DNA of 5 (7%) isolates (Figure 3B-C). PCR also indicated that 4 (7%) strains simultaneously harbored B/O and FIA replicons (Figure 3C). The oligonucleotide primers failed to amplify any of other 13 plasmid families from the template DNA of the isolates.
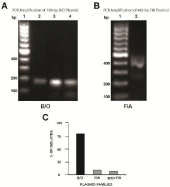
Figure 3: Profiles of plasmids of the 55 E. coli strains isolated from imported
shrimp. Lane 1, strain EC120, lane 2, strain EC126, lane 3, strain EC221,
lane 4, strain EC123, lane 5, strain EC338, lane 6, strain EC827, lane 7, strain
EC803, lane 8, strain EC01, lane 9, strain ECIS801, lane 10, strain EC503,
lane 11, strain EC510.
Characterization of plasmids isolated from β-lactam and tetracycline-resistant E. coli isolated from shrimp
Seventeen of the fifty-five isolates did not contain any plasmids. Eleven distinct plasmid profiles were observed in the 38 isolates; the plasmids varied in size from 2.0 to greater than 16.0 kb (Figure 4). Strain EC 120 (lane 1) differed from other strains by harboring two distinct plasmid; one plasmid measured ca. 14.0 kb while the other was a Mega Plasmid (M3) measuring above 16.0 kb. Strains EC126 and EC 221 had distinct multiple plasmids measuring 6.0 kb to above 16.0 kb (Figure 4, Lanes 2-3). Strain EC126 had plasmids measuring 6, 9, 14 kb and 2 mega plasmids above 16.0 kb. Strain EC221 (lane 3) had plasmids measuring 6, 10 and 14 kb and two mega plasmids measuring above 16.0 kb. Strain EC123 had two plasmids measuring 5.0 and 6.0 kb and a mega plasmid measuring above 16.0 kb (Figure 4, lane 4). Strain EC338 (lane 5) had two distinct plasmids. One plasmid measuring 14.0 kb and the other a mega plasmid measuring above 16.0 kb. Strains EC827, EC803 and EC01 contained plasmids of various sizes (lanes 6-8). Strains EC827 and EC803 had two identical plasmids measuring ca. 8.0 kb, 12.0 kb and mega plasmids of different sizes measuring above 16.0 kb. Strain EC803 had plasmids measuring 9.0 kb, 12.0 kb and a mega plasmid. Strains ECIS801, EC503 and EC510 had plasmids of variable sizes (Figure 4; lanes 9-11). Strain ECIS801 (lane 9) had multiple plasmids measuring 8.0, 10.0, and 16.0 kb and 2 mega plasmids measuring above 16.0 kb. Strain EC503 had 2 plasmids measuring 13.0 and 16.0 kb and one mega plasmid. Strain EC510 had 2 plasmids measuring 13.0 and 16.0 kb.
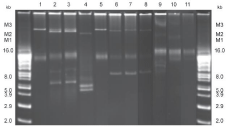
Figure 4: Detection and quantification of the various plasmids families by
the PCR-based replicon typing. (A) lane 1, 100-bp molecular weight marker;
lanes 2-4, 159-bp B/O plasmid. (B) lanes 1; 100-bp molecular weight marker;
lane 2, 462-bp FIA plasmid (C) quantification of the different plasmid families
occurring in the template DNA of the isolates.
Horizontal transfer of β-lactam and tetracycline resistance phenotypes and genotypes
Four strains (E. coli strain 120, 123, 126 and 338) that were resistant to 256 μg/mL of tetracycline but sensitive to 800 μg/mL of sodium azide were selected as donors. Our preliminary analysis indicated that these strains harbored blatem, blactx-m and tetB genes. In addition, these strains also carried B/O plasmid. Transconjugants were only obtained when mating donor strains E. coli 126 and 338 with the recipient strain J53 for 24 h. No transconjugants were obtained when using strains 120 and 123 as donors. Our experiments also indicated higher rates of transconjugants were obtained when using E. coli strain 338 than E.coli strain 126. Serial dilution techniques indicated the rates of transconjugation to be 4.9 × 104 when using E. coli 126 when compared to the conjugation rates of 2.46 × 105 when using E. coli 338. All transconjugants were resistant to 256 μg/mL of tetracycline, ampicillin and 800 μg/mL of sodium azide, respectively. Template DNA of transconjugants from both strain 126 and 338 were positive for the presence of the 550-bp region of the blaCTX-M, 436-bp region of the tetB and the 159-bp region of the B/O plasmid.
