
Research Article
J Bacteriol Mycol. 2021; 8(2): 1166.
Molecular Characterization and Antibiogram Profiling of Multidrug Resistant Staphylococcus haemolyticus Isolated from Patients with Urinary Tract Infection in Bangladesh
Haque MH1*, Miah ML1, Sarker S2, Shamsuzzaman M3 and Shiddiky MJA4
1Department of Veterinary and Animal Sciences, Faculty of Agriculture, Rajshahi University, Rajshahi-6205, Bangladesh
2Department of Physiology, Anatomy and Microbiology, School of Life Sciences, La Trobe University, Melbourne VIC 3086, Australia
3Specialty doctor in Acute Medicine, Leighton Hospital, Middlewich Rd, United Kingdom
4School of Environment and Science (ESC), and Queensland Micro- and Nanotechnology Centre (QMNC) Griffith University, Nathan Campus, 170 Kessels Road, QLD 4111, Australia
*Corresponding author: HaqueMd. Hakimul, Department of Veterinary and Animal Sciences, Faculty of Agriculture, Rajshahi University, Rajshahi-6205, Bangladesh
Received: January 25, 2021; Accepted: March 09, 2021 Published: March 16, 2021
Abstract
The emergence of antibiotic-resistant bacteria in human is a potential global public health concern. Profiling of antibiotic-resistant bacteria with their antimicrobial susceptibility patterns from Urinary Tract Infections (UTIs) is crucial to guide antibiotic therapy. Herein we report a detailed bacteriological and molecular analysis of Staphylococcus haemolyticus and their antibiogram typing from UTIs. A total of 100 human urine samples of patients with UTIs were collected between January and December 2019 and were subjected to the conventional characterization of bacteria using the standard protocol. Molecular characterization was performed via sequencing followed by phylogenetic analysis. All bacterial strains were examined against thirteen commonly used antibiotics for susceptibility using the Kirby-Bauer disk diffusion method. The overall prevalence of S. haemolyticus in UTI was 26% with female patients having a higher prevalence of UTI (21 out of 26 or 80.76%) than male patients (5 out of 26 or 19.24%). The isolated S. haemolyticus showed 100%, 100%, 88.46%%, 76.93%, 73.08% and 65.39% resistant to penicillin, ampicillin, amoxicillin, erythromycin, ciprofloxacin and tetracycline, respectively. Importantly, S. haemolyticus demonstrated the highest sensitivity to vancomycin (100%), followed by azithromycin (80.76%), amikacin (84.61%), gentamycin (69.23%), levofloxacin (73.08%), ceftriaxone (80.76%) and doxycycline (61.54%). Overall, six variations were noted in S. haemolyticus in which most (5/6) modifications were substitutions and one (1/6) was deletion. These findings imply that mutations in the 16S rRNA gene sequence are the dominant source for species identification and variation in the drug sensitivity pattern against the S. haemolyticus. Phylogenetic analysis of the resultant 16S rRNA indicated that the isolated S. haemolyticus in this study belonged to genus Staphylococcus, but was different from the rest of the available S. haemolyticus isolates in other countries. Multidrug-resistant pathogenic S. hemolyticus is commonly found in urine samples of UTI in human in Bangladesh, which warrants a one-health approach for controlling this emerging ailment.
Keywords: Multidrug-resistance; Staphylococcus hemolyticus; Urinary tract infection; Antibiotic-resistant bacteria; Antimicrobial susceptibility
Introduction
Antimicrobial Resistance (AMR) is a global public, animal and poultry health ailment, which results from injudicious use of antimicrobial agents in all sectors since the early 1940s [1]. Globally, increasing incidence rate of AMR towards several antimicrobial groups has been reported in various bacterial pathogens [2]. Indeed, Urinary Tract Infection (UTI) is a chronic health problem caused by a variety of bacterial species affecting millions of individuals worldwide, including Bangladesh [3]. UTIs affect people in all age groups and sex, and are diagnosed in both hospitalized and unhospitalized patients. UTIs easily influence women than men due to the anatomical structure of their genitourinary tract. One-third of all women suffer from UTI at some point during their lifetime [4]. This type of infection causes a severe socioeconomic burden on affected individuals and leads a high rate of all types of antibacterial drug usages [5]. Importantly, the risk factors in the pathogenicity of UTI varies among countries owing to geography variation and antibiotic use. Furthermore, the rapid onset of antibiotic therapy with broad-spectrum antibiotics is crucial to treatment achievement. Still, the repeated antibiotic use often ensues in the emergence of antibiotic-resistant bacteria. The rate of antimicrobial resistance to antibiotics among community-acquired UTIs is increasing and shows significant geographical variations [6]. Deep understanding of the diverse etiology of UTIs and the resistance pattern against antibiotics of the causative organisms is crucial to physicians while managing such patients.
