
Research Article
J Bacteriol Mycol. 2020; 7(8): 1159.
Plant Rhizosphere Growth-Promoting Bacterium with Root-Knot Nematode Inhibition and Its Effect on the Tomato Rhizosphere Microbial Community Structure
Jianfeng Du1,2, Qixiong Gao1, Zhaoyang Liu1, Chaohui Li1, Xin Song1, Ruiping Xu1, Yanyan Zhou1, Yue Liu1, Huying Li1, Rui Zheng1, Xunli Liu1*
1College of Forestry, Shandong Agriculture University, China
2State Key Laboratory of Crop Biology, Shandong Provincial Key Laboratory for Biology of Vegetable Diseases and Insect Pests, College of Plant Protection, Shandong Agricultural University, China
*Corresponding author: Xunli Liu, College of Forestry, Shandong Agriculture University, No. 61, Daizong Street, Taian, Shandong 271018, China
Received: November 17, 2020; Accepted: December 15, 2020; Published: December 22, 2020
Abstract
The purpose of this study was to evaluate the ability of Bacillus aryabhattai P-3 to control Meloidogyne incognita and its influence on the tomato rhizosphere microbial community. When the P-3 strain was used to treat the J2s of Meloidogyne incognita for 24 hours, the corrected mortality of the J2s of Meloidogyne incognita was 81.23%± 1.23b%. When the P-3 strain was used to treat the J2s of Meloidogyne incognita for 48 hours, the corrected mortality rate of the J2s of Meloidogyne incognita was 83.56%±2.56 % for in vitro tests. The P-3 was identified as Bacillus aryabhattai by 16srDNA and physiological biochemical tests. In the pot experiment, the control effect of Bacillus aryabhattai on Meloidogyne incognita was 41%. Bacillus aryabhattai P-3 was proven to control Meloidogyne incognita. MiSeq sequencing and bioinformatics analysis verified that the P-3 can change the composition of the microbial community in the tomato rhizosphere and reduce the number of plant pathogens, increase the complexity of the bacterial microbial community, and make the bacterial community structure more stable. The P-3 as the ability to control Meloidogyne incognita. Meanwhile, the P-3 can be developed as a microbial agent. This research hopes to contribute to the development of microbial inoculants. We demonstrated Bacillus aryabhattai P-3 efficacy and value to control Meloidogyne incognita. We also clarified the effect of Bacillus aryabhattai P-3 on tomato rhizosphere microbial community.
Keywords: PGPR; Meloidogyne incognita; Microbial Community; Microbial Inoculant
Introduction
Root-knot nematode disease is a common plant disease that seriously endangers world agricultural production [1] and affects many plants such as tomatoes [2]. It is mainly caused by Meloidogyne incognita [3]. The disease commonly occurs in tomato plants based in greenhouses and open fields [4]. Particularly in greenhouses, it may occur all the year-round, making it a serious threat to tomato production [5]. Root-knot nematode disease can decrease crop yields by 10%-20% that can reach more than 75% in severe cases [6]. With the continuous development of facility horticulture in China, the production area of vegetables grown in greenhouses is increasing. In China, Shandong Province is an important vegetable planting area especially for tomatoes [7,8]. Currently, the methods of controlling Meloidogyne incognita in agriculture mostly involve chemical control [9]. However, chemical control can lead to Meloidogyne incognita developing a resistance, which can also damage the ecological balance [10]. Some chemical pesticides can cause environmental pollution [11,12]. With the increase in awareness of environmental protection and increasing concern for food safety [13], strengthening the exploitation of microbial resources is of great significance for future agricultural production [14,15].
After years of research, many microbial resources have been screened for controlling Meloidogyne incognita, including fungi, bacteria, and actinomycetes. For example, Paecilomyces lilacinus is currently widely used in the agricultural field [16]. Rhizosphere bacteria are important to help the control of Meloidogyne incognita [17,18]. Studies have shown that many rhizosphere bacteria can control Meloidogyne incognita, such as Bacillus subtilis, Bacillus licheniformis, Bacillus megaterium, Bacillus coagulans, and Pseudomonas fluorescens [19]. Plant-growth-promoting rhizobacteria are important biological resources [20]. They can increase crop yields and help plants resist pathogenic microorganisms [21]. [22] Evaluated the effects of 662 rhizobacteria on Meloidogyne incognita and found Bacillus to be causing the highest Meloidogyne incognita mortality [22]. Bacillus can not only directly stimulate plant growth by enhancing nutrient acquisition or stimulating the host plant's defense mechanism but also by inhibiting the growth of pathogenic microorganisms [23]. Antoun demonstrated that approximately 2-5% of rhizobacteria can promote plant growth [24]. Moreover, growth, and single or multiple rhizosphere bacteria can control root-knot nematodes. A study shows that rhizobacterial can be used to prevent parasitic nematodes of grapevine [25]. A study used Phanerochaete chrysosporium to inhibit J2s and eggs of Meloidogyne incognita [26]. Liu identified Bacillus halotolerans, B. kochii, B. oceanisediminis, B. pumilus, B. toyonensis, B. cereus, Pseudomonas aeruginosa, and B. pseudomycoides as rhizobacteria effective at controlling Meloidogyne incognita [27]. Rhizosphere bacteria can also induce resistance in plants to Meloidogyne incognita [28]. The combination of Bacillus amyloliquefaciens and Bacillus subtilis strains can reduce the number of Meloidogyne incognita in the soil [29]. Studies have shown that rhizobacteria not only have the ability to control Meloidogyne incognita [30,31] but also have the ability to improve soil fertility [32] and reduce the number of plant pathogens in the soil [33,34]. Bacillus aryabhattai is an important component of rhizobacteria [35]. It can not only synthesize biological hormones or active organic matter but also promote plant growth [36]. For example, Bacillus aryabhattai AB211 can dissolve inorganic phosphate, synthesize iron carriers, and produce hormones such as Indole Acetic Acid (IAA) [37]. This species also controls root-knot nematodes. For example, Bacillus sneb517 controls Heterodera glycines through seed coating ichinohe to promote plant growth Bacillus aryabhattai SRB02 can increase the yield of crops such as rice and soybean [38]. At the same time, Bacillus aryabhattai can control plant pathogens in the soil. For example, Bacillus aryabhattai inhibits Pyricularia oryzae and Fusarium moniliforme to increase rice yield [36]. It is well known that the composition and function of microbial communities in the rhizosphere in soil play a vital role in the healthy growth of plants [39]. In the underground ecosystem, the soil rhizosphere microbial community is a key component, which can directly or indirectly affect the growth of plants and change the soil’s functional performance [40]. In fact, some studies have shown that rhizosphere bacteria are able to prevent and control soil-borne diseases and increase available phosphorus in the soil [41], but there are few studies investigating the effect of Bacillus aryabhattai on underground microbial communities. This study used Miseq sequencing technology and bioinformatics methods to comprehensively analyze and compare the microbial community composition of tomato rhizosphere. It is expected to contribute to the development of the microbial inoculum.
