
Research Article
J Bacteriol Mycol. 2021; 8(2): 1168.
Candida albicans Activation of Human Monocytes Toward M2 Profile is Reversed by Amphotericin B and Fluconazole
Icely PA1,2, Vigezzi C1,2, Rodriguez E1,2, Miró MS1,2, Salido Renteria B1 and Sotomayor CE1,2*
1Department of Clinical Biochemistry, National University of Cordoba, Argentina
2Center for Research in Clinical Biochemistry and Immunology, CIBICI-CONICET, Argentina
*Corresponding author: Claudia E Sotomayor, Haya de la Torre y Medina Allende, Ciudad Universitaria, 5000, Cordoba, Argentina
Received: February 20, 2021; Accepted: March 22, 2021 Published: March 29, 2021
Abstract
Phagocytes, including monocytes/macrophages, play an important role in the host defense during Candida albicans infections. In the L-arginine metabolism, the balance between the activation of two enzymes, inducible Nitric Oxide Synthase (iNOS) and arginase, promotes in the macrophages two alternative metabolic states, while M1 profile is related with host protection, M2 favored the fungal growth and evasion. Our aim was to evaluate the effect of Amphotericin B (AMB) and Fluconazole (FLC) on polarization of human monocytes to M2 profile induced by C. albicans. The human monocytic (Mo) cell line U937 was co-cultured with viable yeast of C. albicans, or Lipopolysaccharides (LPS) or Phorbol-12-myristate-13-acetate (PMA). Nitric Oxide (NO), cytokines production and arginase activity were evaluated. The effect of AMB or FLC on these metabolic pathways in immune cells and on fungus intrinsic arginase activity was studied. C. albicans inhibits NO production in human-monocyte and induces strong host arginase activity (p<0.0001). AMB and FLC inhibited C. albicansinduced arginase activity in immune cells (p<0.001), reaching a percentage of inhibition of 90% for AMB and 78% for FLC. Arginase intrinsic activity of the fungus was blocked by nor-NOHA (arginase inhibitor) and AMB (p<0.05). These results show that C. albicans drives human Mo toward M2 profile and that both antifungal drugs evaluated have the ability to revert C. albicans-induced M2 profile. In a relevant manner, it also provides data about additional effect of AMB as inhibitor of C. albicans endogenous arginase activity. Here in we provide new evidence for the effect of these drugs over the immune cells and the yeast.
Keywords: Amphotericin B; Fluconazole; Candida albicans; Monocytes; L-arginine pathway; Candida albicans arginase activity
Introduction
During the last decades the medical importance of the fungus as pathogens have increased dramatically, as a result of intensive use of antibiotics, immune suppressor therapies and HIV epidemic [1]. Candida albicans is a component of the human oral and gastrointestinal flora and this opportunistic fungus can in certain circumstances switch between the yeast to the mycelial form increasing its invasiveness, entering to host tissues and causing a local or systemic disease [2,3]. The host´s resistance to C. albicans is determined by a coordinated interaction between the innate and the adaptive mechanisms of defense [4].
In the innate immune response, the phagocytic cells such as macrophages (MF) and neutrophils are considered the effector cells [1]. In neutropenic patients, the mononuclear cells (Monocytes/ Macrophages) (Mo/MF) contribute to the protection against the infection, although the molecular mechanisms of the macrophagefungus interaction have not been completely established. In the last years, a relevant role has been assigned to this cell population in the host defense against this opportunistic pathogen [4]. The Mo/ MF have the capacity of killing C. albicans with the production of metabolites such as H2O2, superoxide anion, Nitric Oxide (NO) and peroxidenitrites that have a candidacidal activity more powerful than their progenitors [4-6]. The NO and other intermediaries of oxygen are important effectors molecules that directly act on the yeast and the hyphal form of fungus [6].