Urinary tract infections in human are caused by a wide range of Gram-negative and Gram-positive bacterial species, but most commonly by Escherichia coli, Klebsiella pneumoniae, Pseudomonas spp., Proteus mirabilis, Citrobacter spp., Enterococcus faecalis, and coagulase-negative staphylococci [7,8]. Staphylococus haemolyticus is a coagulase-negative bacterium and a member of the genus Staphylococcus. Although coagulase-negative staphylococci represent significant causative microorganisms in nosocomial infections, they typically account for fewer than 10% of all UTIs [9]. Most of these coagulase-negative staphylococcal UTIs comprise of the three species, namely Staphylococcus epidermidis, Staphylococcus haemolyticus and Staphylococcus saprophyticus. S. haemolyticus is commonly present on the human skin and can be isolated from axillae, perineum, and inguinal areas of humans and is dismissed as culture contaminants, for example in urine cultures [10]. This opportunistic bacterium species colonize the urethra or periurethral of males and females. S. haemolyticus with other coagulase-negative staphylococcal species often exceed those of Gram-negative bacilli, which cause 80% of all UTIs [11]. Staphylococcus haemolyticus present an average of 10% of clinical coagulase-negative staphylococcal isolates in infections [12]. Strains of S. haemolyticus produce a haemolysin, cytolysin, and enterotoxin, and are frequently resistant to antibiotic [13]. Species determinations of coagulase-negative staphylococci are not typically done in most clinical microbiology laboratories. Therefore, the mechanisms of antimicrobial resistance in S. haemolyticus and the extent to which it shares resistance genes with other staphylococci are also unknown. The possibility of unique arrangements of resistance in this poorly understood species led us to investigate its characteristics further.
Besides, in developing countries, antibiotics can be bought by the public from various sources including hospitals and pharmacies, licensed medicine stalls, and drugstores, roadside stalls and peddlers. Despite restrictive laws, antibiotics can be purchased by people without a prescription. This widespread availability has resulted in inappropriate use of antibiotics by patients, pharmacists, health-care providers and public themselves. Furthermore, antibiotic therapy is mainly empirical due to the relative lack of appropriate laboratory facilities for culture and susceptibility testing of bacteria. The heavy use of antibiotics and the absence of susceptibility testing have led to a steady increase in antimicrobial drug resistance. In addition, humans may be exposed to antibiotics indirectly, i.e. through food, water and herbal medicines. The antimicrobial agents are also used in livestock to treat, prevent, and control diseases and to enhance feed efficiency and weight gain. Like human medicine, the use of antimicrobial agents in veterinary medicine creates selective pressure for the emergence and dissemination of antimicrobial-resistant bacteria in livestock. These resistant bacteria can usually be transmitted to humans either by direct contact with animals or through the food. In this way, Staphylococcus haemolyticus could receive various resistance genes through the same species of staphylococci or other bacteria. However, people in developing countries are more prone to be affected due to the inappropriate use of antimicrobial agents, nonhuman antibiotic use, poor quality of drugs, inadequate surveillance, and factors associated with individual and national poverty including poor health-care standards, malnutrition, chronic and repeated infections, and unaffordability of more effective and costly drugs [14].
Increasing antimicrobial resistance poses a regional and global threat to the developing country of South Asia, including Bangladesh. It was previously reported that the widespread availability of antimicrobials without prescription in Bangladesh is responsible for the habit of self-medication among patients, and the indiscriminate utilization of antibiotics in humans and food animals and fisheries, and consequently for spreading resistant strains through environmental contaminations [5]. In addition, very few studies investigated coagulase-negative staphylococci bacteria associated with UTI in Bangladesh. However, no study has investigated the isolation, molecular detection, and antibiogram profiling of multidrug-resistant S. haemolyticas isolated from human urine in Bangladesh. The present study was therefore conducted for the first time to isolate, characterize and determine antimicrobial resistant of S. haemolyticas isolated from the urine of UTI in Bangladesh using molecular techniques.
Materials and Methods
Collection of samples
A total of 100 patients with urinary tract infection have been recruited randomly without any bias from a private diagnostic centre, Bangladesh during the period from January to December 2019. Informed consent was taken from all patients. Clean-catch midstream morning urine samples were collected in sterile widemouth glass containers using the standard protocol. The samples were then transported immediately to the Department of Veterinary and Animal Sciences, Rajshahi University, for bacteriological analysis while maintaining sterile and cold chain conditions. The time between sample collection and sample analysis did not go beyond one hour. Clinical and pathological parameters were documented for each patient, including age, sex, colour, and appearance of urine, presence of blood or pus, pH, marital status, pregnancy, residence, UTI acquired from a hospital or as an outpatient and season. Ethical permission was obtained from the Institutional Animal, Medical Ethics, Biosafety, and Biosecurity Committee (IAMEBBC) of Institute of Biological Science (IBSc), the University of Rajshahi for experimentations on the animal, human, Microbes and living natural sources (Memo no:144/320/I.A.M.E.B.B.C./IBSc). All methods were performed following relevant guidelines and regulations.
