
Case Report
J Bacteriol Mycol. 2021; 8(1): 1162.
Metabolic and Microbial Characterizations for the Gastrointestinal Digesta of the Zoo Colobus guereza
Toyoda A1,2,3,*, Shionome N1, Kohari D1,2,3, Iida S4, Masato H4, Namae N4, Nakamura S5 and Tsukahara T5
1Department of Food and Life Sciences, Ibaraki University, College of Agriculture, Japan
2Tokyo University of Agriculture and Technology, United Graduate School of Agricultural Science, Japan
3Ibaraki University Cooperation between Agriculture and Medical Science (IUCAM), Japan
4Hitachi Kamine Zoo, Japan
5Kyoto Institute of Nutrition & Pathology, Japan
*Corresponding author: Toyoda A, Department of Food and Life Sciences, Ibaraki University, College of Agriculture, 3-21-1 Chuo, Ami-machi, Inashiki, Ibaraki 300-0393, Japan
Received: January 11, 2021; Accepted: February 05, 2021 Published: February 12, 2021
Abstract
Abyssinian black-and-white colobus (Colobus guereza) inhabits west, central, and east Africa and lives in social groups in the rain forests. Colobus guereza has a unique foregut digestive system similar to that of ruminants and absorbs organic acids, including Short-Chain Fatty Acids (SCFAs), as energy sources derived from microbial fermentation of plant materials in the gastrointestinal tract. In this study, the gastrointestinal metabolic and microbial features of a male Colobus guereza singly housed in a zoo were characterized, and each digesta from each segment of the digestive tract was collected and subjected to biochemical, microbiome, and metagenome analyses. In this case report, high levels of acetate and propionate were observed in the foregut, while a relatively high level of lactate was detected in the small intestine. Moreover, in the hindgut and the feces, acetate was dominant compared to the other SCFAs. SCFAs analysis indicated that Colobus guereza obtains energy via SCFAs, especially acetate, fermentation in the foregut and hindgut. A metagenome analysis revealed that each part of the digestive tract of Colobus guereza has a unique microbiota. Similar to ruminants, Prevotella and Selenomonas were the dominant genera in the foregut, which may indicate microbial fermentation of plant materials in the foregut of Colobus guereza.
Keywords: Colobus Guereza; Gastrointestinal Digesta; Metabolome; Microbiome; Organic Acid
Introduction
Abyssinian black-and-white colobus (Colobus guereza), one of the major colobine monkeys, inhabits west, central, and east Africa and lives in social groups in the rain forests [1]. A previous study reported that Colobus guereza spends 52-63 % of its time resting and 19-26 % feeding [2] and has an abnormally low basal metabolic rate, which may be linked to their lower activity level [3]. Moreover, they choose valuable food habitats and consume various plant materials, including leaves, seeds, and fruits, depending on their location and the season [4-8]. Colobus guereza consumes mainly leaves and fruits [2]. Therefore, they obtain their dietary energies and nutrients mainly from plant materials, including cellulosic fibers via a foregutfermentation digestive system similar to that of ruminants [9]. The Colobus monkey has a forestomach consisting of three regions, namely the cardiac gland, proper gastric and pyloric gland regions [10]. The cardiac gland region of the stomach is larger than the other regions and provides the environment for microbial fermentation of plant materials. Actually, microbial fermentation in the foregut of the colobus is similar to rumen fermentation in terms of bacterial counts, digestive enzymes, organic acids, and pH [11-14]. Recently, a metagenome analysis of the feces of the colobus was performed using the next-generation sequencing system [15,16]; however, the whole composition of their digestive microbiome is unclear. Colobus guereza is captured and displayed in zoos, and their care manual [17] provide the appropriate and practical information on the nutrients and feeds for zoo Colobuses based on several basic research [7,18-21]. However, knowledge of the microbial and metabolic features of the digestive tracts of zoo Colobuses is limited. In this study, we obtained digesta samples from the zoo Colobus guereza, which died during anesthesia, and elucidated the general digestive features by analyzing the organic acids, metabolome, and metagenome of the digesta.