The ability to release NO can be modulated by different stimulus, including bacterial products, tumor cells and cytokines [6]. Its production depends on a cytosolic enzyme, the inducible Nitric Oxide Synthase (iNOS) and the availability of the substrate L-arginine [5]. Besides iNOS, the other major L-arginine consuming enzyme, arginase, hydrolyzes the L-arginine producing L-ornithine and urea. Both metabolic pathways depend on the extracellular supply of L-arginine. The competition between arginase and iNOS is more evidently when the availability of L-arginine is compromised as it occurs during sepsis [7,8]. Several lines of investigation suggest that the induction of one of the enzymes undergo with the suppression of the other, indicating two competitive states in murine macrophages [9]. The polarization of the metabolism of the L-arginine is associated with two types of response: the induction of the iNOS linked to MFM1 profile and the T-helper 1 type (Th1) response, denominated “classic pathway”, and the arginase activation associated to MFM2 profile and the Th2 response known as “alternative pathway” [5,9]. While M1 profile is associated with host protection, M2 favored the fungus resistance, making the M1 vs. M2 balance, a relevant event in the development of infection.
Our group showed evidence about the induction of both metabolic pathways in an experimental model of candidemia [10,11]. In the present work we evaluated the activation of the L-arginine pathway in a human monocytic cell line exposed to C. albicans and the effect of two antimicrobial agents frequently used in Latin America, Fluconazole (FLC) and Amphotericin B (AMB). We also provide novel evidence about the effect of these antifungal agents on C. albicans intrinsic arginase activity. Our results support an interesting perspective about the complex relation between pathogen, host and the antifungal drugs, extending the properties of the last ones beyond its well-known classic effects.
Materials and Methods
Microorganism
The C. albicans strain ATCC36801 was used in all the experiments [12,13]. Yeast cells were kept by serial subculture on Sabouraud Dextrose Agar (SDA) (Britania, Argentina) at Room Temperature (RT). For the experiments, yeast cells were grown on SDA for 48h at RT, harvested in sterile Phosphate-Buffered Saline (PBS) and centrifuged at 1000×g; after two washes in PBS, the yeast were counted and diluted to the desired concentration [12,13].
Cells
The human monocytic cell line U937 (ATCC-CRL-1593.2, USA) was cultured in RPMI1640 medium (GIBCO, USA) supplemented with 10% SBF (NATOCOR, Argentina) heat inactivated (56°C, 30min), 2mM L-glutamine, 50Uml penicillin, 50μg/ml streptomycin at 37°C in a 5% CO2/95% air atmosphere. Cell viability was determined by using trypan blue exclusion test (≥96%) [11].
Antifungal agents
A stock solution of each antifungal (FLC or AMB) was prepared using sterile distilled water. For FLC the stock solution of 2mg/ml from Lazar Laboratory (Argentina) was diluted with RPMI1640 medium (GIBCO-USA) to a final concentration of 1μg/ml in the well [14]. For AMB the stock solution of 5mg/ml from Fada Laboratory (Argentina) was diluted to a final concentration of 2.5μg/ml with RPMI1640 medium (GIBCO-USA) [14]. Both concentrations were below the Minimum Inhibitory Concentration (MIC) and the fungal viability after incubation with FLC or AMB was checked by the MTT assay [12].
Experimental design
The U937 cells (3x105cell/ml) were co-cultured in 96 multiwell plates with viable yeast of C. albicans, or Lipopolysaccharides (LPS) or Phorbol-12-myristate 13-acetate (PMA) and incubated for 48h at 37°C, 5% CO2/95% air atmosphere; U937 cells cultured alone (without stimuli) were used as control. C. albicans was used in a relation of 5:1 yeast/cell; LPS from Escherichia coli serotype 055:B5 (Sigma-Aldrich, USA) was used at a final concentration of 10μg/ml in the well and PMA (Sigma-Aldrich) was used at a final concentration of 1μg/ml.