Isolation and identification of Staphylococcus spp
Initially, a sterile loopful sample was used to inoculate into the nutrient broth, followed by culturing on different selective media such as blood agar, soybean casein digest agar and mannitol salt agar to observe specific colony characteristics. The broth and agar plates were incubated overnight at 37°C aerobically. The colony was counted to measure the significant growth of bacteria. Gram staining was performed from discrete colonies, and further subculturing was done to obtain pure cultures. The morphological and biochemical characterization of Staphylococcus haemolyticus was done through carbohydrate fermentation test, catalase test, coagulase test, Methyl Red test, Voges-Proskauer test, indole test and reaction in TSI agar tests, as previously described [15,16].
Extraction of genomic DNA
Bacterial genomic DNA was extracted from pure cultures of Staphylococcus haemolyticus by the boiling method as described previously by Mahmud et al. [17]. Briefly, initially 200 μL deionized ultrapure water was added into an Eppendorf tube. A loop full pure bacterial colony of Staphylococcus haemolyticus from the overnight culture grown at 37°C on mannitol salt agar plate and mixed gently to make a homogenous cell suspension. Then the tube was transferred into boiling water and incubated for 10 min, transferred immediately into ice for a cold shock for about 10min. Finally, the tubes with bacterial cell suspension were centrifuged at 10,000 rpm for 10 min. 100 μL of supernatant containing bacterial DNA from each tube was collected. The quality and quantity of purified DNA were checked via Nanodrop Spectrophotometer (BioLab, Ipswich, MA, USA), and the DNA was stored at -20°C until use.
Polymerase Chain Reaction (PCR)
All isolates were confirmed as Staphylococcus haemolyticus using a previously established PCR method and a set of designed primers (Sense27F, 5'-AGAGTTTGATCMTGGCTCAG-3' and antisense1492R, 5'-CGGTTACCTTGTTACGACTT-3') targeted for 16S rRNA gene [18]. PCR reaction was achieved in a total volume of 20 μL reaction mixture comprising 10 μL of Hot Start Green Master Mix (Promega, USA), 1 μL of every ten picomoles/μL primer, 1 μL of bacterial genomic DNA at 50 ng/μL and 7 μL of Nuclease-free water. Thermal cycling consisted of initial denaturation at 95°C for 3 min followed by 35 cycles of 95°C for 30 seconds (denaturation), 48°C for 30 seconds (annealing), and 72°C for 90 seconds (extension) and a final extension at 72°C for 5 min. Amplified products were analyzed by electrophoresis in 1.5% agarose gel. Ethidium bromide was used, and PCR products were visualized under ultraviolet transilluminator (Biometra, Germany). The 1 KB DNA ladder (Thermo Fisher Scientific, MA, USA) was used as molecular weight markers.
Purification of PCR products and sequencing
Successfully amplified specific PCR bands were cut and purified according to the manufacturer’s protocols from the NucleoSpin Gel and PCR Clean-up kit (Macherey- Nagel, Bethlehem, PA, USA). The purified DNA was mixed with the primer (10-40 ng of DNA + 1 μL of 3.2 pmol primers in 10 μL of H2O) and sequenced by Sanger sequencing to further confirm the detected Staphylococcus haemolyticus. The Sanger sequencing was performed and analyzed using an ABI PRISM 3730×l Capillary sequencer (Applied Biosystems, USA) under standardized cycling PCR conditions. The sequences were trimmed for primers, aligned to construct contigs using a minimum overlap of 35 bp and a minimum match percentage of 95%, and the construction of consensus sequence was carried out in Geneious 10.2.2 (Biomatters, New Zealand).
Nucleotide Sequence Accession Numbers
The nucleotide base sequences of the gene 16S rRNA reported in this paper were submitted to the GenBank using the National Centre for Biotechnology Information (NCBI, Bethesda, /MD, USA) under the accession numbers MT622589 and MT622590, respectively.
Phylogenetic tree analysis
The base sequence of the PCR product was matched and aligned with known 16S ribosomal RNA gene sequences in the same genus, which was randomly selected from the GenBank database via multiple sequence alignment. Also, the 16S rRNA gene sequences of the isolates of the same species from 14 different countries, and at least two strains of each reference genera with their GenBank accession numbers were recorded for phylogenetic analysis including Escherichia, Shigella, Salmonella, Citrobacter, Yersinia, Acinetobacter, Pseudomonas, Legionella, Bartonella, Ochrobactrum, Streptomyces and Staphylococcus. The evolutionary relationship of the bacterial isolates analyzed was inferred using the neighbourjoining method [19]. The evolutionary distances were computed by the Maximum Composite Likelihood method as the number of base substitutions per site [20]. All steps involved in evolutionary analysis were conducted in MEGA6 software.
