
Research Article
J Bacteriol Mycol. 2021; 8(5): 1181.
Comparing Quantitative and Qualitative Methods for Detecting the In Vitro Activity of Colistin against Different Gram-Negative Bacilli
Shams N1#, AlHiraky H1#, Moulana N1, Riahi M1, Alsowaidi K1, Albukhati K1, Zughaie SM2,3, Eltai NO4*
1Department of Health Sciences, Biomedical Sciences, QU Health, Qatar University, Doha, Qatar
2College of Medicine, QU Health, Qatar University, Doha, Qatar
3Biomedical and Pharmaceutical Research Unit, QU Health, Qatar University, Doha, Qatar
4Biomedical Research Centre, Microbiology Unit Qatar University, Doha, Qatar
#These authors contributed equally to work
*Corresponding author: Nahla O. Eltai, Biomedical Research Centre, Microbiology Unit Qatar University, Doha, Qatar
Received: June 24, 2021; Accepted: July 15, 2021; Published: July 22, 2021
Abstract
The surge in the prevalence of Multidrug-Resistant (MDR) Gram-negative bacterial infections with limited treatment led to colistin reusing to treat MDR infections. This study aimed to determine economical, simple, and reliable colistin susceptibility testing methods as an alternative to the microdilution technique. We compared seven colistin susceptibility testing methods, including quantitative and qualitative, namely: Disk diffusion, E-test, ComASPTM SensiTest Colistin, Colistin broth disk elution, and colistin agar test CHROMagarTM COL-APSE, and BD Phoenix ID/AST automated identification and susceptibility testing system to the gold standard Broth Microdilution (BMD). Whole-genome sequencing was performed on all isolates to determine if the genetic resistant factors affect the phenotypic profile of the colistin resistance. Our results revealed that disk diffusion is still an ineffective method for measuring colistin susceptibility in Gram-negative Bacilli with the highest major error (31.75%), the lowest Kappa 0 (0%), and categorical agreement (68.25%) values. Phoenix, and CompASPTM SensiTest colistin methods have remained superior in reproducibility, sturdiness, and simplicity of use, similar to the currently recommended broth microdilution procedure; with high sensitivity of 95.56%, and 97.73%, specificity of 95.24, and 100%, and Kappa values of 0.89 and 0.95, respectively. This study revealed that Phoenix, and ComASPTM SensiTest colistin methods are recommended for routine microbiology laboratories with a large workload.
Keywords: Colistin; Gram-negative bacilli; Resistance; Microdilution; MIC
Introduction
Colistin is a cationic polypeptide antibiotic that belongs to the family polymyxin, including polymyxin B and polymyxin E (colistin). It is the last resort for treating multidrug resistance Gramnegative Bacilli (MDR-GNB), mainly against Escherichia coli (E. coli), Klebsiella pneumoniae (K. pneumoniae), and Pseudomonas aeruginosa (P. aeruginosa) [1,2]. It is synthesized naturally by Paenibacillus polymyxa and is rapidly bactericidal to Gram-negative bacteria. Colistin targets lipid A of Lipopolysaccharide (LPS) in the Gram-Negative Bacilli (GNB) outer layer cell membrane. Its central role is to inhibit cell membrane function by increasing its permeability to absorb polymyxins substances. This increase in permeability leads to the destruction of inner macromolecules and cell ions, essential for cell survival [2,3]. However, several MDR-GNB bacteria developed their defense mechanisms against colistin. This resistance development could be either chromosomal or plasmid-mediated. The chromosomal resistance can occur due to mutation/insertion in LPS biosynthesis genes (lpx, pmrA/B, mgrB, and phoP/Q). On the other hand, the plasmid-mediated resistance is acquired by the horizontal gene transmission, Encoding Phosphoethanolamine Transferase (pEtN) enzymes [4]. As a result, the cell membrane permeability will be reduced, and polymyxin binding will be suppressed [1,5-7].