Materials and Methods
Determination of the effect of P-3 strain poisoning the J2s of Meloidogyne incognita
The P-3 strain was inoculated in Lysogeny Broth (LB) medium, cultured at 30°C, 200 r/min in the dark for 48 hours, centrifuged at 1073 × g for 10 minutes, filtered through a 0.22 μm bacterial filter, and 0.8 mL of the filtrate was placed in a 1.5 centrifuge tube. One hundred J2s of Meloidogyne incognita were added to each centrifuge tube, and their corrected mortality was calculated at 24 hours and 48 hours. And each treatment had 9 duplicates.
Identification and phylogenetic analysis of the P-3 strain
According to the common bacterial system identification manual, the bacterial morphology and physiological and biochemical indicators of P-3 strains were measured [42]. The 16S rDNA fragment was amplified [43], and the sequencing results were analyzed with BLAST from the NCBI's GenBank database. The neighbor-joining phylogenetics were then analyzed with MEGA of multiple sequence homology [44].
Pot test
The tomato Micro-Tom of the tested variety was sown in a nursery tray, and when the tomato seedlings grew to four true leaves, they were transplanted into a plastic pot with a diameter of 20-cm containing diseased soil. Two days after transplanting, each tomato was watered with P-3 strain 10×109 CFUs, and the same amount of sterile water was used as a control. After inoculation, pots were randomly placed on the operating platform of a glass greenhouse. After 60 days of cultivation, the plants were taken out of the pots. Each treatment is repeated 3 times, each time 10 tomato seedlings are replicated. The incidence index was recorded and the effect of controlling southern root-knot nematodes was calculated according to the method of Liu [27]. The rhizosphere soil collected from each duplicated 10 pots of tomato seedlings was thoroughly mixed as a duplicate, so each treatment had 3 duplicates. Rhizosphere soil was collected around the tomato rhizosphere and stored at -80°C for microbial community structure analysis.
DNA extraction and Illumina MiSeq high-throughput sequencing
The BIO-TEK OMEGA Soil DNA Kit method (Omega Bio-tek, Norcross, GA) was used to extract the total DNA from the soil. At the same time, the bacterial 16S rDNA V3–V4 region and the fungal rDNA-ITS gene were amplified. Polymerase Chain Reaction (PCR) amplification was performed according to a method previously described [45], and Illumina MiSeq was used for sequencing. All reads were clustered with a 97% similarity cut-off using UPARSE (ver. 7.1, http://drive5.com/uparse/), and chimeric sequences were identified and removed using UCHIME [46]. The taxonomy of each 16S rRNA and ITS rDNA gene sequence was analyzed using the RDP Classifier against the Silva (SSU123) 16S rRNA database [47] and the UNITE 7.0/ITS database [48] using a confidence threshold of 70%. Bacterial population functions were performed using the PICRUSt 2 database. The fungal ecosystem analysis was performed using the FUNGuild database [49].
Statistical analysis
In the strain function test and pot test, the data was tested using the Duncan multi-pass test, and the difference was significant. The "Vegan" software package was also used in PCoA to determine community composition differences and community succession based on Bray-Curtis sums. All statistical analyses were performed using R software v. 3.5.2. Using the psych package, the abundance matrix of the top 50 species in the bacterial microbial community and the top 49 species in the bacterial microbial community were calculated at the genus level. Using Gephi 0.9.2, the topological properties of the co-occurring network graph were calculated and drawn.
Results and Analysis
P-3 with controlling J2s of Meloidogyne incognita and phosphorus-dissolving property
In this study, long-term preservation of rhizosphere growth-promoting bacteria in the laboratory was used to screen for the prevention and control of Meloidogyne incognita. Subsequently, these bacterial strains were evaluated against Meloidogyne incognita. However, only the P-3 strain has a 24-hour corrected mortality of Meloidogyne incognita greater than 80%. When the Meloidogyne incognita J2s were treated with the P-3 fermentation supernatant for 24 hours, the mortality rate of the Meloidogyne incognita J2s was 81.23%± 1.23b%. The Meloidogyne incognita J2s had a mortality of 83.56%±2.56% after 48 hours (Table 2). The controlling of Meloidogyne incognita of P-3 strains is 41%. The results showed that the P-3 strain had functions that controlled Meloidogyne incognita. Thus, it can be concluded that P-3 had a strong ability to kill Meloidogyne incognita J2s.