Also U937 cells were cultured during 48h with AMB or FLC at established concentration, in the absence or presence of C. albicans. In other set of experiments, the specific arginase inhibitor, Nω- Hydroxy-nor-L-arginine (nor-NOHA, Roche- Switzerland) was used at 50μM with the purpose to evaluate it effects on: a) C. albicans alone, b) U937 cells co-cultured with C. abicans and c) U937 cells co-cultured with C. abicans in presence of AMB or FLC. After incubation, multiwell plates were centrifuged 10min at 2000rpm, the supernatants were removed and used to determine NO and cytokines production. The cells monolayer was used to determine arginase activity. Each experiment conditions consisted of three replications and at least three individual experiments were carried out.
Cell viability
Eukaryotic cell viability was determined after 48h of incubation using the 3-(4,5-dimethyl-2 thiazoyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay (Sigma-Aldrich). The MTT assay is a colorimetric method that detects mitochondrial activity present in living cells and the signal generated is directly proportional to the number of viable cells. This method was validated to detect fungus and cell viability [12,15]. MTT solution was added to each well at final concentration of 5mg/ml and incubated 4hat 37°C. After wards 200μl of DMSO was added into each well to dissolve the formed darkblue formazan crystals. Absorbance was measured at 570nm on a microplate reader (Bio-Rad, Hercules, USA).
Nitric oxide production
NO production by U937 cells was determined by measuring the nitrite concentration in the supernatant following a microplate assay method as previously described [16]. Briefly, 100μl of the culture supernatants were collected and mixed with 200μl of Griess reagent (sulphanylamide 1.5% in 1N HCl and N-(1-naphtyl) ethylenediamine-dihydrochloride 0.13% in H2O). Plates were incubated for 15min at RT in the dark and absorbance was measured at 540nm in a microplate reader (Bio-Rad). NO concentration was calculated by means of a calibration curve made between 2 and 125 μM of nitrite using sodium nitrite (Sigma-Aldrich) as standard. The results were expressed as μM of NO.
Arginase activity
To assess arginase activity, cell monolayers were washed with PBS and treated with 0.15ml of 0.1% Triton X-100 containing protease inhibitors [16]. Cell lysate (50μl) was added to 50μl of Tris-HCl (50nM; pH=7.5) containing 10nM MnCl2. Monocytes arginase was then activated by heating this mixture at 55-60°C for 10min. The hydrolysis reaction of L-arginine by arginase was carried out by incubating the mixture containing activated arginase with 50μl of L-arginine (0.5M; pH=9,7) at 37°C for 1h and was stopped by adding 400μl of the acid solution mixture (H2PO4:H3PO4:H2O=1:3:7). For colorimetric determination of urea, a-isonitroso-propiophenone (25μl, 9% in absolute ethanol) was then added, and the mixture was heated at 100°C for 45min. After placing the samples in the dark for 10min at RT, the urea concentration was determined spectrophotometrically by the absorbance at 550nm measured with a microplate reader (Bio-Rad). The amount of the urea produced was referred as arginase activity and expressed in mU/106 cells.
Cytokine immune assays
The level of cytokines in U937 culture supernatants was quantified using IL-1β, IL-8 and TNF (OptEIA BD, USA) ELISA tests as previously described [13]. Absorbance was measured using a microplate reader (Bio Rad), and concentrations extrapolated from standard curves. Data were expressed as pg/ml.
Statistical analyses
Data were expressed as means ± SEM. Differences between group means or ratios were assessed using one-way or two-way ANOVA followed by Bonferroni test for multiple comparisons. A p-value<0.05 was considered statistically significant. GraphPad Prism version 5.0 for Windows (GraphPad Software, USA) was employed. All experiments were repeated at least three times to test the reproducibility of results.
Results
C. albicans upregulates the alternative L-arginine pathway in human monocytes
It was reported that human Mo are able to express the iNOS, the limitant enzyme for the NO production, after the activation by different stimulus [17]. In order to evaluate if C. albicans induces the production of this nitric derivative product in human Mo, the U937 cell line was cultured with yeast, o with different stimulus for 48h and the production of NO was measured in the cell culture supernatants. We observed in Figure 1A, that the fungus induces a significant decrease in the basal production of the toxic nitrogen derivative (p<0.0001). In this system, the human Mo were refractory to the E.coli LPS and PMA activation. We also tested the Mo viability exposed to different stimuli using MTT assay directly in the U937 cell cultured in basal condition and stimulated with LPS, PMA, or C. albicans. The results demonstrate that U937 cells were alive at all different treatments (Figure 1B).