Case Presentation
Animal and digesta samples
Abyssinian black-and-white colobus (Colobus guereza, male, 7 years old) was singly reared and housed at the Kamine Zoo (Hitachicity, Ibaraki, Japan), and died unexpectedly during anesthesia for a regular health examination. After the post-mortem examination by the veterinarian, the complete gastrointestinal tract was removed and the regions were numbered (Nos.1-12) as shown in (Figure 1A); the digesta was collected immediately from each region and stored at -80 °C until analyses. Before the sampling, Colobus guereza was normally kept in a cage during the daytime and in the next closed bedroom during nighttime. The feed contents for Colobus guereza are shown in (Table S1). Feed was provided three times every day, and water was provided ad libitum. During the daytime, visitors could nearly approach but not directly touch the Colobus guereza in the cage.
Organic acid analysis using ion-exclusion HPLC
Organic acid concentrations in the digesta were measured using ion-exclusion High-Performance Liquid Chromatography (HPLC) as described previously [22]. Organic acids were detected in all the digesta samples, but the digesta from the Pars Pylorica (PP9) had a relatively low concentration of organic acids compared to the digesta from the other regions. Obviously, higher levels of acetate were detected in both the foregut (PS12, TG11, and S10) and the hindgut (C4, PC3, DC2, and R1). Conversely, the concentrations of succinate, formate, and n-valerate were lower than those of acetate and n-butyrate. Furthermore, lactate concentrations were higher in the digesta from the small intestine (D8, PJ7, DJ6, and I5) than the digesta from other regions (Figure 1B, Table S2).
Microbiota analysis using the miseq platform
Bacterial DNA was extracted from the digesta as described previously [23]. The microbiota composition of the digesta was analyzed using Next-Generation Sequencing (NGS) and the MiSeq platform (Illumina, CA, USA) as previously described [24]. The alpha diversity in each gastrointestinal digesta was different: the hindgut digesta showed higher alpha diversity than the forestomach. Weighted UniFrac principal coordinates analysis showed that the forestomach, small intestine, and large intestine had different microbiota. The top 10 abundant genera in the stomach, small intestine, and large intestine are shown in (Table 1), respectively. As shown in (Tables 2), each gastrointestinal digesta had a unique microbiota. In the forestomach (PS12, TG11, and S10), the genera Prevotella and Selenomonas were dominant, while the genus Clostridium was dominant in PP9 (Table 1). In the small intestine, each digesta (D8, PJ7, DJ6, and I5) also had a unique microbiota, the genus Turicibacter was predominant in I5 compared with the other regions (Table 2). In the large intestine, the various digesta (C4, PC3, DC2 and R1) had similar microbiota, relatively the family Ruminococcaceae was dominant (Table 3).
Phylum
Class
Order
Family
Genus
PS12
TG11
S10
PP9
Stomach mean
Bacteroidetes
Bacteroidia
Bacteroidales
Prevotellaceae
Prevotella
43.2
38.0
28.2
12.6
30.5
Firmicutes
Clostridia
Clostridiales
Veillonellaceae
Selenomonas
24.7
26.9
22.7
2.8
19.3
Cyanobacteria
Chloroplast
Streptophyta
unclassifled
unclassifled
4.5
7.5
10.1
2.0
6.0
Finnicutes
Clostridia
Clostridiales
Clostridiaceae
Clostridium
0.0
0.0
0.0
18.3
4.6
Finnicutes
Clostridia
Clostridiales
Laclmospiraceae
Butyrivibrio
3.9
3.8
4.4
2.4
3.6
Finnicutes
Clostridia
Clostridiales
Veillonellaceae
l'v1egasphaera
5.0
4.7
2.6
0.7
3.3
Finnicutes
Clostridia
Clostridiales
Veillonellaceae
unclassifled
4.4
4.8
0.6
0.1
2.4
Finnicutes
Clostridia
Clostridiales
Veillonellaceae
Other
3.9
3.6
1.7
0.3
2.4
Bacteroidetes
Bacteroidia
Bacteroidales
p-2534-18B5
unclassifled
0.5
0.5
3.8
2.7
1.9
Proteobacteria
Alphaproteobacteria
Rhizobiales
Rhizobiaceae
Rhizobium
0.0
0.0
0.0
7.3
1.8
Table 1: Top 10 abundant genus in the stomach of Colobus guereza.