Antimicrobial Susceptibility Testing
The Kirby-Bauer disk diffusion method was used to determine the antibiogram phenotype against thirteen commonly used antibiotics classes, namelyfluoroquinolones (levofloxacin-5 μg), tetracycline (doxycycline-30 μg), aminoglycoside (Amikacin-30 μg), glycopeptides (Vancomycin-30 μg), cephalosporin (ceftriaxone-30 μg), macrolides (erythromycin-15 μg), macrolides (azithromycin-15 μg), aminoglycoside (gentamycin -10 μg), penicillin (amoxicillin -30 μg), penicillin (penicillin -30 μg), penicillin (ampicillin -25 μg), fluoroquinolones (ciprofloxacin-5 μg) and tetracycline (tetracycline-30 μg) (Hudzicki, 2009). The antimicrobial susceptibility test by the disk diffusion method was done on Mueller-Hinton agar (Hi-Media, India) plates with a concentration of bacteria equivalent to 0.5 McFarland standard and aerobic incubation at 37°C for 18- 24 h. Vancomycin screen agar (Mueller-Hinton agar with six μg/ ml of vancomycin) was used to determine its minimum inhibitory concentration. The results of the antimicrobial susceptibility profiling were documented as sensitive or resistant based on the diameters of the zones of inhibition as per the guidelines provided by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [21]. Staphylococcus haemolyticus isolates found to be resistant to at least three classes of antibiotics were categorized as multidrug resistance (MDR) [22].
Statistical analysis
All data were incorporated in the Excel sheet (Microsoft-2010) and analyzed by SPSS software version-24 (IBM Corp., Armonk, NY, USA). Descriptive analysis was performed to calculate prevalence. Chi-square (Χ2) test was done to evaluate the significant relationship of the clinicopathological parameter with the bacterial isolates and the difference among proportions of antibiotic resistance. A p-value <0.05 was considered significant.
Results
Identification of Staphylococcus haemolyticus in human urine
Among the 100 samples bacteriologically analyzed, a total of 26 (26%) was found to be positive for the presence of Staphylococcus haemolyticus from human urine samples collected from patients with UTI. This is evidenced by the isolation on selective media, followed by identification via Gram staining, and biochemical tests. Cultural, microscopic, and various biochemical methods for positive growth of coagulase-negative S. haemolyticus are presented in Table S1 and Figures S1, S2. Of these 26 positive cases, 5 (19.24%) were from male and the rest 21 (80.76%) were from female. This result indicated that female patients had a higher prevalence of Staphylococcus haemolyticus in UTI than in males. As can be seen in Table S2, the most susceptible age group of patients to UTI was 31-45 years (42.30%) followed by 16-30 years (26.92%), 46-60 years (19.23%), > 60 years (7.70%) and 0-15 years (3.85%) (Figure 1). This study suggests that UTI is overall common in the age group between 16-60 years. The bacterial isolates of S. haemolyticus in UTI showed no significant statistical correlations with the clinicopathological parameters of the patients (p>0.05).
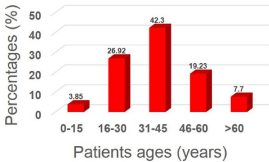
Figure 1: Percentage distribution of Staphylococcus haemolyticus isolated
from patients with UTI according to age.
Genetic Detection of Staphylococcus haemolyticus by 16S rRNA
All S. haemolyticus isolates identified by conventional methods were further confirmed by using a well-established PCR method followed by Sanger sequencing. In these isolates, 1465 base pair fragments amplified by targeting the 16S RNA gene for S. haemolyticus, as shown in Figure S3. Sanger sequencing revealed the congruence of the chromatogram with the isolation of S. haemolyticus of amplified product of the 16SrRNA gene, which had 99.58% nucleotide identity with a S. haemolyticus strain VB19458 isolated from human blood in India (GenBank accession no. CP045187). Overall, 6 novel mutations were found in S. haemolyticus. Of these, five mutations were found to be single substitution (A1G, G2C, T5A, C986T and T1432A), whereas the other was deletion mutation (T1414T) (Figure 2).
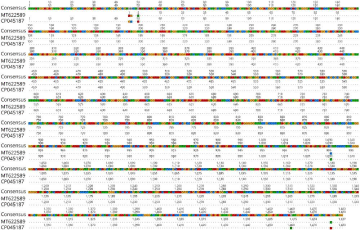
Figure 2: Representative Sanger sequencing results of 16S rRNA gene in Staphylococcus haemolyticus. The alignment of 16S rRNA sequence of the isolate with
the sequences of representative Staphylococcus haemolyticus from GenBank in the database showing different mutations.
Phylogenetic analysis
To understand the evolutionary relationship of the newly sequenced 16S rRNA gene of S. haemolyticus from Bangladeshi UTI patients, two phylogenetic trees were constructed using the isolates of the same species from 14 different countries and other selected strains of each reference genera. The genetic distance of the isolates from Bangladesh with other countries is noted according to the Phylogenetic tree and the comparison can be seen in Figure 3 and Table S3. These alignments showed partial sequence similarity in the 16S ribosomal RNA gene for translating specific regions. Hierarchical cluster analysis showed four clusters that can be seen in Figure 3. Firstly, the large Cluster divided into several necks such as first root Peru, Belgium, China, Pakistan, Japan, India, South Korea, Hungary, Mexico, Egypt, Brazil where the genetic distance was by 0.0000 (Figure 3). The second Cluster where the root was the USA, Bangladesh and Thailand by the genetic distance was 0.0007. The third Cluster where the root was the Chile, which supported by the genetic distance was 0.0011. According to the phylogenetic analysis, S. haemolyticus from Bangladesh is different from the isolates of the same species from 14 different countries. Furthermore, a phylogenetic tree constructed with the inclusion of other various bacterial species indicated that the isolated S. haemolyticus strain in this study was different (Figure S4).