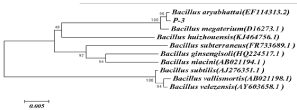
Figure 1: Phylogenetic tree based on 16S rDNA sequence of the P-3.
A) Family richness table of tomato rhizosphere bacterial;
B) Family richness table of tomato rhizosphere fungi;
C) PCoA analysis based on Bray Curtis distance of bacterial community;
D) PCoA analysis based on unweighted UniFrac distances of bacterial community.
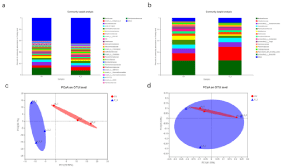
Figure 2: Composition and succession of microbial communities.
A) Family richness table of tomato rhizosphere bacterial;
B) Family richness table of tomato rhizosphere fungi;
C) PCoA analysis based on Bray Curtis distance of bacterial community;
D) PCoA analysis based on unweighted UniFrac distances of bacterial community.
a) CoG function classification analysis;
b) FUNGuild functional classification statistical histogram.
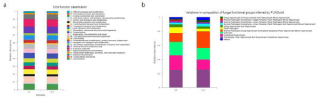
Figure 3: Analysis of rhizosphere bacterial community function and functional guild of fungi.
a) CoG function classification analysis;
b) FUNGuild functional classification statistical histogram.
Identification of bacterial strain P-3
The P-3 strain was round, white, and transparent and had a moist surface, regular edges, rod-shaped bacteria, no spore production, and a positive gram stain. Details of their physiological and biochemical properties are summarized in Table 1. The homology of 16S rRNA between the P-3 strain and the known strain Bacillus aryabhattai (EF114313) reached 99% (Figure 1). Combined with the observation results of its morphology and colony characteristics, and the determination of physiological and biochemical indicators, P-3 strain was identified as Bacillus aryabhattai.
Effect of P-3 on the composition and structure of tomato rhizosphere microbial community
The composition and cluster analysis of the microbial community of tomato rhizosphere microorganisms are shown in Figure 2. P-3 treatment can change the composition of bacterial communities in tomato rhizosphere. Among all identified families, the relative abundance of 27 families in all samples was >1%. As shown in Figure 1, the dominant families in the control (relative abundance in at least one sample was >3%) were Bacillaceae, norank_c_Subgroup_6, Nocardiodaceae, and JG30-KF-CM45. The dominant family in the P-3 processing were Bacillaceae, norank_c_Subgroup_6, Nocardiodaceae, JG30-KF-CM45, and Gemmatimonadaceae. In general, there were differences in the composition of the dominant gates of the two groups of samples. From PCoA analysis based on Bray–Curtis distance, it was also confirmed that the P-3 can change the composition of bacterial communities in the tomato rhizosphere. PCoA analysis based on unweighted UniFrac distances showed that compared with CK, the use of P-3 microbial inoculants did not cause significant succession for a tomato rhizosphere bacterial community. Bacillus aryabhattai can also alter the composition of tomato rhizosphere fungi. Of all the identified families, the relative abundance of 15 families in all samples was more than 1%. As shown in Figure 2, the dominant families (relative abundance > 3% in at least one sample) were Mortierellaceae, Chaetomiaceae, unclassified-c-omycetes, Aspergellaceae, Plectosephaerellaceae, Nectriaceae, Acodesmidaceae, unclassified-o-Sordarales, unclassified-fungi, and Gymnoasccae. The dominant families in P-3 processing were Mortierellaceae, Chaetomiaceae, unclassified_c_sordariomycetes, Aspergillaceae, Plectosaphaerellaceae, Nectriaceae, Ascodesmidaceae, and unclassified_o_sordariales. In general, the composition of the dominant families of the two groups of samples is different. PCoA analysis based on Bray-Curtis distance distances showed that compared with CK, the use of P-3 microbial inoculants did not significantly change the composition of tomato rhizosphere fungi. PCoA analysis based on unweighted UniFrac distances showed that compared with CK, the use of P-3 microbial inoculants caused tomato rhizosphere bacterial community succession. In summary, the P-3 can change the composition of bacterial and fungal microbial communities in the tomato rhizosphere and allow fungal communities in the tomato rhizosphere to undergo directional succession.
(a) Tomato rhizosphere bacterial species correlation network diagram of CK;
(b) Tomato rhizosphere bacterial species correlation network diagram of P-3;
(c) Tomato rhizosphere fungi species correlation network diagram of CK;
(d) Tomato rhizosphere fungi species correlation network diagram of P-3.
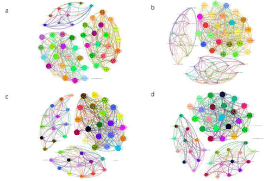
Figure 4: Tomato rhizosphere fungi and bacterial species correlation network.
(a) Tomato rhizosphere bacterial species correlation network diagram of CK;
(b) Tomato rhizosphere bacterial species correlation network diagram of P-3;
(c) Tomato rhizosphere fungi species correlation network diagram of CK;
(d) Tomato rhizosphere fungi species correlation network diagram of P-3.