Natural resistance to colistin has been reported on several bacteria, mainly chromosomal mutations in some Gram-negative species. However, recently plasmid-mediated colistin resistance emerged in clinically significant Gram-negative bacilli. The emergent resistance was first reported in China in November 2015 through the mcr-1 gene [8, 9]. Since then, strains carrying the mcr-1 gene, especially E. coli, have been isolated worldwide from humans [10], food-producing animals [11], and the environment [12]. Another plasmid-carried colistin-resistant gene (mcr-2) was detected in E. coli isolates from pigs in Belgium [13]. To date, ten mobile mcr genes have been described [14,15]. Colistin was first introduced as an antibiotic in 1952 and was used until the early 1980s to treat infections caused by GNB. Its use was discontinued because of problems with toxicity, such as nephrotoxicity and neurotoxicity [16]. The increase of multidrug resistance among clinically significant GNB renewed interest in colistin as a last resort therapeutic option for treating MDR-GNB bacteria. In clinical practice, there are some situations where clinician needs to use colistin; therefore, it is critical for clinical laboratories to determine colistin resistance to prevent prolonged administration to the patient for whom it would not be effective.
The poor diffusion and binding of colistin to plastics lead to its complicated phenotypic resistance testing [17]. In addition, some studies noted discrepancies in phenotypic colistin resistance results obtained by different methods. Besides, no reference method has been demarcated to compare colistin susceptibility testing results [4]. As such, both the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommend only Broth Microdilution (BMD) [17]. However, the method is impractical for clinical laboratories due to the large sample load in routine microbiology laboratories [18]. Thus, an alternative, more practical method is needed. Several commercial methods are available for colistin Minimum Inhibitory Concentration (MIC) measurement, but only a few studies describe their efficacy compared to each other. In this context, we conducted this study to evaluate and compare the different available quantitative and qualitative colistin susceptibility testing methods on some GNB to find the feasible colistin test. These methods include disk diffusion (Kirby- Bauer test), gradient diffusion method (E- test), CHROMagarTM COL-APSE, ComASPTM colistin broth microdilution, colistin broth disk elution, agar dilution, BD Phoenix ID/ AST automated identification, and susceptibility testing system.
Additionally, we performed Whole-Genome Sequencing (WGS) to identify the genetic determinants of colistin resistance and find if it affects the phenotypic testing method. The study outcomes will provide a framework for clinical laboratories to select the appropriate method to test colistin resistance.
Materials and Methods
Bacterial strains
Two sets of previously collected bacterial isolates (n=63) were employed in this study. The first set comprises isolated human pathogens collected from Hamad Medical Corporation (HMC), Doha, Qatar, during routine diagnostic testing (n=37). The second set was obtained from poultry fecal samples (n=26). Non-duplicate bacterial strains were characterized and identified as GNB via Biomic V 3 (Giles Scientific, USA) or Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MAL-DI-TOF MS) (Bruker Daltonik GmbH, Leipzig, Germany). These isolates include 43 strains of E. coli, 18 strains of K. Pneumoniae, and two strains of P. aeruginosa. Clinical Isolates were selected because of their resistance to third-generation cephalosporins and/or colistin. E. coli isolates from poultry fecal samples were obtained from farms using colistin as a prophylaxis and growth promotor. The reference strains E. coli ATCC® 25922, K. pneumoniae ATCC® 700603, and P. aeruginosa ATCC® 27853 were used as quality control organisms in all experiments. This study was approved by Qatar University Institutional Biosafety Committee (QU-IBC) number QU-IBC-2020/030.
Phenotypic methods to detect colistin resistance
The subsequent techniques were applied to all 63 isolates to determine their susceptibility to colistin, and readings were interpreted according to the CLSI [19], NCCLS [20], and Gales et al. [21] (Supplementary Table S1).
Disk diffusion method (Kirby-Bauer test)
Bacterial isolates were cultured on nutrient agar plates for 18- 24hrs at 37°C. After incubation, 2 to 3 pure colonies were suspended in a phosphate buffer solution (Atom Scientific, UK) to achieve an inoculum equivalent to 0.5 McFarland Standard as measured by DensiCHEK PLUS (bioMérieux, France). The suspension was then swabbed onto a Mueller-Hinton (MH) agar plate (Himedia, Mumbai, India) and allowed to dry completely. Next, antibiotic disks impregnated with 10μg colistin (Liofilchem®, Roseto Degli Abruzzi, Italy) were applied to the agar surface and incubated at 37°C for 24hrs. The zone of inhibition was measured in mm (Figure 1). These values were interpreted according to the CLSI [19].