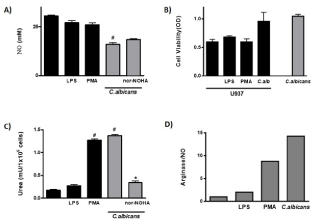
Figure 1: NO Production and Arginase Activity in human monocytes.
A: NO production in U937 cells stimulated with LPS, PMA and C. albicans
with or without the arginase enzyme inhibitor nor-NOHA.
B: U937 cells viability was tested by the mitochondrial enzyme activity (MTT
technique) and expressed as O.D.
C: Activation of arginase pathway in U937 cells stimulated with LPS, PMA
and C. albicans with or without the arginase enzyme inhibitor nor-NOHA.
D: The balance of the metabolic pathway of L-arginine was determined by the
relative relationship between products of both metabolic pathways. The data
shown are representative of three independent experiments and results are
expressed as mean ± SEM. *p<0.05, **p<0.01, #p<0.001.
The evaluation of alternative L-arginine pathway showed that the contact of U937 cells with C. albicans induced its activation with a significant increase in the urea production compared with basal condition (p<0.0001) (Figure 1C). PMA activated this metabolic pathway in Mo (pvs basal<0.0001) and interestingly, arginase activation by fungus reached similar value (Figure 1C); LPS exposition did not stimulate this pathway (p=NS). The competition between arginase and iNOS has been suggested in different conditions [18]; in order to test this balance during fungus-monocytes interaction, we cultured U937 cells and C. albicans in presence of nor-NOHA, a classic inhibitor of arginase pathway, and the impact in the iNOS and arginase activity was evaluated (Figure 1A and 1C). As it is shown in Figure 1C, the level of arginase activity was significantly diminished when U937 cells were cultured with C. albicans in presence of nor-NOHA (pvs C. albicans<0.05), and the levels of iNOS enzyme activation has not been modified (Figure 1A), indicating the independence between these metabolic pathways after fungus stimulation.
As the polarization between these pathways has consequences in the outcome of immune response, we analyzed the balance between iNOS and arginase activity in human monocytic cell line co-cultured with the different stimuli. The results indicated that PMA favored de arginase pathway activation and that fungus clearly promotes the alternative activation of this cellular population (Figure 1D).
Modulation of intrinsic arginase activity of C. albicans
In order to establish if nor-NOHA could also modulate C. albicans intrinsic arginase activity, we evaluated the levels of end metabolite in cultures of fungus alone (Figure 2A). We found that urea production was down-regulated in the presence of nor-NOHA (p<0.05). With the purpose to evaluate the effect of antifungal drugs on C. albicans endogen arginase activity, we co-cultured the fungus with AMB or FLC (concentration<MIC). Interestingly AMB was able to significantly decreased intrinsic fungus arginase activity (53%) (p<0.05) reaching a value of inhibition similar to the one observed with nor-NOHA (60%) (Figure 2B); on the other hand, FLC did not show this effect. The Figure 2C shows that the C. albicans co-cultured with different stimuli (antifungal drugs or arginase inhibitor) did not modify fungus viability, suggesting that the decreased in enzyme activity observed was due to selective effect of nor-NOHA or AMB on arginase activity.
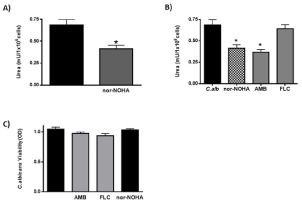
Figure 2: Effect of different antifungal agents in C. albicans arginase
production.
A: Arginase activity in C. albicans and effect of arginase enzyme inhibitor
nor-NOHA.
