
Research Article
J Bacteriol Mycol. 2019; 6(6): 1118.
Construction of T169V and P172H Mutations in the Bile Salt Hydrolase (BSH) from Lactobacillus plantarum B14 and Biochemical Analysis of the Mutant BSHs
Öztürk M* and Türker F
Department of Biology, Faculty of Arts and Science, Bolu Abant Izzet Baysal University, Bolu, Turkey
*Corresponding author: Öztürk M, Department of Biology, Faculty of Arts and Science, Bolu Abant Izzet Baysal University, Bolu, Turkey
Received: November 21, 2019; Accepted: December 23, 2019; Published: December 30, 2019
Abstract
Bile Salt Hydrolase (BSH), synthesized by some intestinal bacteria, has an important role in the host metabolism relating with lipid absorption, energy homeostasis and glucose metabolism. Glycine and taurine in bile salts can be liberated by BSH and this process is called deconjugation. Deconjugation property of BSH exerts health benefits to the host such as reduction of blood cholesterol level. Moreover, using BSH active stains as an alternative to Antibiotic Growth Promoter (AGP) is under research. Because of the link between BSH and human health, the functional and biochemical features of BSHs necessitate an intensive study. Although BSHs exhibit variations in their catalytic activities, in addition to the five conserved amino acids, threonine-169 (T169) and proline-172 (P172) amino acids are also strictly conserved in all members of the N-terminal nucleophile (Ntn) hydrolase superfamily. However, their functions remain to be elucidated. In order to analyze the correlation between these two conserved amino acids and catalytic activity or stability of BSH, the small T169 and P172 amino acids of BSH enzyme were substituted for the aliphatic valine-169 (V169) and aromatic histidine-172 (H172) amino acids respectively by PCR-based site directed mutagenesis. Then the mutant recombinant BSHs were expressed in Escherichia coli BLR (DE3). While the effects of the mutations on catalytic activity were detected by the ninhydrin assay, the effects of the mutations on formation of the BSHs were visualized by SDS-PAGE analysis. The results showed that T169V mutation decreased the activity of BSH. Distinctly; inactive enzyme was obtained from P172H mutant. Meanwhile, assembly and folding of the BSH proteins were not affected by these mutations. Our finding demonstrated that these two amino acids might be responsible for catalytic activity or substrate binding of BSH enzyme but not for stability of it.
Keywords: Bile Salt Hydrolase; Catalytic Activity; Lactobacillus plantarum; Probiotics; Substrate Preference
Abbreviations
AGP: Antibiotic Growth Promoter; Bi: Bifido Bacterium; BS: Bile Salt; BSH: Bile Salt Hydrolase; C: Cystein; CA: Cholic Acid; Cl: Clostridium; D: Aspartate; GCA: Glycocholic Acid; GCDCA: Glycochenodeoxycholic Acid; GDCA: Glycodeoxycholic Acid; GIT: Gastrointestinal Tract; H: Histidine; Lb: Lacrtobacillus; LB: Luria- Bertani; N: Asparagine; Ntn: N-Terminal Nucleophile; R: Arginine; rBSH: Recombinant Bile Salt Hydrolase; SDS-PAGE: Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis; TCA: Taurocholic Acid; TCDCA: Taurochenodeoxycholic Acid; TDCA: Taurodeoxycholic Acid
Introduction
Bile Salts (BSs), a diverse group of amphipathic steroids, play an important role in lipid digestion in mammals [1]. The primary bile acids in human, mostly Cholic Acid (CA) and Chenodeoxycholic Acid (CDCA), are synthesized from hepatic cholesterol followed by conjugation with either glycine or taurine which are stored in the gall bladder, and then secreted into the small intestine [2]. Following secretion of the BSs into the intestinal lumen, BSs can be deconjugated into the amino acids and free bile salts by the BSH expressed in many bacterial genera residing in the human Gastrointestinal Tract (GIT). Expression of functional BSHs plays a vital role in bacterial survival and colonization in the human GIT. Therefore, the presence of an active BSH has been considered an important criterion for selection of the candidate bacterial strains that would be used as a probiotics.