Phylum
Class
Order
Family
Genus
D8
PJ7
DJ6
15
Small intestine mean
Firmicutes
Clostridia
Clostridiales
Other
Other
14.5
1.1
36.8
29.5
20.5
Firmicutes
Clostridia
Clostridiales
Clostridiaceae
Clostridium
15.1
5.1
6.1
14.4
10.2
Firmicutes
Clostridia
Clostridiales
Veillonellaceae
Selenomonas
8.2
17.4
6.5
3.5
8.9
Firmicutes
Bacilli
Turicibacterales
Turicibacteraceae
Turicibacter
0.2
0.0
9.4
24.2
8.4
Firmicutes
Clostridia
Clostridiales
Lachnospiraceae
Butyrivibrio
10.2
13.5
4.6
2.5
7.7
Firmicutes
Clostridia
Clostridiales
Peptostreptococcaceae
unclassified
0.4
0.2
15.9
6.8
5.9
Cyanobacteria
Chloroplast
Streptophyta
unclassified
unclassified
9.3
7.1
1.1
0.6
4.5
Firmicutes
Clostridia
Clostridiales
Peptostreptococcaceae
[Clostridium]
9.5
0.1
2.8
2.0
3.6
Actinobacteria
Coriobacteriia
Coriobacteriales
Coriobacteriaceae
unclassified
2.1
7.5
1.7
1.0
3.1
Proteobacteria
Alphaproteobacteria
Rhizobiales
Rhizobiaceae
Rhizobium
3.1
4.7
l.3
1.3
2.6
Table 2: Top 10 abundant genus in the small intestine of Colobus guereza.
Phylum
Class
Order
Family
Genus
C4
PC3
DC2
R1
F13
Large intestine mean
Firmicutes
Clostridia
Clostridiales
Ruminococcaceae
unclassified
34.8
33.8
42.3
37.7
32.3
37.1
Finnicutes
Clostridia
Clostridiales
unclassifled
unclassified
7.2
7.3
9.1
12.2
10.7
8.9
Finnicutes
Clostridia
Clostridiales
Lachnospiraceae
unclassified
5.9
6.7
5.5
4.6
8.3
5.7
Firmicutes
Clostridia
Clostridiales
Ruminococcaceae
Ruminococcus
5.3
7.0
6.0
5.0
5.1
5.8
Firmicutes
Clostridia
Clostridiales
Lachnospiraceae
Coprococcus
6.6
5.8
4.3
2.7
4.1
4.8
Bacteroidetes
Bacteroidia
Bacteroidales
Bacteroidaceae
Bacteroides
4.0
3.5
2.7
3.5
3.1
3.4
Finnicutes
Clostridia
Clostridiales
Ruminococcaceae
Oscillospira
1.9
2.0
3.7
4.8
4.6
3.1
Spirochaetes
Spirochaetes
Spirochaetales
Spirochaetaceae
Treponema
2.2
2.6
1.5
1.3
3.0
1.9
Finnicutes
Clostridia
Clostridiales
Veillonellaceae
Phascolarctobacterium
1.8
2.4
2.7
2.9
3.3
2.5
Bacteroidetes
Bacteroidia
Bacteroidales
S24-7
unclassified
2.7
2.6
1.7
1.5
1.3
2.1
Table 3: Top 10 abundant genus in the small intestine of Colobus guereza.
Metabolome analysis using gas chromatography (GC)-mass spectrometry (MS)
Metabolome analyses of the digesta in water-soluble lowmolecular- weight metabolites were performed using GC-MS as described elsewhere [25]. Although statistical analysis was not performed, palmitic acid levels were relatively higher across all the regions of the gastrointestinal tract of Colobus guereza. In the small intestine, the levels of several metabolites such as 2-aminobutyric acid, 2-aminoethanol, threonine, and tryptophan were higher than those of the other regions.