Property
Result
Property
Result
Morphology
Rod-shaped
Glucose oxidative fermentation
Fermented
Swelling spore
-
Aerobic
+
Gram staining
+
Nitrate reduction
+
Colony color
white
2?NaCl
+
Colony morphology
Round, transparent
5?NaCl
+
Colony edge
neat
Starch hydrolysis
+
Colony surface
Moist
Methyl red test
-
Bulge
Raised
(+) = positive; (-) = negative
Table 1: Physiological and biochemical properties of the P-3 strain.
Inhibition effect on Meloidogyne incognita
24h
48h
Corrected mortality%
81.23±1.23b
83.56±2.56 a
Effect of controlling of Meloidogyne incognita
-
41%
24 h and 48 h represent J2 mortality of (Bacillus aryabhattai) P-3 in 24 h and 48 h in vitro exposure. Means in each column followed by the same letter do not differ significantly according to ANNOVA's multiple range test at P= 0.05.
Table 2: Identifying the ability of the P-3 to kill the Meloidogyne incognita and dissolve phosphorus.
Effect of the P-3 on rhizosphere bacterial community function and functional guild of fungi
Co-occurrence analysis of the tomato rhizosphere bacteria microbiome by CK and P-3 demonstrated in the gate-level correlation network diagram revealed that tomato rhizosphere bacteria had significant interactions between different gates. In the bacterial microbial community of the tomato rhizosphere, the total number of edges in CK treatment is 456, of which the number of positively and negatively correlated edges accounts for 55.26% and 44.74%, respectively. Nonomuraea, Agromyces, and Bacillus are the key flora. The total number of edges in P-3 processing is 494, of which the number of positively and negatively correlated edges accounts for 57.89% and 42.11% of the total edges, respectively. Nocardioides and norank_c__Subgroup_6 are the key flora. In the tomato fungal microbial community, the total number of edges in CK treatment is 436, the number of positively and negatively correlated edges accounts for 65.37% and 34.63%, respectively. Cephaliophora, Mortierella, Conocybe are the key flora. The total number of edges in the P-3 treatment is 424. The number of positively and negatively correlated edges accounts for 57.78% and 42.22% respectively. Mortierella, unclassified_c__Sordariomycetes, and Myceliophthora are the key flora (Table 3). The results showed that the P-3 treatment increased the complexity of the bacterial microbial community and reduced the complexity of the fungal microbial community, and the P-3 treatment also changed the key flora of the tomato rhizosphere microbial community by increasing the number of positive and negative correlation edges, which indicates that P-3 treatment makes the bacterial microbial community more stable. However, in the tomato rhizobacterial fungal microbial community, P-3 strains reduce the number of positive correlation edges and increase the number of negative correlation edges. Combining the data from the FUNGuild database, this phenomenon may be related to the P-3 strain reducing the number of plant pathogens in the soil. The average degree and clustering coefficient in the topological properties of the co-occurrence network graph also verified that the P-3 treatment increased the complexity of the bacterial microbial community and reduced the complexity of the fungal microbial community (Figure 4).
Test treatment
Average degree
Total number of edges
Positive correlation edge
Number of negative correlation edges
Clustering coefficient
Bacterial
CK
18.24
456
55.26%
44.74%
0.976
P-3
19.76
494
57.89%
42.11%
0.98
Fungus
CK
17.8
436
65.37%
34.63%
0.977
P-3
17.31
424
57.78%
42.22%
0.977
Table 3: Topological properties of co-occurring network graphs.
Discussion
In China, high-value crops such as tomatoes are easily attacked by root-knot nematodes, leading to reduced yields [50,51]. Many biological resources have been used to control root-knot nematodes including several PGPRs, which play important role in plant protection. Serious ecological problems are caused by the long-term use of pesticides and fertilizers in developing countries. S. proteamaculans has been reported to be related to the control of Meloidogyne incognita. It has also been reported that Pseudomonas kills Meloidogyne incognita; and Bacillus subtilis, Bacillus amyloliquefaciens, and Bacillus cereus can all inhibit egg hatching. It is reported that Bacillus cereus UW85 can control disease occurrence, including Meloidogyne incognita. A study used Bacillus cereus, Bacillus licheniformis, Lactobacillus sphaeroides, Pseudomonas fluorescens, and Pseudomonas brassicae to conduct a greenhouse test against Meloidogyne incognita, and the results showed that Bacillus licheniformis and Pseudomonas fluorescens significantly reduced the infection of tomato roots by second stage juveniles of Meloidogyne incognita [52]. Endophytic bacteria Bacillus cereus BCM2 can affect rhizosphere secretions (2,4-di-tert-butylphenol, 3,3-dimethyloctane, and n-tridecane secretions). They can inhibit the J2s of Meloidogyne incognita and reduce the number of Meloidogyne incognita in soil [53]. A study found that B. aryabhattai A08 can reduce the number of Meloidogyne incognita in the soil [54]. It is difficult to prevent and control Meloidogyne incognita in the soil because it easily interacts with plant parasitic nematodes and pathogens, forming a complex disease environment. Therefore, it is of great significance to screen multifunctional strains to control Meloidogyne incognita and increase soil nutrient content. The in vitro results showed that the P-3 strain has a strong toxic effect on Meloidogyne incognita (Table 2). The pot experiments indicated that the P-3 strain significantly reduced the abundance of Meloidogyne incognita and plant pathogens (Figure 3). This shows that the P-3 strain has the function of preventing Meloidogyne incognita and plant pathogens.