B: Arginase activity in C. albicans co-cultured with AMB, FLC or nor-NOHA.
C: C. albicans viability co-cultured with AMB, FLC or nor-NOHA. The data
shown are representative of three independent experiments and results are
expressed as mean ± SEM, *p<0.05.
Effect of Amphotericin B and Fluconazole on the polarization of the L-arginine metabolic pathway in human monocytes exposed to C. albicans
With the purpose to evaluate if antifungal drugs could modulate the metabolic pathway of L-arginine in human monocytes exposed to C. albicans, U937 and C. albicans were co-cultured with two antifungal agents, AMB and FLC. We found that the basal levels of NO production in U937 cells weren´t modified by AMB or FLC (p=NS) (Figure 3A). In the presence of C. albicans, both antifungal agents were unable to reverse the effect caused by the fungus. The use of the selective inhibitor nor-NOHA showed similar levels of NO production as the observed with the antifungal agents over the fungus-monocytes culture. As a control, U937 viability was evaluated after treatment with AMB and FLC and values were similar to the basal, showing that they weren´t harmful at the concentrations used in these experiments (Figure 3B).
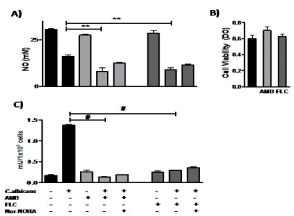
Figure 3: NO production and arginase activity by human monocytes exposed
to antifungals in the presence or absence of C. albicans.
A: U937 cells viability after treatment with AMB and FLC.
B: NO production by the U937 cells, in the presence of the C. albicans and
both antifungal drugs.
C: Arginase activity induced by the fungus, with or without AMB and FLC. The
data shown are representative of three independent experiments and results
are expressed as mean ± SEM. *p<0.05, **p<0.01, #p<0.001.
Besides, we found that both antifungal agents per se didn´t trigger the arginase activity on U937 cells (Figure 3C) (pvs U937=NS). Interestingly, we observed a significant decrease of the total enzymatic activity when U937 cells co-cultured with C. albicans and AMB or FLC (p<0.0001). This reduction was in 90% and 78% respectively.
Effect of Amphotericin B and Fluconazole on cytokines released by human monocytes exposed to C. albicans
Finally, we evaluated the production of proinflammatory cytokines in human Mo. After U937 cells were incubated with C. albicans, neither IL-1β nor TNF was detected, and only a decreased level of IL-8 was observed (p<0.001) (Figure 4). The antifungal agents didn´t modify the basal production of these cytokines in U937 cells, but when these cells were cultivated with C. albicans in presence of AMB and FLC, both blocked the down regulation of IL-8 produced by the fungus.
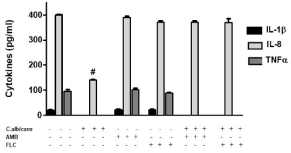
Figure 4: AMB blocks the inhibition of C. albicans in the release of IL-8 by
U937 cells. Evaluation of IL-1β, IL-8 and TNF concentration by ELISA test
on supernatants. The data shown are representative of three independent
experiments and results are expressed as mean ± SEM. #p<0.001.
Discussion
Mo/MF, play an important role in the defense mechanism against C. albicans [2,4,19]. These cells in response to proinflammatory cytokines or pathogen stimulation, undergo dramatic changes in the metabolism of the L-arginine. Many studies show that enzymatic activation of iNOS or arginase, conditions the MF polarization to either M1 or M2 phenotype [7-10,20]. In opposition to the available information on murine systems, the knowledge about the role of this metabolic pathway in humans is partial and its implication in host response has not yet been completely elucidated.