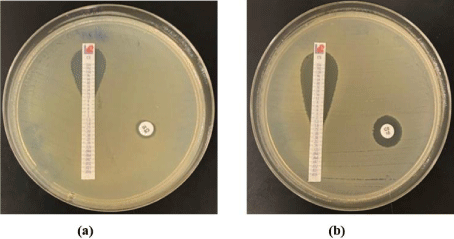
Figure 1: Colistin susceptibility testing by the disk diffusion and E-test
methods (Liofilchem®). (a) The isolate’s disk diffusion test showed an
inhibition zone of 8 mm around the colistin disk (right) and MIC of 12μg/mL
(left). (b) disk diffusion test of the isolate showing inhibition zone of 12 mm
around the colistin disk (right), inhibition zone with MIC of 1.5 μg/mL (left).
Gradient diffusion method (E-Test)
The isolates were processed similarly to the disk diffusion. The E-test strip (Liofilchem, Zona Ind. le, Italy), containing a gradient of antibiotic concentrations, was placed onto the prepared MH agar plate. The next day, the MIC (the point where the edge of the ellipse intersects the strip) in μg/mL was recorded (Figure 1), and susceptibility was interpreted according to CLSI [19].
CHROMagarTM COL-APSE (selective colistin bacterial culture medium)
The CHROMagarTM COL-APSE (CHROMagar, Paris, France) medium was briefly prepared in-house according to the manufacturer’s recommendation. The medium was challenged with 63 different GNB recovered from human and animal samples. The media’s ability to detect colistin Gram-negative resistant isolates was assessed by direct streaking of a single colony, taken from overnight nutrient agar culture onto the prepared CHROMagar plate. Then the plate was incubated at 37°C for 18-24 hrs. The growth of typical single colonies appearance, E. coli (pink), Pseudomonas (natural pigmentation cream to green), coliforms (metallic blue) was considered as colistin-resistant (Figure 2). If the growth was inhibited, the isolate was considered sensitive.
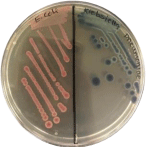
Figure 2: Resistant E. coli and K. pneumoniae growing on CHROMagar
COL-APSE after overnight incubation at 37°C. Colistin-resistant E. coli are
dark pink to reddish (Left), colistin-resistant K. pneumonia is metallic blue (right).
ComASPTM SensiTest Colistin
ComASPTM Colistin (Liofilchem, Italy) was used as a simple version of the BMD and following the manufacturer’s recommendations. Succinctly, suspensions equivalent to the 0.5 McFarland of the isolates were prepared. Further dilutions were obtained by diluting 50μL of the bacterial suspensions in 950μL of saline. Then 100μL of the diluted suspension was dispensed in the panel’s wells. The panel was covered with a lid and incubated in ambient air at 37°C for 16-20 hrs. The first clear well was considered the MIC of the examined sample. Concentrations in the ComASPTM colistin plate range from 0.25μg/mL to 16μg/mL as depicted in Figure 3.

Figure 3: Colistin susceptibility testing by ComASP. Concentrations in the
ComASPTM colistin plate range from 0.25μg/mL to 16μg/mL. The well on the
right of the red line indicates the value of the MIC for the isolate.