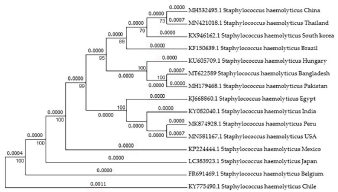
Figure 3: Phylogenetic tree analysis estimated from Staphylococcus haemolyticus 16S ribosomal RNA gene in the same species from different countries.
Antibiogram profiling
Thirteen different drugs were used for the disk diffusion test to determine the antibiotic susceptibility pattern. Determination of antibiotic susceptibility pattern revealed that all bacterial isolates tested were resistant to various antibiotics. Figure 4 shows the proportions of coagulase-negative staphylococci resistant to individual antimicrobial agents (Figure 4 and Table S4). Among the antibiotics tested, remarkably penicillin and ampicillin were found to be 100% resistant because all the isolates of S. haemolyticus showed resistant to penicillin and ampicillin. The resistant percentages of the isolates to other antibiotics included amoxicillin (88.46%), erythromycin (76.93%), ciprofloxacin (73.08%) and tetracycline (65.39%). Conversely, among the tested antibiotics vancomycin seems to be the most effective for treating UTI patients (S. haemolyticus susceptibility being 100%) followed by azithromycin (80.76%), amikacin (84.61%), gentamycin (69.23%), levofloxacin (73.08%), ceftriaxone (80.76%) and doxycycline (61.54%).
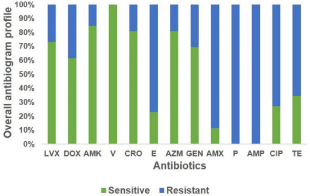
Figure 4: Antibiotic sensitivity pattern of Staphylococcus haemolyticus
against thirteen different antibiotics. LVX, Levofloxacin; DOX, Doxycycline;
AMK, Amikacin; V, Vancomycin; CRO, Ceftriaxone; E, Erythromycin; AZM,
Azithromycin; GEN, Gentamycin; AMX, Amoxicillin; P, Penicillin; AMP,
Ampicillin; CIP, Ciprofloxacin; TE, Tetracycline.
Discussion
The identification of bacterial pathogens with their antibiogram in human infection plays an essential role in the management of patients with disease in both the local community and hospital practices. Staphylococcus haemolyticus is known as critical nosocomial pathogens, accounting for up to 90% of all Coagulase- Negative Staphylococci (CNS) strains isolated from clinically significant specimens with chronic diseases [11,23]. In this study, we found 26% of patients with UTI were positive for S. haemolyticus, which is consistent with the previous findings [9]. Surprisingly, a study conducted by Lerbech et al. in Ghana reported that S. haemolyticus was the predominant pathogen (75%) in human urine collected from patients with UTI; whereas, others in India found a much lower incidence rate of S. haemolyticus in patients with UTI ranging from 15-18% [24,25,26,27]. The variation in the incidence rate of S. haemolyticus might be due to the factors influencing the variability of collected samples, lack of species identification opportunity in most laboratories, time, antibiotic use, and lack of advanced technologies in most developing countries like Bangladesh. However, these findings are in line with studies done in Indonesia, Ghana, South-west Ethiopia, and Iran; where, they suggested that the prevalence rate variation of causative bacteria in UTI is partly due to the sample size, the difference in the use of UTI guidelines, financial burdens, hospitalization duration, and lack of hospital training staff [7,24,28,29].
Similar to other studies [30,31,32], our findings showed that female patients had a higher prevalence of UTI than in males. There are a large number of factors associated with a high incidence of infection in females, such as shorter and broader urethra in females than in males, lack of antimicrobial properties of the prostatic fluid as in males, hormonal changes affecting the bacterial mucosal adherence and trauma of urethra during sexual intercourse. This result is consistent with other studies reported in Turkey and Iran [31,32].The rate of UTI mainly caused by S. haemolyticus was common in patients of age group between 16-60 years. In addition, the age group 31-45 years of patients had 15% and 22% high infection rate compared to the age groups 46-60 years and 16-30 years, respectively. Other investigator demonstrated that S. haemolyticus is the most susceptible (32.13%) at age group of 31-50 years of patients, which is almost in line with our findings [26]. The high occurrence of UTI at this age may be due to frequent sexual intercourse, use of contraceptive spermicidal agents, diaphragms, and menopause for women and enlargement of the prostate gland for men and a lack of sufficient knowledge of hygiene practices [30]. These findings correlate with other reports in India and Italy, which showed that patients’ age and gender might influence the prevalence of causative pathogens in UTI [33,34].