Discussion
The isolation of multiple antibiotic-resistant E. coli strains, resistant to high concentrations of β-lactam and tetracycline antibiotics, from imported shrimp suggest the usage of these antibiotics in commercial aquaculture ponds. The usage of these drugs may have played a key role in the selection of these resistant strains. These strains, as a reservoir of multiple antibiotic-resistance genes, may transfer their resistance markers to other ecosystems and thus become a potential public health hazard [17,6].
The detection, characterization and classification of β-lactamases have been well documented [18-20]. These enzymes include the Extended Spectrum β-Lactamases (ESBLs), observed as TEM- 1, TEM-2 or SHV-1; more than 150 different TEM and SHV-type ESBLs have been reported [21,19,9]. TEM-1 is the most commonly encountered β-lactamase in Gram-negative bacteria; up to 90% of ampicillin resistance in E. coli is due to the synthesis of TEM-1 [19,9]. This enzyme is also responsible for ampicillin and penicillin resistance in Haemophilus influenzae and Neisseria gonorrhoeae [19]. Unlike TEM-ESBLs, SHV-ESBLs are predominantly found in Klebsiella pneumoniae and are known to regulate resistance to ampicillin and penicillin [19]. Very few variants of SHV-lactamases have been reported. Both TEM and SHV-ESBLs are borne on transmissible plasmids that frequently co-transfer resistance to other classes of antibiotics [19,9,22]. Recently, both TEM and SHV-derived ESBLs have been joined by the CTX-M family of β-lactamases in E. coli [19,20]. The CTX-M lactamase has less than 40% similarity to TEM and SHV-lactamases. Results from our investigations indicate that the blaCTX-M gene is the predominant ESBL gene in the template DNA of the 55 aquaculture isolates. It occurred in 56.0% of the isolates, followed by blaTEM, which was found in a minority (16.0%) of the isolates. However, these results contrast with an earlier report from India [17] that indicated the predominant occurrence of blaTEM (100%) and low occurrence (16%) of blaCTX-M ESBLs in the template DNA of E. coli isolated in aquatic environments. A report from aquaculture from China [23] indicated the predominant occurrence of blaTEM followed by blaSHV. Few isolates in these previous studies harbored blaCTX-M. Additionally, results from our investigation indicate that 9 of the isolates had an MIC of 256 μg/ml for both ampicillin and penicillin. These isolates were found to harbor both blaCTX-M and blaTEM, indicating that the concomitant occurrence of these two genetic determinants may be required for the higher MICs of these isolates. Results from all of these investigations clearly indicate that aquaculture ecosystems are reservoirs of ESBLs.
The occurrence and prevalence of tetracycline resistance in aquaculture ecosystems has been well documented. Furushita et al., [24] studied the prevalence of tet determinants in Gram-negative bacteria isolated from seafood. They reported that 43% of the isolates harbored tetB determinants, followed by tetC and tetD determinants. Another study [10] indicated that 77% of the E. coli isolates from catfish (Ictalurus punctatus) harbored tetB, followed by tetA (25%) and tetC (5%). An Australian investigation of Gram negative bacteria isolated from aquaculture sediments [25] indicated that tetM (50%) was the predominant tetracycline resistance determinant followed by tetE (45%). Results from our current investigation indicated that tetB (71%) was the dominant tet determinant, followed by tetA. Findings from all these investigations indicate that aquaculture may be a reservoir of a variety of tet genes with wide regional and continental differences in prevalence and distribution patterns of tet determinants.