Rhizosphere bacteria can inhibit the development of soil-borne diseases. Research shows that the Serratia.spp strain is an important resource for controlling soil-borne diseases. Soil microbial communities play an important role in disease control, and beneficial soil microorganisms may help to suppress plant pathogens. Han reported that Bacillus amyloliquefaciens B1408 can promote plant growth and reduce the damage induced by Fusarium oxysporum f. Sp. Cucumerinum (FOC) by altering the composition of cucumber rhizosphere microbial communities [55]. Rhizosphere bacteria can control plant pathogens [56,57], but few people use the FUNGuild database to analyze rhizosphere plant pathogen flora. The FUNGuild database shows that the P-3 strain can significantly reduce the number of plant pathogens, which means that the number of tomato rhizosphere plant pathogens is significantly reduced. Therefore, the P-3 strain can reduce the risk of plant diseases and improve the health of the tomato rhizosphere ecosystem.
PGPR is an important component of beneficial rhizosphere microorganisms [58]. Luo reported that the application of Sphingomonas sp. Cra20 changed the rhizosphere native bacterial community and could promote the growth of Arabidopsis thaliana by driving the developmental plasticity of the roots, thereby stimulating the growth of lateral roots and root hairs [59]. Rhizosphere microorganisms have significant importance because they can manage nutrient transformation, nutrient acquisition and use, and crop sustainability [60]. Rhizosphere microflora enhances plant growth under abiotic stress through nitrogen fixation, plant hormone production, mineral solubilization, and iron carrier and HCN production, and triggers plant defense mechanisms against different bacterial and fungal pathogens [61]. The composition of rhizosphere microbial communities plays an important role in the stability of plants, soil, and rhizosphere microbial communities [62]. Previous research indicates that farming practices may affect the composition of rhizosphere microbial communities [63]. Syringic acid changes occur in the community composition of bacteria and fungi in the cucumber rhizosphere, which may have a negative impact on the growth of cucumber seedlings by inhibiting plant beneficial microorganisms [64]. PGPR can affect microbial community succession and increase plant yield [51]. Rhizosphere microorganisms play an important role in most ecosystem processes, and different crop management strategies applied to agricultural production can change microbial composition. Rhizosphere bacteria have the ability to increase available phosphorus content in soil. Studies show that rhizosphere bacteria can be used as biological fertilizers to provide nutrients for plant growth PGPR can live in the rhizosphere of plants, thereby improving the control of nematodes and promoting plant growth. Bacillus cereus can promote plant growth. Studies have shown that underground microbial communities play a vital role in plant growth [65]. Beneficial underground microbial communities allow the healthy growth of plants. However, the microbial community structure in the rhizosphere is most closely related to plant growth [66]. Plant root exudates can affect the microbial community structure in the rhizosphere, and changes in the microbial community structure in the rhizosphere may affect the growth of plants [67]. At present, in the development process of microbial inoculants, indicators such as reducing incidence and increasing yield are used as standards for measuring the effect of microbial inoculants [18]. Similarly, rhizosphere bacteria can alter the composition of rhizosphere microbial communities [50] and control plant pathogens, improve soil health, and promote plant growth. Rhizosphere bacteria can change key flora, increase the abundance of beneficial flora, and promote plant growth. Studies have shown that key bacteria in rhizosphere communities are related to soil health [68,69]. Pseudonocardiaceae is an important biological control resource which can produce a variety of antibiotics [70]. For example, strain A. pretiosum can produce ansamitocin [71]. Ceratobasidiaceae has important ecological functions as saprophytes, non-mycorrhizal endophytes, orchid mycorrhizal, and ectomycorrhizal symbiotic bacteria [72]. Additionally, Ceratobasidiaceae have been demonstrated as ECM fungi [73,74]. Univariate correlation network analysis revealed that the P-3 strain can make the Pseudonocardiaceae and Ceratobasidiaceae to the most critical flora. The P-3 strain can reduce the risk of plant diseases and improve the health of the tomato rhizosphere ecosystem. Therefore, the P-3 strain changed the composition of key flora in the microbial community, and strengthened the role of beneficial microorganisms in the underground rhizosphere microbial community. Rhizosphere microorganisms participate in soil nutrient cycling, plant protection, and induce plant disease resistance [75-77]. P-3 strain is able to control Meloidogyne incognita, improve the rhizosphere microbial environment. The P-3 strain can be developed as a microbial inoculant. This research hopes to contribute to the development of microbial inoculants.
Author Contributions
Jianfeng Du and Qixiong Gao conceived and designed the experiments; Jianfeng Du and Qixiong Gao performed the experiments; Jianfeng Du, Qixiong Gao, Zhaoyang Liu, Chaohui Li, Xin Song, Ruiping Xu, Yanyan Zhou, Yue Liu, Huying Li and Rui Zheng analysed the data; Jianfeng Du wrote the paper. Xunli Liu guided the research work and revised the manuscript.
Acknowledgment
This research was funded by Special research project for public welfare industry (Agriculture) (201503112-10), Shandong forestry science and technology innovation project (2019ly003-5), Major scientific and technological innovation project of Shandong Province (2019jzzy020614), and Special project for scientific and technological innovation and development of Linyi City (2019zdyf013).
References
- Sirca S, Urek, G, Karssen G. First Report of the Root-Knot Nematode Meloidogyne ethiopica on Tomato in Slovenia. Plant Disease. 2004; 88: 1262-1262.