In the present study, S. haemolyticus species identified from urine samples of patients with UTI using the conventional and molecular methods. The results of traditional methods showed that the isolate was Gram-positive cocci, haemolytic, MR, V-P test and catalasepositive but coagulase-negative. In developing countries, including Bangladesh, the most traditional way of bacteria identification is to isolate the organism and to study its phenotype through culture, biochemical tests, and Gram staining. Similar studies were conducted in India, Iran, Turkey and Indonesia, where they used the conventional method for the identification of causative bacteria from the chronic disease [25-27,31-34]. Although this method is very informative, yet it is time-consuming, inapplicable for non-cultivable bacteria, biochemical tests are not suited for many known genera and species of bacteria and need a skilled person. In that case, 16S rRNA sequencing has been adopted as a new gold-standard for identification of bacteria because of the 16S ribosomal RNA gene is highly conserved for bacteria even among species of the same genus. With the advent of this method, many non-cultivable bacteria and bacteria with non-characteristic biochemical patterns can be categorized and re-classified into new families. PCR amplification and phylogenetic tree via 16S rRNA gene sequencing indicated that the isolate in this study belongs to the genus Staphylococcus, a conclusion supported by the comparison and alignment of the isolate 16S rRNA with other genus 16S rRNAs in the GenBank. In addition, we found 6 novel mutation sites including 5 substitution (A1G, G2C, T5A, C986T and T1432A) and 1 deletion mutation (T1414T). This mutation in 16S ribosomal RNA gene might be liable for identifying species-specific bacteria detection and various pattern on antibiotics resistance. Therefore, monitoring reservoir and distribution of S. haemolyticus should be performed in nosocomial infection by determining this causal pathogen at species level, and antibiogram profiling will be helpful in choosing the appropriate therapy [35]. In Bangladesh, the identification of the bacterial species is not made generally before doing the antimicrobial susceptibility test. This study will pave the way to detect S. haemolyticus from urine samples in patients with UTI in human.
This study on antibiogram profiling showed that there is a high prevalence of multidrug resistance in S. haemolyticus particularly penicillin (100%), ampicillin (100%), amoxicillin (88.46%), erythromycin (76.93%), ciprofloxacin (73.08%) and tetracycline (65.39%), which correlates with a previous study conducted by Barros et al. [36]. Furthermore, pattern of antibiotic resistant such as penicillin (91.3%), amoxicillin (88.9%), and erythromycin (77.8%) against S. haemolyticus was consistent with our findings, and the study carried out by Parashar, whereas other antibiotic does not correlate with susceptibility pattern reported by others [27,36]. In the present study, resistance to erythromycin (76.93%) and tetracycline (65.39%) was found to be almost similar with the study conducted by Fogget et al.-; the resistant pattern of erythromycin and tetracycline were 79% and 75%, respectively [9]. The susceptibility to ampicillin and erythromycin reported in this study is also in agreement with previous research conducted by other investigators [35]. The variation of different antibiotic resistance pattern in S. haemolyticus might be due to the different geographical distribution of strain as well as discordant finding between phenotypic test and genotypic characterization of various strains of S. haemolyticus. Another study by Pain et al., revealed that the widespread use of antibiotics might promote the development of antimicrobial resistant in S. haemolyticus through acquisition of mobile genetic elements or beneficial point mutations and rearrangements in surface associated genes [37].The mechanism of antibiotic-resistant in S. haemolyticus is still under investigation. A study done by Fogget et al. found that these nosocomial isolates were resistant to multiple antimicrobial agents more frequently than other coagulases negative staphylococcus. They suggest genetic evidence that S. haemolyticus shares a shared antimicrobial resistance gene pool with S. epidermidis [9]. On the other hand, in the study among the tested antibiotics, vancomycin was found to be most effective for treating UTI patients against S. haemolyticus [24,25,38,39]. In our study, S. haemolyticus was 100% sensitive to vancomycin followed by azithromycin (80.76%), amikacin (84.61%), gentamycin (69.23%), levofloxacin (73.08%), ceftriaxone (80.76%) and doxycycline (61.54%), respectively. Nonetheless, with the escalating use of these agents, we may encounter resistance to these drugs in future. It is essential that large appointment hospitals in Bangladesh keep continuous survey on the antimicrobial susceptibility pattern and the physicians use these agents judiciously to prevent antimicrobial resistance.
Because of the widespread accessibility of antimicrobial drugs, UTIs have become increasingly challenging to treat, due to increasing resistance to antimicrobial agents. The potential for species to acquire multi-antibiotic resistance has made it a severe threat to worldwide health care facilities. Whole-genome sequencing of S. haemolyticus will not only allow further examinations of the species’ characteristic but also enables the development of novel immunotherapeutic and chemotherapeutic approaches to mitigate this chronic health problem. Clinical investigations combined with genomic and genetic methods are necessary for understand more effective strategies against the development of S. haemolyticus antibiotic-resistant strains.