On the other hand, BSH plays an important role on cholesterol level, body weight and host lipid metabolism because of its signaling functions in lipid metabolism of the host [3]. Therefore, there is strong relationship between BSH activity and some human diseases such as bowel syndrome, colon cancer [4] and irritable gallstone formation [5]. Considering the implications between deconjugation of conjugated salts and negative or positive health consequences [6,7], further functional studies of BSH proteins and site directed mutagenesis of strictly conserved amino acids are required.
Even though the crystal structures of BSHs are available for Clostridium (Cl.) perfringens [8], Bifidobacterium (Bi.) longum [9], Lactobacillus (Lb.) salivarius [10] and Enterecoccus (En.) faecalis [11], to date, the structural bases of such an important enzyme is not known very well. As described by Dong and Lee [12], there are six strictly conserved residues in the active site of BSH, including C2, R18, D21, N82, N173 and R226 (equivalent to C2, R16, D19, N79, N170 and R223 in BSH from Lb. plantarum B14). From 3D structure, x-ray crystallography and amino acid alignment of all of the known BSHs, it was predicted that these six strictly conserved amino acids might be responsible for the catalytic activity and stability of the BSH. Recent site-directed mutagenesis studies on some of these amino acids [9,13,14], supported these prediction obtained from in silico analysis of BSHs. Despite a vast diversity in amino acid sequence, in addition to these six strictly conserved amino acids, T169 and P172 amino acids are also conserved in all members of the N-terminal nucleophile (Ntn) hydrolase superfamily.
However, there is no experimental investigation on these amino acids. This study aims to understand biochemical and structural functions of the strictly conserved T169 and P172 amino acids of BSH enzyme from Lb. plantarum B14 strain.
Materials and Methods
Reagents, preparation of standards, bacterial strains and growth conditions
All enzymes and other chemicals used in this study were purchased from Thermo Scientific (Fermentas, Europe) or Sigma- Aldrich (St. Louis, MO) and were used according to the supplier’s instructions. Bile acids were purchased from Calbiochem (San Diego, CA). The plasmid DNAs and bacterial strains used in this study were listed in Table1. Cultures were inoculated in sterile Luria-Bertani (LB) broth from frozen 30% glycerol stocks and were incubated aerobically for two consecutive passages. The cultures of transgenic E. coli XL1- Blue and BLR(DE3) strains harboring pBluescript (Stratagene, USA) or pET22b (Novagen, USA) plasmids were inoculated in LB medium supplemented with ampicillin (100 μg/ml) and incubated at 37°C with 175 RPM shaking.
Strains
Genotype - Origin
Phenotype
Reference
Lb. plantarum B14
Human origin
This work
E. coli XL1-Blue
rec A end A 1 gyr A986 thi-1hsdr17supE44 rel A1 lac
Ampr
Stratagene
E. coli BLR (DE3)
F-ompT gal dcm lon hsdSB (rB- mB-) λ(DE3)
Novagen
Plasmids
Genotype
Phenotype
Reference
pBluescript
SKII+
Ampr
Fermentas
pCON1
pBluescript SKII+ with 1.0 kb bsh ORF insert
Ampr
This work
pET22b(+)
peL., T7 lac, ApR, pBR322 ori (C-terminal His-tagged
Ampr
Novagen
pCON2
protein) pET22b(+) with 1.0 kb bsh ORF insert
Ampr
This work
pCON1/T169V
Substitution of threonine to valine in pCON1
Ampr
This work
pCON1/P172H
Substitution of proline to histidine in pCON1
Ampr
This work
pFAT/T169V
Substitution of threonine to valine in pCON2
Ampr
This work
pFAT/P172H
Substitution of proline to histidine in pCON2
Ampr
This work
Table 1: Bacterial strains and plasmids used in this study.
In silico analysis of recombinant BSH from Lb. plantarum B14 strain
The amino acid sequence of the BSH from Lb. plantarum B14 strain was aligned with the amino acid sequences of BSHs belonging to different Lactobacillus species using Jalview 2.8.0b1 program. The 3-dimentional structure homology modelling of BSH of Lb. plantarum B14 (B9V401) was generated using online Automated Build Homology Models at SWISS-MODEL Homology Modelling (http://swissmodel.expasy.org/) with the crystal structure of bile salt hydrolase from En. faecalis (PDB accession code 4WL3.1.B, resolution 2.01 Å) as the template. The generated structure of BSH protein was analyzed by the PyMOL program (DeLano Scientific LLC, South San Francisco, California, USA).