- Siddiqui IA, Shaukat SS, Sheikh IH, Khan A. Role of cyanide production by Pseudomonas fluorescens CHA0 in the suppression of root-knot nematode, Meloidogyne javanica in tomato. World Journal of Microbiology and Biotechnology. 2006; 22: 641-650.
- Bourne JM, Kerry BR, De Leij FAAM. The Importance of the Host Plant on the Interaction Between Root-knot Nematodes Meloidogyne spp. and the Nematophagous Fungus, Verticillium chlamydosporium Goddard. Biocontrol Science and Technology. 1996; 6: 539-548.
- Anwar S, McKenry M. Incidence and Reproduction of Meloidogyne incognita on Vegetable Crop Genotypes. Pakistan Journal of Zoology. 2010; 42.
- Terefe M, Tefera T, Sakhuja P. Effect of formulation of Bacillus firmus on root-knot nematode Meloidogyne incognita infestation and the growth of tomato plants in the greenhouse and nursery. Journal of invertebrate pathology. 2008; 100: 94-99.
- Humbert JF, Dorigo U, Cecchi P, Berre BL, Bouvy M. Comparison of the structure and composition of bacterial communities from temperate and tropical freshwater ecosystems. Environmental Microbiology. 2009; 11: 2339-2350.
- Dong S, Qiao K, Zhu Y, Wang H, Xia X, Wang K. Managing Meloidogyne incognita and Bemisia tabaci with thiacloprid in cucumber crops in China. Crop Protection. 2014; 58: 1-5.
- Parvatha RP. Recent advances in crop protection. 2013.
- Katooli N, Moghadam EM, Taheri A, Nasrollanejad S. Management of root knot nematode (Meloidogyne incognita) on Cucumber with the extract and oil of nematicidal plants. 2010; 5: 582-586.
- Kiewnick S, Sikora RA. Biological control of the root-knot nematode Meloidogyne incognita by Paecilomyces lilacinus strain 251. Biological Control. 2006; 38: 179-187.
- Kumari NS, Sivakumar CV. Biological control potential of the obligate parasite Pasteuria penetransagainst the root-knot nematode, Meloidogyne incognita infestation in Brinjal. 2005; 70: 905-908.
- Sharma P, Pandey R. Biological control of root-knot nematode; Meloidogyne incognita in the medicinal plant; Withania somnifera and the effect of biocontrol agents on plant growth. African Journal of Agricultural Research. 2009; 4: 564-567.
- Briggs J, Dickinson G, Murphy K, Pulford I, Mekki AM. Sustainable Development and Resource Management in Marginal Environments: Natural Resources and Their Use in the Wadi Allaqi. Applied Geography. 1993; 13: 259-284.
- Cordell D, Drangert J-O, White S. The story of phosphorus: Global food security and food for thought. Global Environmental Change. 2009; 19: 292-305.
- Singh RB. Environmental consequences of agricultural development: a case study from the Green Revolution state of Haryana, India. Agriculture Ecosystems & Environment. 2000; 82: 97-103.
- Wei LH, Xue QY, Wei BQ, Wang YM, Guo JH. Screening of antagonistic bacterial strains against Meloidogyne incognita using protease activity. Biocontrol Science & Technology. 2010; 20: 739-750.
- Collange B, Navarrete M, Peyre Gl, Mateille T, Tchamitchian M. Root-knot nematode (Meloidogyne) management in vegetable crop production: The challenge of an agronomic system analysis. Crop Protection. 2011; 30: 1251-1262.
- Li XZ, Chen S. Screening and identification of cucumber germplasm and rootstock resistance against the root-knot nematode (Meloidogyne incognita). Genetics & Molecular Research Gmr. 2017; 16.
- Xiang N, Lawrence KS, Donald PA. Biological control potential of plant growth-promoting rhizobacteria suppression of Meloidogyne incognita on cotton and Heterodera glycines on soybean: A review. Journal of Phytopathology. 20108; 166: 449-458.
- Bhattacharyya PN, Jha DK. Plant Growth-Promoting Rhizobacteria (PGPR): emergence in agriculture. World Journal of Microbiology and Biotechnology. 2012; 28: 1327-1350.
- Liu K, Garrett C, Fadamiro H, Kloepper JW. Induction of systemic resistance in Chinese cabbage against black rot by plant growth-promoting rhizobacteria. Biological Control. 2016; 99: 8-13.
- Xiang N, Lawrence KS, Kloepper JW, Donald PA, McInroy JA, Lawrence GW. Biological Control of Meloidogyne incognita by Spore-forming Plant Growth-promoting Rhizobacteria on Cotton. Plant Disease. 2017; 101: 774-784.
- Kumar A, Prakash A, Johri BN. Bacillus as PGPR in crop ecosystem. Crop Ecosystems. 2001; 37-59.
- Antoun H. Plant-Growth-Promoting Rhizobacteria. Brenner's Encyclopedia of Genetics (Second Edition). 2013; 353-355.
- Aballay E, Prodan S, Correa P, Allende J. Assessment of rhizobacterial consortia to manage plant parasitic nematodes of grapevine. Crop Protection. 2020; 131: 105103.
- Du BXY, Dong H, Li Y, Wang J. Phanerochaete chrysosporium strain B-22, a nematophagous fungus parasitizing Meloidogyne incognita. PLoS One. 2020; 15: e0216688.