Discussion
The organic acid concentrations in the forestomach (PS12, TG11, and S10) were higher than those of the Pars Pylorica (PP9), although the profiles of organic acids in PS12, TG11, and S10 were similar to each other (Figure 1B & Table S2). SCFAs, including acetate, propionate, and n-butyrate were the major organic acids in the forestomach of the Colobus guereza, which is consistent with previous observations in the wild colobus [11,12]. In addition, the hindgut digesta contained higher levels of SCFAs, similar to those of the forestomach; however, the ratio of acetate-to-propionate was higher in the hindgut than that in the forestomach (Figure 1B & Table S2). These results indicate that microbial digestion and fermentation of plant materials occurred in both the forestomach and hindgut of the zoo Colobus guereza as described previously [13]. Interestingly, high concentrations of SCFAs were also detected in the hindgut that may implicate the hindgut in the microbial digestion and fermentation of undigested fibrous feed residues after foregut digestion. Previously, high levels of SCFAs, especially acetate, were detected in in vitro cultures of the feces of zoo Colobus guereza, showing a high acetateto- propionate ratio [26]. Collectively, Colobus guereza might absorb SCFAs as energy sources in both the forestomach and the hindgut fermentation.
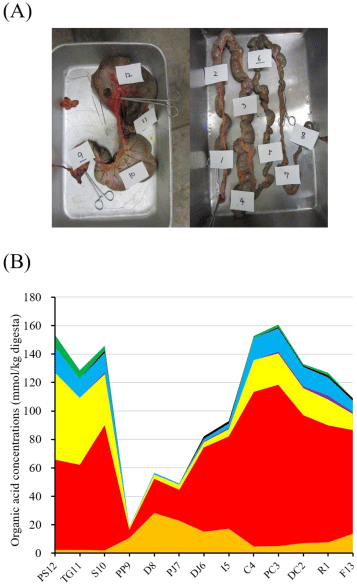
Figure 1A, 1B: The gastrointestinal tract and organic acid concentrations
of the digesta in Colobus guereza (A). No.12, presaccus (designated as
PS12); No.11, tubus gastricus (TG11); No.10, saccus (S10); No.9, Pars
Pylorica (PP9). No.8, duodenum (D8); No.7, proximal jejunum (PJ7); No.6,
distal jejunum (DJ6); No.5, ileum (I5); No.4, cecum (C4); No.3, proximal colon
(PC3); No.2, distal colon (DC2); No.1, rectum (R1). Orange area, lactate;
red area, acetate; yellow area, propionate; purple area, iso-butyrate; lightblue
area, n-butyrate; black area, iso-valerate; and green area, n-valerate.
Succinate and formate were not detected, therefore, we omitted these acids
from the graphs.
The metagenome analysis revealed that each gastrointestinal digesta had different microbiota. Genera Prevotella and Selenomonas were detected as the dominant genera in the forestomach; Butyrivibrio and Megasphaera were also detected (Table 1). These are popular genera in the rumen microorganisms and play an important role in rumen metabolism [27,28]; therefore, the foregut microbial ecosystem of Colobus guereza may be similar to the rumen ecosystem. Genus Prevotella comprised 42-60 % of the bacterial rRNA gene copies in the bovine rumen [28], which was similar to the abundance of Prevotella in the forestomach of Colobus guereza (Table 1). In the large intestine, the family Ruminococcaceae was the most abundant, although the major genus was not classified (Table 3). Genera Ruminococcus and Oscillospira were also identified in the rumen [29-31], especially Ruminococcus favefaciens and R. albus are wellcharacterized cellulolytic rumen bacteria. In a previous metagenome study, Oscillibacter and Faecalibacterium were predominant in the feces of the wild Colobus guereza [15]. These microbiome in wild colobus were different from our metagenome results in the zoo Colobus guereza (Table 3). A previous report also showed that gut microbiota was different between wild and captive monkeys, Rhinopithecus brelichi [32].