Colistin broth disk elution
This method was performed as described by CLSI [19]. Three to five colonies were picked, and turbidity was adjusted to the equivalent of a 0.5 McFarland standard, then further diluted to 1:10 in PBS. Four tubes of MH Broth (Liofilchem®, Roseto degli Abruzzi, Italy) were labelled 0 (for control), 1, 2 and 4μg/mL for each isolate, corresponding to the concentration of colistin disks (Liofilchem®, Roseto degli Abruzzi, Italy) that contains. One colistin disk was added to the tube labeled (1μg/mL), two colistin disks to tube labeled (2μg/ mL), and four colistin disks to tube labeled (4μg/mL). The prepared inoculum (50μL) was added to all tubes to attain a final concentration of approximately 7.5x 105 CFU/mL. The tubes were incubated in a shaking incubator (Innova®44, incubator shaker series, Eppendorf, Germany) at 33-35°C in ambient air for 16-20 hrs. MIC was read as the lowest agar concentration that completely inhibits the growth of the tested isolate (Figure 4).
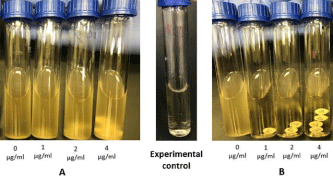
Figure 4: Colistin Broth-Disk Elution (CBDE). (A) Resistance, with a colistin
MIC of >4μg/mL (B) Sensitive, with colistin MIC of 1μg/mL.
Colistin agar test (CAT)
Colistin stock antibiotic solution was made by adding 15.7mg of colistin sulfate salt powder (Potency 19000 U/mg) to 1mL of distilled water to obtain a final concentration of 10mg/mL, according to the following equation: Weight (mg)=Volume (mL) X colistin concentration (μg/mL)/potency (μg/mg) [22]. Colistin agar plates with different colistin concentrations were prepared as follows: the autoclaved MH agar was allowed to cool to 45-50°C in a water bath before aseptically adding colistin (15μL, 30μL, and 60μL to 150mL warm MH agar to achieve concentrations of 1, 2, and 4μg/ mL, respectively. Subsequently, the prepared MH agar was poured into plates labeled with appropriate concentrations. After agar solidification, each colistin agar plate of specific concentration was divided into six parts labeled with a marker to test up to 6 isolates per plate. Then 10μL of ten-fold dilution of McFarland inoculum, which was prepared as mentioned above, were streaked onto the appropriate portion of each colistin agar plate and incubated at 33- 35°C in ambient air for 16-20 hrs. MIC was read as the lowest agar concentration that completely inhibits the tested isolate’s growth (Figure 5).
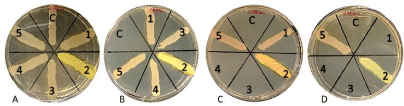
Figure 5: Colistin agar dilution test indicates the MIC for five isolates and a
susceptible Control (C) in four different concentrations. A) 0μg/mL, B) 1μg/
mL, C) 2μg/mL and D) 4μg/mL. Inhibition of the growth indicating the MIC of
the isolates. The plates results: C = susceptible control, 1 = resistance, 2 =
resistance, 3 = sensitive, “4 = sensitive,” 5 = resistance.
Micro-broth dilution (BMD)
This method involves using small volumes of MH broth distributed in sterile, round-bottom 96-plastic well plates described by [22,23]. Briefly, two-fold colistin serial dilutions (50μL each), starting with the lowest concentration of 0.125μg/mL to secondhighest concentration up to 250μg/mL, were added to the wells in each row. Thus, one plate contains seven samples, including E. coli ATCC 25922 as a quality control organism, while the last 8th row contains each tested isolate’s growth control (Figure 6). 50μL of the standardized inoculum (prepared as described above) was added to each well within 15 minutes of its preparation. The inoculated 96 well plates were incubated at 35±2°C for 16-18 hrs. MIC was recorded as the lowest colistin concentration that completely inhibits bacteria growth as detected by the unaided eye.
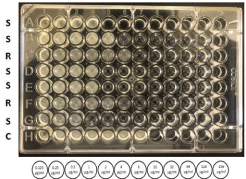
Figure 6: A plate showing MIC of Gram-negative bacilli to colistin by broth
microdilution method. The isolates from A to D are different strains of E. coli,
from E to G are different strains of K. pneumonia, and H is a growth control of
all tested isolates. S is sensitive (MIC ≤2μg/ml). R is resistance (>2 μg/ml). C
is 0 μg/mL (experimental control for all samples).