Our study demonstrate that C. albicans induces a strong activation of arginase and a significant reduction of NO production in human Mo, showing that the metabolic balance favors the M2 profile after fungal contact. Interestingly, the use of a specific arginase inhibitor, suppressed in human Mo the activation of this enzyme without promoting iNOS activity, thus revealing the independency between these two metabolic pathways after fungus stimulation. According to these results, other stimuli such as LPS and Galectin-1 have a similar effect on this pathway [5,21]. We previously reported that iNOS activation in rat peritoneal MF is crucial to limit C. albicans growth [10], and for the control of intrahepatic fungal burden during systemic infection [9]. In this model, strong arginase activity also was observed in isolated and tissue-resident MF [10,11]. Interestingly, purified fungal lipase modulate the L-arginine pathway in C. albicansprimed in vivo or in vitro MF [5]. These previous evidences show the ability of the fungus and its released virulence factor to modulate the L-arginine pathway in different manners, depending on the microenvironment where the interaction host-pathogen takes place [5,10,11]. The final consequence of this balance can deeply modify the outcome of the infection process.
Chitin is a well-known polysaccharide component of fungus cell wall. Wagener et al. [22] tested the ability of purified fungal chitin to induce arginase protein expression and arginase activity in IFN-γ and LPS-activated human monocyte-derived MF. They showed that chitin exposed during C. albicans-macrophage interactions is a strong inducer of arginase-1 activity in human MF. By blocking the C. albicans-induced arginase macrophage activity with nor-NOHA or chitinase inhibitor (bisdionin F), NO production is restored and the efficiency of fungal killing increases. This observation obtained from human MF and our results in a human monocytic cell line reveal the importance of the shifts of classically activated Mo/MF toward an alternative activated phenotype during their first interaction with pathogen, and fungal strategies to guarantee its own survival. With the aim of interfering this pathway, we checked the effect of two classical antifungal drugs used for treatment of systemic C. albicans human infections in Latin America, FLC and AMB [23]. Multiple studies performed in vitro and in vivo have demonstrated that AMB has an effect on host, in the presence of not only the pathogen, but also when uninfected cell lines or animals are treated with this antifungal. This drug stimulates transcription and production of multiple mediators of the immune system such as cytokines and chemokines [24-27]. Here we report that both antifungal agents reverse C. albicansinduced polarization to the alternative pathway of L-arginine in U937 cells by inhibition of arginase activity. We observed an important reduction of arginase activity, 90% for AMB and 78% for FLC. In this system, when both antifungal agents were cultured with cells alone, they did not promote NO production, and were unable to restore NO decreased levels after fungus stimulation. In relation with proinflammatory cytokines, the constitutive expression in human monocytes was down regulated after C. albicans exposure, suggesting that immune cells could be exhibited a weak functional competence after exposure to strongest inflammatory stimulus or in response to invasive pathogens [28]. Neither AMB nor FLC modified the basal levels of both IL-1β and TNF in U937 cells, and had any effect on these cytokines after 48h of culture with C. albicans, but they restored IL-8 production, in agreement with previously effects described on this pathway [24,29,30]. The trade-off between fungal signaling and drug effect on cytokine pathway remains unknown, in part due to the complexity of the inflammatory response. In a relevant manner, an important conclusion of this set of experiments is that both antifungal drugs reversed the activation of C. albicans-induced Mo toward M2 profile, thus adding a new capacity to the above described, and also extending their spectrum of immunomodulatory properties.
The induction of both intracellular arginase (Car1p) and urea amidolyase is a very fascinating strategy used by Candida to achieve hyphal switching and escape from macrophages after being ingested. Candida induces two other endogenous arginases, Arg1 and Arg3 that are involved in its survival [31]. An important finding of the present work is that beside the effect of the fungus in the balance of L-arginine in the host cells, its intrinsic arginase activity could be inhibited by AMB. Interestingly, the effect reached a value similar to observed after fungus treatment with Nor-NOHA. This fact sheds light on the notion that this antifungal agent can also interfere in the endogenous arginase activation with a relevant impact on Candida dimorphism and evasion. More studies are needed in order to establish the molecular target of this effect.