In the small intestine and the pars pylorica, lactate was a major product of organic acid (Figure 1B & Table S2); however, genera Lactobacillus and Bifidobacterium were almost undetectable in the small intestine, therefore, other microorganisms might produce lactate in the intestine of Colobus guereza. Obviously, Clostridium was predominant in the small intestine and the pars pylorica (Table 2). Previously, Clostridium thermolacticum was identified as a thermophilic anaerobe producing high amounts of lactate [33]; therefore, Clostridium might be linked to lactate fermentation in Colobus guereza. Furthermore, methane producers, such as Methanosphaera and Methanobrevibacter, were found in the small intestine but not in the foregut. Genera Methanosphaera and Methanobrevibacter were identified in the large intestine of mammals [34-36], and also in the rumen [37]. Methane metabolism in the small intestine of mammals has not been well investigated; therefore, the research about small intestinal methanogens should be focused on to understand the physiology of methane production in the future.
Acknowledgement
We are grateful to Drs. K Ohashi and T Kawase (Kyoto Institute of Nutrition & Pathology, Kyoto, Japan) for their technical assistance.
References
- Oates JF. The social life of a black-and-white Colobus monkey, Colobus guereza. Z Tierpsychol. 1977; 45: 1-60.
- Fashing PJ. African colobine monkeys: their behavior, ecology, and conservation. Campbell CJ, Fuentes A, MacKinnon KC, Bearder S, Stumpf R, editors. In: Primates in Perspective, 2nd edn. Oxford University Press. 2011; 203-229.
- Muller EF, Kamau JMZ, Maloiy GMO. A comparative study of basal metabolism and thermoregulation in a folivorous (Colobus guereza) and an omnivorous (Cercopithecus mitis) primate species. Comp Biochem Physiol. 1983; 74: 319-322.
- Clutton-Brock TH. Feeding behaviour of red colobus and black and white colobus in East Africa. Folia Primatol. 1975; 23: 165-207.
- Oates JF. The Guereza and its food. Clutton-Brock TH, editor. In: Primate Ecology: Studies of Feeding and Ranging Behaviour in Lemurs, Monkeys and Apes. Academic Press. 1977; 276-319.
- Dunbar RIM. Habitat quality, population dynamics, and group composition in Colobus monkeys (Colobus guereza). Int J Primatol. 1987; 8: 299-329.
- Wasserman MD, Chapman CA. Determinants of colobine monkey abundance: The importance of food energy, protein and fibre content. J Anim Ecol. 2003; 72: 650-659.
- Harris TR, Chapman CA. Variation in diet and ranging of black and white colobus monkeys in Kibale National Park, Uganda. Primates. 2007; 48: 208- 221.
- Bauchop T, Martucci RW. Ruminant-like digestion of the langur monkey. Science. 1968; 161: 698-700.
- Stevens CE, Hume ID. Comparative physiology of the vertebrate digestive system. Cambridge, UK: Cambridge University Press. 1995.
- Kay RN, Hoppe P, Maloiy GMO. Fermentative digestion of food in the Colobine monkey, Colobus polykomos. Experentia. 1976; 32: 485-486.
- Ohwaki K, Hungate RE, Lotter L, Hofmann RR, Maloiy G. Stomach fermentation in East African colobus monkeys in the natural state. Appl Microbiol. 1974; 27: 713-723.
- Stevens CE, Hume ID. Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol Rev. 1998; 78: 393- 427.
- Schienman JE, Holt RA, Auerbach MR, Stewart CB. Duplication and divergence of 2 distinct pancreatic ribonuclease genes in leaf-eating African and Asian colobine monkeys. Mol Biol Evol. 2006; 23: 1465-1479.
- Yildirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL, et al. Characterization of the fecal microbiome from non-human wild primates reveals species-specific microbial communities. PLoS One. 2010; 5: e13963.
- McCord AI, Chapman CA, Weny G, Tumukunde A, Hyerora D, Klotz K, et al. Fecal microbiomes of non-human primates in Western Uganda reveal species-specific communities largely resistant to habitat perturbation. Am J Primatol. 2014; 76: 347-354.
- Guereza AZA. Angola Colobus Monkey Species Survival Plans in association with the AZA Old World Monkey Taxon Advisory Group, Colobus monkey (Colobus) care manual. 2012.