BD Phoenix ID/AST automated identification and susceptibility testing system
BD Phoenix™ M50 (New Jersey, US) was used to determine Antibiotic Susceptibility Testing (AST) following the manufacturer’s recommendations. Briefly, few overnight bacterial colonies were suspended in the provided BD phoenix broth to achieve an inoculum equivalent to 0.5 McFarland Standard. Next 25μl of this prepared inoculum solution was transferred to a tube containing Phoenix AST broth and a drop of the AST indicator solution. After sample preparation was completed, 4.5 mL was added to the BD phoenix Gram-negative panel. This panel has a wide range of two-fold doubling dilution of several antibiotics (including colistin) and one growth control well. The result was read within 16-18 hrs. The Phoenix system measures the MIC by microdilution and utilizes a redox indicator to detect organism growth in the presence of an antimicrobial agent. Organism identification is used in the interpretation of the MIC values of each antimicrobial agent.
DNA extraction, sequencing, and bioinformatics
According to the manufacturer’s protocol, genetic DNA was extracted from all isolates using a QIAamp® UCP Pathogen mini kit (Qiagen, Düsseldorf, Germany). Each isolate’s genomic DNA was purified and quantified using a Qubit dsDNA high sensitivity assay (Thermo Fisher, Waltham, United States). Then DNA was sent to BGI Genomics Co., Ltd., China, for whole-genome sequencing. The received raw sequence reads were subjected to a quality check and analyzed using CLC genomics workbench v20.0.4 (https:// digitalinsights.qiagen.com). Briefly, the reads were quality assessed, trimmed, followed by de novo assembly. Colistin resistance genes were identified using ResFinder v 4.1 [24] and the comprehensive antibiotic resistance database CARD [25].
Data analysis
The MIC and breakpoint results obtained from the examined methods were interpreted according to CLSI [19], NCCLS [20], Gales, et al. [21] guidelines and then compared to those obtained by the BMD reference method. A Very Major Error (VME) is defined as the error indicating a false susceptible result in the tested method and resistance in the BMD method’s reference method. In contrast, a Major Error (ME) denoted a false resistant result in the tested method and susceptible in the reference method. Categorical Agreement (CA) percentage is the percentage of isolates with the same results in both reference and test methods. Sensitivity, which measures the correctly identified positive portion, is characterized as true positive. On the other hand, specificity measures the correctly identified negative portion, characterized as true negative [26]. Cohen’s Kappa (CK) statistics measure inter-rater agreement for categorical items to check the test reliability. According to CK interpretation [27], the agreement level is none if Kappa value ranges between 0-0.20, minimal; if Kappa value (0.21-0.39), weak; if Kappa value (0.40-0.59), moderate; if Kappa value (0.60-0.79), strong; if Kappa value (0.80-0.9), and almost perfect if the Kappa value is above 0.90. The Positive Predictive Value (PPV) is the proportion of bacterial strains giving positive resistant test results, which are true resistant. On the other hand, the Negative Predictive Value (NPV) is the proportion of bacterial strains giving negative resistant test results, which are true sensitive as measured by BMD [26]. The sensitivity, specificity, PPV, NPV, CA, ME, VME, and CK of each test were calculated using the Equations in Supplementary Table S2, based on the BMD reference method.
Results
Overall, 43 (68%) of the studied Gram-negative isolates were resistant to colistin by gold standard Broth Microdilution (BMD) with resistance detected in 13 (72%) of K. pneumoniae, 2 (100%) P. aeruginosa, and 28 (65%) E. coli isolates (Table 1). Quality control findings were in line with published standards [19].
Organism
Number of isolates
Colistin MIC (μg/mL)
0.25
0.5
1
2
4
8
16
32
64
128
E. coli
43
5
10
3
15
9
1
K. pneumonia
18
1
4
1
1
1
3
4
3
P. aeruginosa
2
1
1
Total
63
0
0
6
14
5
17
10
4
4
3
Table 1: Colistin MIC distributions by broth microdilution for the 63 Gram-negative Bacilli isolates.