This work demonstrates the fungus´s capacity to promote the human Mo toward M2 profile and the ability of AMB and FLC to interfere in this pathway. In a relevant manner, it also provides data about additional effect of AMB as inhibitor of C. albicans endogenous arginase activity. Here in we provide new evidence for the effect of these drugs over the immune cells and the yeast.
Acknowledgments
This work was supported by grants from Agencia Nacional de Promoción Científica y Tecnológica-FONCyT, (PICT-2015-1393); SECyT-UNC (30720150100934CB) and CONICET (PIP-112 201501-00652CO).
References
- Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. Invasive candidiasis. Nature Reviews Disease Primers. 2018; 4: 18026.
- Su C, Yu J, Lu Y. Hyphal development in Candida albicans from different cell states. Current Genetics. 2018; 64: 1239-1243.
- Vigezzi C, Icely PA, Dudiuk C, Rodríguez E, Miró MS, Castillo GDV, et al. Frequency, virulence factors and antifungal susceptibility of Candida parapsilosis species complex isolated from patients with candidemia in the central region of Argentina. Journal de Mycologie Médicale. 2019; 100907.
- Lionakis MS, Netea MG. Candida and Host Determinants of Susceptibility to Invasive Candidiasis. Heitman J, editor. PLoS Pathogens. 2013; 9: e1003079.
- Paraje MG, Correa SG, Albesa I, Sotomayor CE. Lipase of Candida albicans induces activation of NADPH oxidase and L-arginine pathways on resting and activated macrophages. Biochemical and biophysical research communications. 2009; 390: 263-268.
- MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annual review of immunology. 1997; 15: 323-350.
- Corraliza IM, Soler G, Eichmann K, Modolell M. Arginase induction by suppressors of nitric oxide synthesis (IL-4, IL-10 and PGE2) in murine bone-marrow-derived macrophages. Biochemical and biophysical research communications. 1995; 206: 667-673.
- Jahani M, Noroznezhad F, Mansouri K. Arginine: Challenges and opportunities of this two-faced molecule in cancer therapy. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018; 102: 594-601.
- Locati M, Mantovani A, Sica A. Macrophage activation and polarization as an adaptive component of innate immunity. Advances in immunology. 2013; 120: 163-184.
- Correa SG, Rodríguez-Galán MC, Salido-Rentería B, Cano R, Cejas H, Sotomayor CE. High dissemination and hepatotoxicity in rats infected with Candida albicans after stress exposure: potential sensitization to liver damage. International immunology. 2004; 16: 1761-1768.
- Rodriguez-Galán MC, Sotomayor C, Costamagna ME, Cabanillas AM, Rentería BS, Masini-Repiso AM, et al. Immunocompetence of macrophages in rats exposed to Candida albicans infection and stress. American journal of physiology Cell physiology. 2003; 284: C111-118.
- de Freitas Araújo MG, Pacífico M, Vilegas W, Dos Santos LC, Icely PA, Miró MS, et al. Evaluation of Syngonanthus nitens (Bong.) Ruhl. extract as antifungal and in treatment of vulvovaginal candidiasis. Medical Mycology. 2013; 51: 673-682.
- Miró MS, Rodríguez E, Vigezzi C, Icely PA, García LN, Peinetti N, et al. Corrigendum: Contribution of TLR2 pathway in the pathogenesis of vulvovaginal [Pathogens and Disease., 76, 2018, (fty050)]. Pathogens and Disease. 2018; 76.
- Hedayati MT, Tavakoli M, Zakavi F, Shokohi T, Mofarrah R, Ansari S, et al. In vitro antifungal susceptibility of Candida species isolated from diabetic patients. Revista da Sociedade Brasileira de Medicina Tropical. 2018; 51: 542-545.
- Hjertstedt J, Hahn BL, Kos WL, Sohnle PG. Comparison of fungal viability assays using Candida albicans yeast cells undergoing prolonged incubation in the absence of nutrients. Mycoses. 1998; 41: 487-492.
- Chang CI, Liao JC, Kuo L. Arginase modulates nitric oxide production in activated macrophages. The American journal of physiology. 1998; 274: H342-348.