- Liu G, Lin X, Xu S, Liu G, Liu F, Mu W. Screening, identification and application of soil bacteria with nematicidal activity against root-knot nematode (Meloidogyne incognita) on tomato. Pest Management Science. 2020.
- Siddiqui IA, Shaukat S. Rhizobacteria-mediated Induction of Systemic Resistance (ISR) in Tomato against Meloidogyne javanica. Journal of Phytopathology. 2002; 150: 469-473.
- Burkett-Cadena M, Kokalis-Burelle N, Lawrence K, van Santen E, Kloepper J. Suppressiveness of root-knot nematodes mediated by rhizobacteria. Biol. Control. 2008; 47: 55-59.
- Siddiqui IA, Shaukat SS. Rhizobacteria-mediated Induction of Systemic Resistance (ISR) in Tomato against Meloidogyne javanica. Journal of Phytopathology. 2002; 150: 469-473.
- Siddiqui IA, Shaukat SS, Sheikh IH, Khan A. Role of cyanide production byPseudomonas fluorescensCHA0 in the suppression of root-knot nematode,Meloidogyne javanicain tomato. World Journal of Microbiology & Biotechnology. 2006; 22: 641-650.
- Bhardwaj D, Ansari M, Sahoo R, Tuteja N. Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microbial Cell Factories. 2014; 13: 66.
- Altieri MA. The ecological role of biodiversity in agroecosystems. Agric. ecosyst.environ. 1999; 74: 19-31.
- Trabelsi D, Mhamdi R. Microbial Inoculants and Their Impact on Soil Microbial Communities: A Review. Journal of Biomedicine & Biotechnology. 2013; 11: 863240:.
- Lee S, Ka J-O, Song H-G. Growth promotion of Xanthium italicumby application of rhizobacterial isolates of Bacillus aryabhattaiin microcosm soil. Journal of Microbiology. 2012; 50: 45-49.
- Park YG, Mun BG, Kang SM, Hussain A, Shahzad R, Seo CW, et al. Bacillus aryabhattai SRB02 tolerates oxidative and nitrosative stress and promotes the growth of soybean by modulating the production of phytohormones. Plos One. 2017; 12: e0173203.
- Bhattacharyya C, Bakshi U, Mallick I, Mukherji S, Bera B, Ghosh A. Genom guided in-sights into the plant growth promotion capabilities of the physiologically versatile Bacillus aryabhattai strain AB211 Frontiers in Microbiology. 2017; 8: 411-426.
- JingZhao, DanLiua, Yuanyuan, Wangb, XiaofengZhua, LijieChena, YuxiDuan. Evaluation of Bacillus aryabhattai Sneb517 for control of Heterodera glycines in soybean. Biological Control. 2020; 142: 104147-104168.
- Xiong J, Wu L, Tu S, Van Nostrand JD, He Z, Zhou J, et al. Microbial Communities and Functional Genes Associated with Soil Arsenic Contamination and the Rhizosphere of the Arsenic-Hyperaccumulating Plant Pteris vittata L. Applied & Environmental Microbiology. 2010; 76: 7277-7284.
- Philippot L, Raaijmakers JM, Lemanceau P, Van dP, Wim H. Going back to the roots: the microbial ecology of the rhizosphere. Nature Reviews Microbiology. 2013; 11: 789-799.
- Hayat R, Ali S, Amara U, Khalid R, Ahmed I. Soil beneficial bacteria and their role in plant growth promotion: a review. Annals of Microbiology. 2010; 60: 579-598.
- Holt JG, K.N., Sneath PHA. Bergey’s Manual of Determinative Bacteriology, (Ed.) B.W.W. Company. 1994.
- Jagoueix S, Bove J, Garnier M. The Phloem-Limited Bacterium of Greening Disease of Citrus Is a Member of the Subdivision of the Proteobacteria. International journal of systematic bacteriology. 1994; 44: 379-86.
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics. 2004; 5: 150-163.
- Sui J, Ji C, Wang X, Liu Z, Sa R, Hu Y, et al. A plant growth-promoting bacterium alters the microbial community of continuous cropping poplar trees’ rhizosphere. Journal of Applied Microbiology. 2019; 126: 1209-1220.
- Zheng J, Xiao X, Zhang Q, Mao L, Yu M, Xu J. The Placental Microbiome Varies in Association with Low Birth Weight in Full-Term Neonates. Nutrients. 2015; 7: 6924-6937.
- Amato K, Yeoman C, Kent A, Righini N, Carbonero F, Estrada A, et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. The ISME Journal. 2013; 7: 1344-1353.
- Abarenkov K, Henrik Nilsson R, Larsson K-H, Alexander IJ, Eberhardt U, Erland S, et al. The UNITE database for molecular identification of fungi--recent updates and future perspectives. New Phytologist, 2010; 186: 281-285.
- Nguyen N, Song Z, Bates S, Branco S, Tedersoo L, Menke J, et al. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecology. 2015; 20.
- Zhou X, Zhang J, Pan D, Xin G, Wu F. p-Coumaric can alter the composition of cucumber rhizosphere microbial communities and induce negative plant-microbial interactions. Biology & Fertility of Soils. 2018; 54: 363-372.
- Zhang Y, Gao X, Shen Z, Zhu C, Jiao Z, Rong L, et al. Pre-colonization of PGPR triggers rhizosphere microbiota succession associated with crop yield enhancement. Plant and Soil. 2019; 439: 553-567.
- Colagiero M, Rosso L, Ciancio A. Diversity and biocontrol potential of bacterial consortia associated to root-knot nematodes. Biological Control. 2017.