Construction of T169V and P172H mutant bsh genes
The “ACA” and “GTG” codons of recombinant bsh (rbsh) gene, encoding T169 and P172, were substituted for the “CCT” and “CAT” codons, respectively encoding valine-169 (V169) and histidine-172 (H172) using pCON1 construct as a template DNA developed by Öztürk and colleagues [13]. These mutations were generated by PCR based Quick Change Site-Directed Mutagenesis Kit (Stratagene, USA) using the appropriate primers pair, as described in Table 2 according to the manufacturer’s instructions. The gel purified PCR products were transformed into the E. coli XL-Blue competent cells using a heat shock method and then the desired mutations on the final constructs were confirmed by DNA sequencing by Macrogen Inc (Seoul, Korea). The new constructs, containing the desired T169V or P172H mutant recombinant bsh (mrbsh) genes, were designated as pCON1/T169V and pCON1/P172H, respectively. These constructs containing the desired mutations were amplified and analyzed in E. coli XL-1 Blue strain.
Primers
Sequences (5’-3’)
Tm
T169V-F
5’-CAGTAGGTGTGTTAGTGAACAATCCTAATTTTG-3’
60 °C
T169V-R
5’-CAAAATTAGGATTGTTCACTAACACACCTACTG-3’
60 °C
P172H-F
5’-GTTAACAAACAATCATAATTTTGACTACC-3’
53°C
P172H-R
5’-GGTAGTCAAAATTATGATTGTTTGTTAAC-3’
53°C
Table 2: List of the primers that were used for substitution of T169 and P172 amino acids for V169 and H172 amino acids respectively.
Expression of mutant BSHs and preparation of the cell lysates
To overexpress rbsh and mrbsh, the rbsh gene from pCON1 and mrbsh ORFs from pCON1/T169V or pCON1/P172H were inserted into EcoRI/NotI sites of pET22b expression vector and then transformed into E. coli BLR(DE3) strain as described by Öztürk and colleagues [13,14]. Transformants were selected based on ampicillin resistance and plasmid size (6.5 kbp). Cell lysates were prepared according to the protocol described by Sambrook and colleagues [15] with some modifications described by Öztürk and colleagues [13]. Protein concentration was measured by the Lowry protein assay [16]. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 0.1% (w/v) SDS and a 12% (w/v) polyacrylamide separating gel described by Laemmli [17] was performed to monitor production of the rBSH and mrBSHs.
Catalytic activity of mutants rBSH
The catalytic activity of the mrBSHs was measured by the ninhiydrin assay developed by Tanaka and colleagues [18] with some modifications described by Öztürk and colleagues [13,14]. The protocol is based on the interaction of the amino acids released from the conjugated bile salts with ninhydrin reagent and the detection of the reaction product (at 570 nm on a U-1900 spectrophotometer (HITACHI, Japan)). The catalytic activities of rBSH and mrBSHs were determined by incubation of the enzyme sample with six different human bile acids, Glycocholic Acid (GCA), Glycodeoxycholic Acid (GDCA), Glycochenodeoxycholic Acid (GCDCA), Taurocholic Acid (TCA), Taurodeoxycholic Acid (TDCA) and Taurochenodeoxycholic Acid (TCDCA). The highest enzyme activity (100%) was recorded with the wild-type enzyme at 37°C and pH 6. The other activities were converted into relative activities, with respect to the wild type. Catalytic activity assay was accomplished in triplicate in individual experiments to produce standard deviations for all experiments. Positive and negative controls were set up simultaneously in each independent experiment. A standard curve using glycine or taurine was performed for each independent assay.
Results
Active site of BSH
The alignment of amino acids residues of BSHs from different organisms in (Figure 1) showed that T169 and P172 amino acids around N170 residue were also strictly conserved. On the other hand, the inferred 3D structure of the BSH revealed that the distance of T169 or P172 side chains was close to the C2 that is structurally important amino acid (Figure 2).