- Schmidt HHHW, Walter U. NO at work. Cell. 1994; 78: 919-925.
- Das P, Lahiri A, Lahiri A, Chakravortty D. Modulation of the Arginase Pathway in the Context of Microbial Pathogenesis: A Metabolic Enzyme Moonlighting as an Immune Modulator. Manchester M, editor. PLoS Pathogens. 2010; 6: e1000899.
- Gao Q, Zhang J, Chen C, Chen M, Sun P, Du W, et al. In Situ Mannosylated Nanotrinity-Mediated Macrophage Remodeling Combats Candida albicans Infection. ACS Nano. 2020; 14: 3980-3990.
- Tate DJ, Vonderhaar DJ, Caldas YA, Metoyer T, Patterson JR, Aviles DH, et al. Effect of arginase II on L-arginine depletion and cell growth in murine cell lines of renal cell carcinoma. Journal of hematology & oncology. 2008; 1: 14-18.
- Rabinovich GA, Sotomayor CE, Riera CM, Bianco I, Correa SG. Evidence of a role for galectin-1 in acute inflammation. European journal of immunology. 2000; 30: 1331-1339.
- Wagener J, MacCallum DM, Brown GD, Gow NAR. Candida albicans Chitin Increases Arginase-1 Activity in Human Macrophages, with an Impact on Macrophage Antimicrobial Functions. mBio. 2017; 8.
- Nucci M, Thompson-Moya L, Guzman-Blanco M, Tiraboschi IN, Cortes JA, Echevarría J, et al. [Recommendations for the management of candidemia in adults in Latin America. Grupo Proyecto épico]. Revista iberoamericana de micologia. 2013; 30: 179-188.
- Mesa-Arango AC, Scorzoni L, Zaragoza O. It only takes one to do many jobs: Amphotericin B as antifungal and immunomodulatory drug. Frontiers in microbiology. 2012; 3: 286-291.
- Roilides E, Lyman CA, Filioti J, Akpogheneta O, Sein T, Lamaignere CG, et al. Amphotericin B formulations exert additive antifungal activity in combination with pulmonary alveolar macrophages and polymorphonuclear leukocytes against Aspergillus fumigatus. Antimicrobial agents and chemotherapy. 2002; 46: 1974-1976.
- Rogers PD. Differential expression of genes encoding immunomodulatory proteins in response to amphotericin B in human mononuclear cells identified by cDNA microarray analysis. Journal of Antimicrobial Chemotherapy. 2002; 50: 811-817.
- Kumar P, Shivam P, Mandal S, Prasanna P, Kumar S, Prasad SR, et al. Synthesis, characterization, and mechanistic studies of a gold nanoparticleamphotericin B covalent conjugate with enhanced antileishmanial efficacy and reduced cytotoxicity. International Journal of Nanomedicine. 2019; 14: 6073-6101.
- Pei X, Fan X, Zhang H, Duan H, Xu C, Xie B, et al. Low frequency, weak MCP-1 secretion and exhausted immune status of peripheral monocytes were associated with progression of severe enterovirus A71-infected hand, foot and mouth disease. Clinical & Experimental Immunology. 2019; 196: 353-363.
- Turtinen LW, Prall DN, Bremer LA, Nauss RE, Hartsel SC. Antibody arraygenerated profiles of cytokine release from THP-1 leukemic monocytes exposed to different amphotericin B formulations. Antimicrobial agents and chemotherapy. 2004; 48: 396-403.
- Ries F, Alflen A, Aranda Lopez P, Beckert H, Theobald M, Schild H, et al. Antifungal Drugs Influence Neutrophil Effector Functions. Antimicrobial Agents and Chemotherapy. 2019; 63.
- Jiménez-López C, Collette JR, Brothers KM, Shepardson KM, Cramer RA, Wheeler RT, et al. Candida albicans induces arginine biosynthetic genes in response to host-derived reactive oxygen species. Eukaryotic cell. 2013; 12: 91-100.