- Chapman CA, Chapman LJ, Bjorndal KA, Onderdonk DA. Application of protein-to-fiber ratios to predict colobine abundance on different spatial scales. Int J Primatol. 2002; 23: 283-310.
- Baranga D. Nutrient composition and food preferences of Colobus monkeys in Kibale Forest, Uganda. Afr J Ecol. 1982; 20: 113-121.
- Rode KD, Chapman CA, Chapman LJ, McDowell LR. Mineral resource availability and consumption by colobus in Kibale National Park. Int J Primatol. 2003; 24: 541-573.
- Fashing PJ, Dierenfeld ES, Mowry CB. Influence of plant and soil chemistry on food selection, ranging patterns, and biomass of Colobus guereza in Kakamega Forest, Kenya. Int J Primatol. 2007; 28: 673-703.
- Tsukahara T, Matsukawa N, Tomonaga S, Inoue R, Ushida K, Ochiai K. Highsensitivity detection of short-chain fatty acids in porcine ileal, cecal, portal and abdominal blood by gas chromatography-mass spectrometry. Anim Sci J. 2014; 85: 494-498.
- Tsukahara T, Inoue R, Nakayama K, Inatomi T. Inclusion of Bacillus amyloliquefaciens strain TOA5001 in the diet of broilers suppresses the symptoms of coccidiosis by modulating intestinal microbiota. Anim Sci J. 2018; 89: 679-687.
- Inoue R, Sakaue Y, Sawai C, Sawai T, Ozeki M, Romero-Perez GA, et al. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci Biotechnol Biochem. 2016; 80: 2450-2458.
- Tomonaga S, Okuyama H, Tachibana T, Makino R. Effects of high ambient temperature on plasma metabolomic profiles in chicks. Anim Sci J. 2018; 89: 448-455.
- Lambert JE, Fellner V. In vitro fermentation of dietary carbohydrates consumed by African apes and monkeys: preliminary results for interpreting microbial and digestive strategy. Int J Primatol. 2012; 33: 263-281.
- Tajima K, Aminov RI, Nagamine T, Matsui H, Nakamura M, Benno Y. Dietdependent shifts in the bacterial population of the rumen revealed with realtime PCR. Appl Environ Microbiol. 2001; 67: 2766-2774.
- Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol. 2007; 75: 165-174.
- Yanagita K, Manome A, Meng X-Y, Hanada S, Kanagawa T, Tsuchida T, et al. Flow cytometric sorting, phylogenetic analysis and in situ detection of Oscillospira guillermondii, a large, morphologically conspicuous but uncultured ruminal bacterium. Int J Syst Evol Microbiol. 2003; 53: 1609-1614.
- Mackie RI, Aminov RI, Hu W, Klieve AV, Ouwerkerk D, Sundset MA, et al. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl Environ Microbiol. 2003; 69: 6808-6815.
- La Reau AJ, Suen GJ. The Ruminococci: key symbionts of the gut ecosystem. J Microbiol. 2018; 56: 199-208.
- Hale VL, Tan CL, Niu K, Yang Y, Zhang Q. Gut microbiota in wild and captive Guizhou snub-nosed monkeys, Rhinopithecus brelichi. Am J Primatol. 2019; 81: e22989.
- LeRuyet P, Dubourguier HC, Albagnac C, Prensier G. Characterization of Clostridium thermolacticum sp. nov., a hydrolytic thermophilic anaerobe producing high amounts of lactate. Syst Appl Microbiol. 1985, 6: 196-202.
- Miller TL, Wolin MJ. “Methanosphaera stadtmaniae gen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen”. Arch Microbiol. 1985; 141: 116-122.
- Biavati B, Vasta M, Ferry JG. Isolation and characterization of “Methanosphaera cuniculi” sp. nov. Appl Environ Microbiol. 1988; 54: 768- 771.
- Miller TL, Wolin MJ. Stability of Methanobrevibacter smithii populations in the microbial flora excreted from the human large bowel. Appl Environ Microbiol. 1983; 45: 317-318.
- Holman DB, Gzyl KE. A meta-analysis of the bovine gastrointestinal tract microbiota, FEMS Microbiol Ecol. 2019; 95: fiz072.