Enterobacteriaceae producing ESBLS (extended spectrum β-lactamases) are a major public health problem and are responsible for triggering many outbreaks as well as sporadic infections worldwide [18,10,20,9]. The extensive prevalence of ESBLs is often due to horizontal gene transfer via plasmids [20,26,27,28,29,23]. Several studies have reported that the replicon types most frequently detected in ESBLs among the Enterobacteriaceae belong to the incompatibility (Inc) group, which includes F, A/C, L/M, I1, H12 and N [26]. IncF and IncI1 are the most frequently reported replicon types associated with the dissemination of ESBLs [29-31]. ESBL blaCTX-M has been found on an IncF plasmid belonging to type FII in combination with FIA [32,33]. These plasmid types with different replicon types have been identified in strains isolated from different origins (environmental, livestock and humans). Results from our investigation indicate that plasmids were present in a majority of isolates and measured from 5.0 to 16.0 Kb with 1-3 megaplasmids. In contrast to several earlier investigations, none of the isolates in our study were found to harbor any of the replicon types F, A/C, L/M, I1 or H12. We for the first time report the occurrence of the B/O replicon type in ESBL strains of E. coli from aquaculture ecosystem. Our results indicate that 80% of the E. coli strains harbored replicon type B/O; 7% of the isolates contained the incompatibility (Inc) group FIA; and some isolates (4%) carried both types. It is possible that B/O type plasmids may play a role in the dissemination of ESBLs but also tet genes in shrimp aquaculture. Since we were unable to amplify the FIA replicon type plasmid from the template DNA of any of the transconjugants, it is possible that these plasmids may not harbor the tet genes or ESBL resistant determinants.
The prevalence of ESBL-blaCTX-M and to a minor extent, blaTEM, along with the presence of tetB and tetA in the template DNA of most isolates, may be a public health concern and highlights the significance of aquaculture ecosystems as reservoir of these resistance determinants. Thus, concerted efforts should be made by all global public health agencies to more closely monitor and introduce control measures to reduce the prevalence of antibiotic resistance in aquaculture ecosystems, as this represents a major pool of resistance determinants likely to pose a threat to the clinical use of these lifesaving drugs to treat human illnesses.
Acknowledgement
We thank Drs. Carl E. Cerniglia, John B. Sutherland, Steve Foley and Huizhong Chen for critical review of the manuscript. This work was supported by the National Center for Toxicological Research, US Food and Drug Administration. Views presented here do not necessarily reflect those of the FDA.
References
- Biao X, Yu KJ. Shrimp farming in China: operating characteristics, environmental impact and perspectives. Ocean Coast Manage. 2007; 50: 538-550.
- Cao L, Naylor R, Henriksson P, Laedbitter D, Metian M, Troell M, et al. China’s aquaculture and the world’s wild fisheries. Science. 2015; 347: 133-135.
- Fisheries and Aquaculture Department: The state of world fisheries and aquaculture. Rome: Food and Aquaculture Organization of the United Nations. 2010; 3.
- Graslund S, Bengtsson BE. Chemicals and biological products used in Southeast Asian shrimp farming and their potential impact on the environment-a review. Sci Total Environ. 2001; 280: 93-131.
- Heuer OE, Kruse H, Grave K, Collignon P, Karunasagar L, Angulo FJ. Human health consequences of use of antimicrobial agents in aquaculture. Clin Infect Dis. 2009; 49: 1248-1253.
- Nawaz M, Erickson BD, Khan AA, Pothuluri JV, Rafii F, Sutherland JB, et al. Human health impact and regulatory issues involving antimicrobial resistance in the food animal production environment. Regulatory Research Perspectives. 2001; 1: 1-8.6.
- Shapiro DJ, Hicks LA, Pavia AT, Hersh AL. Antibiotics prescribing for adults in ambulatory care in the USA, 2007-09. J Antimicrob Chemother. 2014; 69: 234-240.
- Jacoby GA, Medeiros AA. More extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1991; 35: 1697-1704.
- Leonard DA, Bonomo RA, Powers RA. Class D β-lactamases: A re-appraisal after five decades. Acc Chem Res. 2013; 46: 2407-2415.
- Nawaz M, Khan AA, Khan SA, Sung K, Kerdahi K, Steele R. Molecular characterization of tetracycline-resistant genes and integrons from avirulent strains of Escherichia coli isolated from catfish. Foodborne Pathogen Dis. 2009; 6: 553-559.
- Nawaz M, Sung K, Kweon OK, Khan S, Nawaz S, Steele R. Characterization of novel mutations involved in quinolone resistance in Escherichia coli isolated from imported shrimp. Int J Antimicrobial Agents. 2015; 45: 471-476.