Conclusion
Antimicrobial resistance is a significantly growing regional and global threat in Bangladesh where antibiotics are used indiscriminately. AMR has a potential impact on influencing every sustainable development goal, particularly those targeting hunger, health, poverty, and economic growth. Therefore, the National Action Plan for AMR of Bangladesh requires baseline data on the occurrence of resistant bacteria and their genes from various sources to develop effective strategies to minimize AMR-related hazards. The S. haemolyticus associated with UTI patients is a crucial bacterial species showing MDR. In the present study variable antimicrobial resistance susceptibilities shown by all S. haemolyticus isolates against the particular antibiotic, none had a predictable pattern of antibiogram. Therefore, it appears mandatory that single strain should be identified up to species level with their antibiogram in any problematic situation. Conclusively, this study has shown that coagulase-negative S. haemolyticus from UTI patients are multidrugresistant to some commonly used antibiotics. The proper and timely detection of drug-resistant bacteria from UTI patients in hospitals will help to guide therapy management and prevent the emergence and spread of antibiotic-resistant bacteria in both the hospital and the non-hospital environment.
Author Contributions
Conceptualization, M.H.H.; methodology, M.L.M. and M.L.M; software, M.H.H. and S.S.; formal analysis, M.H.H., M.L.M. and S.S.; investigation, M.H.H. and M.S.; resources, M.H.H.; data curation, M.H.H. and M.J.A.S.; writing-original draft preparation, M.H.H. and M.L.M.; writing-review and editing, M.H.H., S.S., M.S. and M.J.A.S.; supervision, M.H.H.; project administration, M.H.H. and M.L.M.; funding acquisition, M.H.H. All authors have read and agreed to the published version of the manuscript.
Funding
pThe study was supported by the research funding from the Government of the People’s Republic of Bangladesh, Ministry of Education, and Bangladesh Bureau of Educational Information & Statistics (BANBEIS) and The World Academy of Sciences (TWAS).Acknowledgment
We thank all patients involved in the study. S.S. is funded by the Discovery Early Career Researcher Award of the Australian Research Council. The author would like to thank Professor M. Sawkat Anwer (Tufts University, USA) for his constructive criticism of the manuscript.
Conflicts of Interest
The authors have declared that no competing interest exist.
References
- Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004; 10: S122-S129.
- Stamm WE, Norrby SR. Urinary tract infections: disease panorama and challenges. J Infect Dis. 2001; 183: 1-4.
- Hossain MD, Sunjukta A, Kabir MS. Antibiotic resistance patterns of uropathogens isolated from catheterized and noncatheterized patients in Dhaka, Bangladesh. Tzu Chi Med J. 2014; 26: 127-131.
- Valiquette L. Urinary tract infections in women. Can J Urol. 2001; 1: 6-12.
- Hoque R, Ahmed SM, Naher N, Islam MA, Rousham EK, Islam BZ, et al. Tackling antimicrobial resistance in Bangladesh: A scoping review of policy and practice in human, animal and environment sectors. PloS one. 2020; 15: e0227947.
- Gupta K, Scholes D, Stamm WE. Increasing prevalence of antimicrobial resistance causing acute uncomplicated cystitis in women. J Am Med Assoc. 1999; 281: 736-738.
- Kitagawa K, Shigemura K, Yamamichi F, Alimsardjono L, Rahardjo D, Kumntaman K, et al. International Comparison of Causative Bacteria and Antimicrobial Susceptibilities of Urinary Tract Infections between Kobe, Japan, and Surabaya, Indonesia. Jpn J Infect Dis. 2018; 71: 8-13.
- Ozturk R, Murt A. Epidemiology of urological infections: a global burden. World J Urol. 2020.
- Froggatt JW, Johnston JL, Galetto DW, Archer GL. Antimicrobial resistance in nosocomial isolates of Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1989; 33: 460-466.
- Ma XX, Sun DD, Hu J, Wang EH, Luo EJ. Epidemiological and molecular characterization of Staphylococcus haemolyticus strains, from a hematology ward, with decreased susceptibility to glycopeptides. Can J Microbiol. 2011; 57: 476-484.
- Kloos WE, Bannerman TL. Update on clinical significance of coagulasenegative staphylococci. Clin Microbiol Rev. 1994; 7: 117-140.
- Gunn BA, Davis CEJr. Staphylococcus haemolyticus urinary tract infection in a male patient. J Clin. Microbiol. 1988; 26: 1055-1057.
- Kunin CM, Steele C. Culture of the surfaces of urinary catheters to sample urethral flora and study the effect of antimicrobial therapy. J Clin Microbiol. 1985; 21: 902-908.
- Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimcrob Resist In. 2017; 6: 47.
- Schleifer KH, Kloos WE. Isolation and characterization of staphylococci from human skin. Amended descriptions of Staphylococcus epidermidis and Staphylococcus saprophyticus and descriptions of three new species: Staphylococcus cohnii, Staphylococcus haemolyticus, and Staphylococcus xylosus. Int J Syst Bacteriol. 1975; 25: 50-61.
- Cheesbrough Monica. Medical Laboratory Manual for Tropical Countries: Vol.ii. Doddington (14 Bevills Close, Doddington, Cambs. PE15 0TT: M. Cheesbrough, 1984.