Figure 1: Multiple sequence alignment of BSH proteins from following organisms; Lb. rhamnosus (WP_014569128.1), Lb. casei (WP_003583296.1), Lb. oris
(WP_062813221.1), Lb. antri (EEW53327.1), Lb. salivarius (YP_535410.1), Lb. fermentum (WP_012390665.1), Lb. oris CNH2 (WP_062812843.1), Lb. hominis
(CCI81875.1), Lb. gasseri (ACL98172.1), Lb. acidophilus (ACL98173.1), Lb. reuteri (PIN31168.1), Lb. antri (EEW53327.1), Lb. salivarius (WP_101898179.1),
Lb. ruminis (WP_003697845.1), Lb. plantarum (AQU14359.1), En. Feacalis (4WL3), Li. monocytogenes (AKI41557.1), Cl. perfringens (WP_003459238.1), Ba.
anthracis (ASE28680.1), Ba. subtilis (Q2HPP6). Frame shows conserved T169 and P172 amino acids around N170.
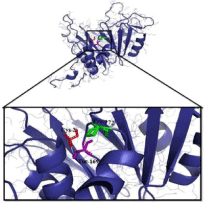
Figure 2: Three-dimentional structure of Lb. plantarum monomer of BSH
protein. The catalytic site of BSH of Lb. plantarum B14 shows strictly
conserved amino acids, cysteine 2, threonine 169 and proline 172. This
Figure was created by pMoL 0.99rc6 program.
Determination of the catalytic activities of the rBSH and mrBSHs
The quantitative catalytic activities of the rBSH and mrBSHs were determined by ninhydrin assay in which six major human bile salts, GCA, GDCA, GCDCA, TCA, TDCA and TCDCA, were used as substrate. The recombinant BSH (rBSH) enzyme from Lb. plantarum B14 strain was able to hydrolyze both the glycine and taurine conjugates. However, the rBSH exhibited high preference for glycineconjugated bile acid over taurine-conjugated bile acids (Figure 3). The highest hydrolysis activity occurred when GDCA was used as the substrate at 37°C and pH 6.0 with wild-type rBSH and this activity was defined as 100% activity. However, The T169V mutation decreased the BSH activity more than 50% against to glycol-conjugated and tauro-conjugated bile acids. On the other hand, P172H mutation almost completely inactivated the BSH enzyme activity especially for glycol-conjugated bile acids (Figure 3).
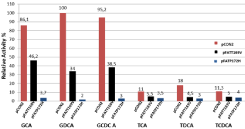
Figure 3: Measurement of the recombinat bile salt hydrolase (rBSH) and
the mutant recombinat bile salt hydrolase (mrBSH) activities. The rBSH and
mrBSHs activities were measured by determining the amount of amino acids
liberated from GCA, GDCA, GCDCA, TCA, TDCA and TCDCA. Relative
activity was defined as the measured BSH activity for each substrate
compared to the highest activity (taken as %100) in the assay. Values are
expressed as the mean of three independent replicates.
Comparison of the expression of the wrBSH and mrBSHs
Mutant rBSHs and rBSH were loaded on 12% SDS-PAGE with a molecular mass of 37 kDa (Figure 4). The theoretical molecular mass of rBSH (36.7 kDa) obtained using BSH amino acid residues by ExPASy compute.pi program is in agreement with that of our experimental results. The replacement of the T169 or P172 amino acids with valine and histidine respectively resulted in presence of desired 36.7 kDa BSH band with the same electrophoretic mobility in the gel. These results indicated that T169 and P172 amino acids are critical for catalytic activity rather than stability of BSH (Figure 4 lanes 3 and 4).
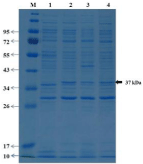
Figure 4: Electrophoretic examination of the recombinant bile salt
hydrolases. SDS-PAGE analysis of wild-type recombinant bile salt hydrolase
(wrBSH) and mutant recombinant bile salt hydrolases (mrBSH), expressed
in E. coli BLR(DE3) strain, was performed on 12% polyacrylamide gels
under denaturing conditions. Gels were stained with Coomassie Brilliant
Blue R250. Lanes: M, molecular weight marker; 1, cell extract from E. coli
pET22b/BLR(DE3) host strain (negative control); 2, cell extract from the E.
coli pCON2/BLR(DE3) host strain encoding wild-type BSH; 3, cell extract
from E.coli pFAT/T169V/BLR(DE3) host strain encoding mrBSHT169V;
4, cell extract from E.coli pFAT/P172H/BLR(DE3) host strain encoding
mrBSHP172H. Similar to wild-type BSH in lane 2, both mutant BSHs are
visible in lanes 3 and 4.