- Li X, Hu H-J, Li J-Y, Wang C, Chen S-L, Yan S-Z. Effects of the Endophytic Bacteria Bacillus cereus BCM2 on Tomato Root Exudates and Meloidogyne incognita Infection. Plant Disease. 2019; 103: 1551-1558.
- Viljoen JJF, Labuschagne N, Fourie H, Sikora RA. Biological control of the root-knot nematode Meloidogyne incognita on tomatoes and carrots by plant growth-promoting rhizobacteria. Tropical Plant Pathology. 2019; 44: 284-291.
- Han L, Wang Z, Li N, Wang Y, Feng J, Zhang X. Bacillus amyloliquefaciens B1408 suppresses Fusarium wilt in cucumber by regulating the rhizosphere microbial community. Applied Soil Ecology. 2019; 136: 55-66.
- Lee B, Lee S, Ryu C-M. Foliar aphid feeding recruits rhizosphere bacteria and primes plant immunity against pathogenic and non-pathogenic bacteria in pepper. Annals of Botany. 2012; 110: 281-290.
- Van Loon LC, Bakker PAHM, Pieterse CMJ. Systemic Resistance Induced by Rhizosphere Bacteria. Annual Review of Phytopathology. 1998; 36: 453-483.
- Babalola OO. Beneficial bacteria of agricultural importance. Biotechnology Letters. 2010; 32: 1559-1570.
- Luo Y, Wang F, Huang Y, Zhou M, Gao J, Yan T, et al. Sphingomonas sp. Cra20 Increases Plant Growth Rate and Alters Rhizosphere Microbial Community Structure of Arabidopsis thaliana Under Drought Stress. Frontiers in Microbiology. 2019; 10: 1221.
- Prasad M, Chaudhary M, Choudhary M, Kumar T, Jat L. Rhizosphere Microorganisms Towards Soil Sustainability and Nutrient Acquisition. 2017; 31-49.
- Mukhtar S, Mehnaz S, Malik KA. Microbial diversity in the rhizosphere of plants growing under extreme environments and its impact on crop improvement. Environmental Sustainability. 2019; 2: 329-338.
- Lu S, Lepo JE, Song H-X, Guan C-Y, Zhang Z-H. Increased rice yield in long-term crop rotation regimes through improved soil structure, rhizosphere microbial communities, and nutrient bioavailability in paddy soil. Biology and Fertility of Soils. 2018; 54: 909-923.
- Wang Z, Li Y, Li T, Zhao D, Liao Y. Tillage practices with different soil disturbance shape the rhizosphere bacterial community throughout crop growth. Soil and Tillage Research. 2020; 197: 104501.
- Wang Z, Zhang J, Wu F, Zhou X. Changes in rhizosphere microbial communities in potted cucumber seedlings treated with syringic acid. Plos One. 2018; 13: e0200007.
- Lanzara A, Saini NL, Brunelli M, Valletta A, Bianconi A. Evidence for onset of charge density wave in the La-based Perovskite superconductors. Journal of Superconductivity. 1997; 10: 319-321.
- Jain A, Chakraborty J, Das S. Underlying mechanism of plant–microbe crosstalk in shaping microbial ecology of the rhizosphere. Acta Physiologiae Plantarum. 2020; 42: 8.
- Marschner P, Crowley D, Yang CH. Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant & Soil. 2004; 261: 199-208.
- De Vries FT, Wallenstein MD, Bardgett R. Below-ground connections underlying above-ground food production: a framework for optimising ecological connections in the rhizosphere. Journal of Ecology, 2017; 105: 913-920.
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science. 2011; 332: 1097-1100.
- Gopalakrishnan S, Srinivas V, Naresh N, Alekhya G, Sharma R. Exploiting plant growth-promoting Amycolatopsis sp. for bio-control of charcoal rot of sorghum (Sorghum bicolor L.) caused by Macrophomina phaseolina (Tassi) Goid. Archives of Phytopathology and Plant Protection. 2019; 52: 543-559.
- Qin S, Xing K, Jiang J-H, Xu L-H, Li W-J. Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Applied Microbiology and Biotechnology. 2011; 89: 457-473.
- Veldre V, Abarenkov K, Bahram M, Martos F, Selosse M-A, Tamm H, et al. Evolution of nutritional modes of Ceratobasidiaceae (Cantharellales, Basidiomycota) as revealed from publicly available ITS sequences. Fungal Ecology. 2013; 6: 256-268.
- Bidartondo MI, Bruns TD, Weiss M, Sergio C, Read DJ. Specialized cheating of the ectomycorrhizal symbiosis by an epiparasitic liverwort. Proc Biol Sci. 2003; 270: 835-842.
- Dearnaley JDW. Further advances in orchid mycorrhizal research. Mycorrhiza. 2007; 17: 475-486.
- Bettenfeld P, Fontaine F, Trouvelot S, Fernandez O, Courty P-E. Woody Plant Declines. What’s Wrong with the Microbiome? Trends in Plant Science. 2020; 25: 381-394.
- Bray RH, Kurtz LT. Determination of Total, Organic, and Available Forms of Phosphorus in Soils. Soil Science. 1945; 59: 39-46.
- Goupry SP, Croguennec T, Gentil E, Robins RJ. Metabolic flux in glucose/citrate co-fermentation by lactic acid bacteria as measured by isotopic ratio analysis. Fems Microbiology Letters. 2000; 182: 207-211.