- Mahmud S, Nazir KHMNH, Rahman MT. Prevalence and molecular detection of fluoroquinolone-resistant genes (qnrA and qnrS) in Escherichia coli isolated from healthy broiler chickens. Vet World. 2018; 11: 1720-1724.
- Senthilraj R, Prasad GS, Janakiraman K. Sequence-based identification of microbial contaminants in non-parenteral products. Braz J Pharm Sci. 2016; 52: 329-336.
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4: 406-425.
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci. USA. 2004; 101: 11030-11035.
- EUCAST. European Committee on Antimicrobial Susceptibility Testing on antimicrobial susceptibility testing: Breakpoint tables for interpretation of MICs and zone diameters. version 10.0. 2020.
- Sweeney MT, Lubbers BV, Schwarz S, Watts JL. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J Antimicrob Chemother. 2018; 73: 1460-1463.
- Nunes APF, Teixeira LM, Bastos CC, Silva MG, Ferreira RBR, Fonseca LS, et al. Genomic characterization of oxacillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus isolated from Brazilian medical centres. J Hosp Infect. 2005; 59: 19-26.
- Lerbech AM, Opintan JA, Bekoe SO, Ahiabu MA, Tersbol BP, Hansen M, et al. Antibiotic exposure in a low-income country: screening urine samples for presence of antibiotics and antibiotic resistance in coagulase negative staphylococcal contaminants. PLoS One. 2014; 9: e113055.
- Kumar SR, Endra J, Das A, Mane P, Sangwan J, Kumari S. “Isolation, Identification and Antibiogram of Coagulase Negative Staphylococcus (CoNS) Isolated from Various Clinical Samples at a Tertiary Care Teaching Hospital, Jaipur, India.” Int J Curr Microbiol Appl Sci. 2018; 7: 3048-3059.
- Roopa C, Biradar Sk. Incidence and Speciation of Coagulase Negative Staphylococcus Isolates from Clinically Relevant Specimens with their Antibiotic Susceptibility Patterns. Int J Curr Microbiol App. Sci. 2015; 4: 975- 980.
- Parashar DS. Staphylococcus haemolyticus; a Nosocomial Pathogen Showing Higher Antimicrobial Resistance. Int J Sci. 2014; 3: 381-382.
- Beyene G, Tsegaye W. Bacterial uropathogens in urinary tract infection and antibiotic susceptibility pattern in Jimma University specialized hospital, southwest Ethiopia. Ethiop J Health Sci. 2011; 21: 141-146.
- Ghashghaee A, Behzadifar M, Azari S, Farhadi Z, Luigi BN, Behzadifar M, et al. Prevalence of nosocomial infections in Iran: A systematic review and meta-analysis. Med J Islam Repub Iran. 2018; 32: 48.
- Haider JS, Hasan A, Bin-Tahir K. Frequency of Urinary Tract Bacterial Infection and their Susceptibility Patterns among Hemodialysis Patients in Zliten Hospital. J Microbiol Exp. 2016; 3: 00093.
- Farajnia S, Alikhani MY, Ghotaslou R, Naghili B, Nakhlband A. Causative agents and antimicrobial susceptibilities of urinary tract infections in the northwest of Iran. Int J Infect Dis. 2009; 13: 140-144.
- Cetin M, Ucar E, Guven O, Ocaks. Community acquired urinary tract infections in Southern Turkey: etiology and antimicrobial resistance. Clin Nephrol. 2009; 71: 30-35.
- Magliano E, Grazioli V, Deflorio L, Leuci AI, Mattina R, Romano P, et al. Gender and age-dependent etiology of community-acquired urinary tract infections. Sci World J. 2012; 2012: 349597.
- Mahajan R, Gupta S, Mahajan B. Antibiotic Susceptibility Pattern of Isolates in Urinary Tract Infection in a Tertiary Care Hospital. J Rational Pharmacother Res. 2014; 2: 44-49.
- Donald EL, Schimidt BK, Kriplani HM. An endemic strain of staphylococcus haemolyticus colonizing and causing bacteremia in neonatal intensive care unit patients. Paediatrics. 1992; 89: 696-700.
- Barros EM, Ceotto H, Bastos MC, DosSantos KR. Giambiagi-Demarval M. Staphylococcus haemolyticus as an important hospital pathogen and carrier of methicillin resistance genes. J Clin Microbial. 2012; 50: 166-168.
- Pain M, Hjerde E, Klingenberg C, Cavanagh JP. Comparative Genomic Analysis of Staphylococcus haemolyticus Reveals Key to Hospital Adaptation and Pathogenicity. Frontiers in microbiology, 2019; 10: 2096.
- Chaudhury A, Kumar AG. In vitro activity of antimicrobial agents against oxacillin resistant staphylococci with special reference to Staphylococcus haemolyticus. Indian J Med Microbiol. 2007: 25: 50-52.
- Goel MM, Singh AV, Mathur SK, Singh M, Singhal S, Chaturvedi UC. Resistant coagulase negative staphylococci from clinical samples. Indian J Med Res. 1991; 93: 350-352.