Discussion
Lambert and colleagues [19] separated members of the Ntn hydrolase superfamily into two groups; a PVA cluster containing penicillin V acylase related proteins and a BSH cluster that contains BSHs. Although the members of Ntn hydrolase superfamily share very limited primary sequence overall similarity, they have strictly conserved critical residues at their active sites. Previous structural and comparative genomics studies [20,21,22] have identified that C2, R16, D19, N170 and R223 amino acids were strictly conserved in all BSHs of Lactobacillus species, PVA of Bacillus sphaericus and CBAH of Cl. perfringens species. Most of these five strictly conserved amino acids verified by site- directed mutagenesis that are responsible for the catalytic activities of BSH enzymes [13,14]. However, there is no experimental study on conserved T169 and P172 amino acids, which are located around N170. Therefore, to discover the critical residues for the catalysis of BSH and to understand the function of these residues and catalytic-binding pocket relationships, comprehensive amino acid substitution mutagenesis was advised by recent articles [23,24,25].
Tanaka and colleagues [26] confirmed the role of Cysteine-2 (C2) by Changing of C2 to Alanine (C2A) by site-directed mutagenesis. The C2A mutation led to complete inactivation of BSH. The more conservative substitutions, C2S and C2T, also inactivated the BSH from Bi. bifidum [27]. The other totally conserved amino acid residues, R16 and D19, in loop I of the BSH from Lb. plantarum B14 strain were substituted for phenylalanine and leucine respectively by Öztürk and colleagues [13]. Their D19L mutation resulted in totally inactivation of BSH enzyme although they observed stable mutant BSH proteins on SDS-PAGE. However, R16F mutation resulted in inexpression of BSH enzyme. On the other hand, N79 of BSH from Enterococcus faecalis (EfBSH) was mutagenized by Chand and colleagues [11] and their mutant analysis showed that the 7a-OH group of substrate glycocholic acid interacted with N79. While N79Y mutant showed good protein expression, it’s enzyme activity retained ~88% of wild-type EfBSH activity. However, their N79W mutant showed drastic decrease in both expression and BSH activity. N79 of BSH from Lb. plantarum B14 was also substituted for the aliphatic and hydrophobic valine by Öztürk and colleagues [14]. They found that although N79V mutation resulted in stable BSH, it reduced the catalytic activity and altered substrate specificity of BSH.
Briefly, our in silico analysis results are consistent with our experimental results. Our in silico analysis results showed that T169 and P172 amino acid residues are physically close to the catalytic site or inside the catalytic site of the BSH. Therefore, T169V and P172H mutant BSHs from Lb. plantarum B14 revealed that these conserved amino acid residues had a significant role on catalytic activity of BSH. However, to better understand the exact functions of the T169 and P172 amino acids and other strictly or partially conserved amino acids, more biochemical investigations and further multiple sitedirected mutagenesis studies are required.
Acknowledgements
This work was supported by funds from The Scientific Technological Research Council of Turkey (TUBITAK) by grant TBAG-113Z130 (to MÖZ).
References
- Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2004; 29: 625-651.
- Pereira DI, Gibson GR. Effects of consumption of probiotics and prebiotics on serum lipid levels in humans. Crit Rev Biochem Mol Biol. 2002; 37: 259-281.
- Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. PNAS of the USA. 2014; 111: 7421-7426.
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic an analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2011; 22: 292-298.
- Thomas LA, Veysey MJ, Bathgate T, King A, French G, Smeeton NC, Murphy GM, Dowling RH. Mechanism for the transit-induced increase in colonic deoxycholic acid formation in cholesterol cholelithiasis. Gastroenterology. 2000; 119: 806-815.
- Ridlon JM, Kang DJ Hylemon, PB. Bile salt biotransformations by human intestinal bacteria. J Lipid
- Res. 2006; 47: 241-259.