- Bauer AW, Kirby WM, Sherris JC, Truck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966; 45: 493-496.
- Clinical and Laboratory Standards Institute. Development of in vitro susceptibility testing, criteria and quality control parameters for veterinary antimicrobial agents; approved guidelines-second edition. Document M37-A2. Wayne, PA: CLSI. 2002.
- Bae D, Cheng CM, Khan AA. Characterization of Extended Spectrum β-Lactamase (ESBL) producing Non-Typhoidal Salmonella (NTS) from imported seafood products. Int J Food Microbiol. 2014: 12-17.
- Bertini CA, Villa A, Falbo LV, Hopkins KL, Threl fall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005; 63: 219-228.
- Tran QT, Nawaz MS, Deck J, Nguyen KT, Cerniglia CE. Plasmid-mediated quinolone resistance in Pseudomonas putida isolates from imported shrimp. Appl Environ Microbiol. 2010; 77: 1885-1887.
- Bajaj P, Singh NS, Kanaujia PK, Virdi JS. Distribution and molecular characterization of genes encoding CTX-M and Amp C β-lactamases in Escherichia coli isolated from an Indian urban aquatic environment. Sci Tot Environ. 2015; 505: 350-356.
- Bardford PA. Extended spectrum β-lactamases in the 21st century: characterization, epidemiology and detection of this important resistance threat. Clin Microbiol Rev. 2001; 14: 933-951.
- Bush K. Alarming β-lactamase-mediated resistance in multidrug resistant Enterobacteriaceae. Curr. Opin. Microbiol. 2010; 13: 558-564.
- Canton R, Gonzalez-Alba JM, Galan JC. CTX-M enzymes: origin and diffusion. Front Microbiol. 2012; 3: 1-19.
- Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrobiol Agents Chemother. 1995; 39: 1211-1233.
- Livermore DM. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995; 8: 557-584.
- Jiang HX, Tang D, Liu YH, Zhang XH, Zeng ZL, Xu L et al. Prevalence and characteristics of β-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J Antimicrob Chemother. 2012; 67: 2350-2353.
- Furushita M, Shiba T, Maeda T, Yahata M, Kaneoka A, Takahashi Y, et al. Similarity of tetracycline resistance genes isolated from fish farm bacteria to those from clinical isolates. Appl Environ Microbiol. 2003; 69: 5336-5346.
- Akinbowale OL, Peng H, Barton MD. Diversity of tetracycline resistance genes in bacteria from aquaculture sources in Australia. J Appl Microbiol. 2007; 103: 2016-2025.
- Carattoli A. Plasmids and spread of resistance. Int J Med Microbiol. 2013; 303: 298-304.
- Cloeckaert A, Praud K, Lefevre M, Doublet B, Pardos M, Granier SA, et al. IncI1 plasmid carrying extended spectrum β-lactamases gene blaCTX-M-1 in Salmonella enterica isolates from poultry and humans in France, 2003-2008. Antimicrob Agents Chemother. 2010; 54: 4484-4486.
- Garcia-Fernandez A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, et al. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum β-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother. 2008; 61:1229-1233.
- Hopkins KL, Liebana E, Villa L, Batchelor M, Threfall EJ, Caratolli A. Replicon typing of plasmids carrying CTX-M or CMY β-lactamases circulating among Salmonella and Escherichia coli Antimicrobiol Agents Chemother. 2006; 50: 3203-3206.
- Marcade G, Deschamps C, Boyd A, Gautier V, Picard B, Branger C, et al. Replicon typing of plasmids in Escherichia coli producing extended spectrum β-lactamases. J Antimicrobial Chemother. 2009; 63: 67-71.
- Onnberg A, Soderquist B, Persson K, Molling P. Characterization of CTX-M producing Escherichia coli by repetitive sequence-based PCR and real-time PCR-based replicon typing of CTX-M-15 plasmids. APMIS. 2014; 122: 1136-1143.
- Carattoli A. Resistance plasmid families in Antimicrobiol Agents Chemother. 2009; 53: 2227-2238.
- Zurfluh K, Glier M, Hachler H, Stephan R. Replicon typing of plasmids carrying blaCTX-M-15 among Enterobacteriaceae isolated at the environment, livestock, human interface. Sci Tot Environ. 2015; 521-522: 75-78.