- Jiang J, Hang X, Zhang M, Liu X, Li D, Yang H. Diversity of bile salt hydrolase activities in different lactobacilli toward human bile salts. Ann Microbiol. 2010; 60: 81-88.
- Rossocha M, Schultz-Heienbrok R, von Moeller H, Coleman P, Sanger W. Conjugated bile acid hydrolase is a tetrameric N-terminal thiol hydrolase with specific recognition of its cholyl but not of its tauryl product. Biochemistry. 2005; 44: 5739-5748.
- Kumar RS, Brannigan JA, Prabhune AA, Pundle AV, Dodson GG, Dodson EJ, Suresh CG. Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V. J Biol Chem. 2006; 281: 32516-32525.
- Xu F, Guo F, Hu X-J, Lin J. Crystal structure of bile salt hydrolase from Lactobacillus salivarius. Acta Cryst. 2016; F72: 376-381.
- Chand DP, Panigrahi N, Varshney S, Ramasamy C, Sures G. Structure and function of a highly active Bile Salt Hydrolase (BSH) from Enterococcus faecalis and post-translational processing of BSH enzymes. BBA - Proteins and Proteomics. 2018; 1866: 507-518.
- Dong X, Byong HL. Bile salt hydrolases: Structure and function, substrate preference and inhibitor development. Protein sci. 2018; 27: 1742-1754.
- Öztürk M, Hacibeyoğlu K, Kiliçsaymaz Z and Önal C. Construction of R16F and D19L mutations in the loop I of Bile Salt Hydrolase (BSH) enzyme from Lactobacillus plantarum B14 and functional analysis of the mutant BSHs. Food Biotechnology. 2019; 33: 125-141.
- Öztürk M and Önal C. Asparagine 79 is an important amino acid for catalytic activity and substrate specificity of bile salt hydrolase (BSH), Molecular Biology Reports. 2019; 46, 4361-4368.
- Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York. 2001.
- Lowry OH, Rosebrough NJ, Farr AL, Randal RJ. Protein measurement with the Folin phenol reagent.
- J Biol Chem. 1951; 193: 265-275.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227: 680-685.
- Tanaka H, Doesburg K, Iwasaki T, Mierau I. Screening of lactic acid bacteria for bile salt hydrolase activity. J Dairy Sci. 1999; 82: 2530-2535.
- Lambert, JM, Bongers RS, de Vos WM, Kleerebezem M. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl Environ Microbiol. 2008; 174: 4719- 4726.
- Begley M, Hill C, Gahan CGM. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol. 2006; 72: 1729-1738.
- Oh HK, Lee JY, Lim SJ, Kim MJ, Kim BG, Kim JH, et al. Molecular cloning and characterization of
- bile salt hydrolase from Lactobacillus acidophilus PH01. J Microbiol Biotechnol. 2008; 18: 449-456.
- Kumar R, Grover S, Mohanty AK, Batish VK. Molecular cloning and sequence analysis of bile salt hydrolase (bsh) gene from Lactobacillus plantarum MBUL90 strain of human origin. Food Biotech. 2010; 24: 215-226.
- Yip KY, Utz L, Sitwell S, Xihao HX, Sidhu SS, Turk BE, et al. Identification of specificity determining residues in peptide recognition domains using an information theoretic approach applied to large-scale binding maps. BMC Biol. 2011; 9: 1-15.
- Chae JP, Valeriano VD, Kim GB, Kang DK. Molecular cloning, characterization and comparison of bile salt hydrolases from Lactobacillus johnsonii PF01. J Appl Microbiol. 2012; 114: 121-133.
- Geng W, Lin J. Bacterial bile salt hydrolase: an intestinal microbiome target for enhanced animal health. Anim Health Res Rev. 2017; 17: 148-158.
- Tanaka H, Hashiba H, Kok J, Mierau I. Bile salt hydrolase of Bifidobacterium longum’s biochemical and genetic characterization. Appl Environ Microbiol. 2000; 66: 2502-2512.
- Kim GB, Yi SH, and Lee BH. Purification and characterization of three different types of bile salt hydrolases from Bifidobacterium strains. J. Dairy Sci. 2004; 87: 258-266.