Comparison of Colistin disk diffusion to the reference BMD method
In all studied isolates, 95.24% (n=60) of the isolates were identified as resistant with disk diffusion compared to 68.25% (n=43) using the reference method BMD (Table 2, Supplementary Figure 1S(A)). The Very Major Error (VME) was not detected 0% (n=0), while 20 isolates showed 31.75% Major Error (ME) (Figure 7). Based on the isolates tested, the disk diffusion had a sensitivity of 100% and specificity of 0% for detecting colistin resistance with a Positive Predictive Value (PPV) of 68.25% (n=43) and a Negative Predictive Value (NPV) 0% (n=0). There was a 63.49% categorical agreement between the disk diffusion test and the BMD method. At the same time, the Kappa value was 0 (0%) (minimal agreement).
Test
Number/ percentage of resistance isolates (R)
Number/ percentage of susceptible isolates (S)
BMD
43 (68.25 %)
20 (31.75%)
Disk
60 (95.24%)
3 (4.76%)
E-test
38 (60.32%)
25 (39.68%)
ComASPTM Sensitest Colistin
42 (66.67%)
21 (33.33%)
Colistin Broth Disk Elution
40 (63.49%)
23 (36.51%)
Colistin Agar
37 (58.73%)
26 (41.27%)
Phoenix
42 (66.67%)
21 (33.33%)
Table 2: BMD, Disk, E-test, ComASPTM Sensitest Colistin, Colistin Broth Disk Elution, Colistin Agar, and Phoenix susceptibility results.
Comparison of E-test to the reference BMD method
There was a 92.06% categorical agreement between the E-test and the gold standard method BMD. E-test showed 60.3% (n=38) colistin-resistant with MICs of >4 μg/mL, whereas 39.68% (n=25) were colistin susceptible with MIC of ≤2 μg/mL. These results were compared to the reference method BMD (Table 2, Supplementary Figure 1S(B)). The test had 7.94% (n=5) VME and no ME 0% (Figure 7). The E-test had a sensitivity of 89.58% and a specificity of 100% in detecting colistin resistance, a positive predictive value of 100%, and a negative predictive value of 80%. The level of agreement by Kappa statistics was 0.83 (83%) (strong agreement).
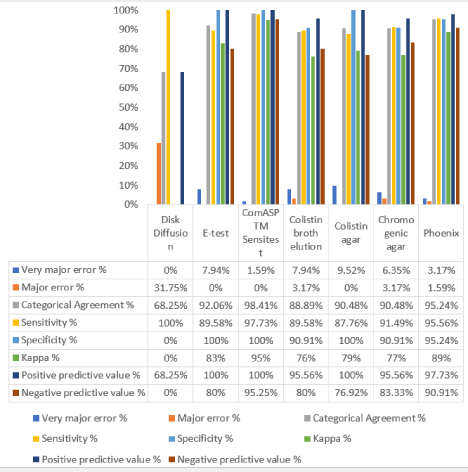
Figure 7: Comparative evaluation of seven colistin diagnostic tests by categorical agreement, Kappa, sensitivity, and specificity values compared to the goldstandard
method (BMD).
Comparison of ComASPTM sensitest colistin to reference BMD method
66.67% (n=42) from isolates were colistin-resistant, and 33.33% (n=21) were colistin susceptible using ComASPTM Sensitest colistin resistance detection method (Table 2, Supplementary Figure 1S(c)). The VME was 1.59% (n=1) while ME was 0% (Figure 7). The ComASPTM Sensitest Colistin had a sensitivity of 97.73% and specificity of 100% in detecting bacterial resistance to colistin with PPV of 100% and NPV of 95.24%. There was a CA 98.4% (n=62) between BMD and ComASPTM Sensitest colistin. (n=62). The Kappa value was 0.95 (95%), which shows an almost perfect agreement between the two methods.
Comparison of colistin broth disk elution to the reference BMD method
A total of 40 (63.49%) of the studied isolates were colistinresistant, and 23 (36.51%) were colistin susceptible, as shown in Table 2 and Supplementary Figure 1S(D). There was CA between Colistin Broth Disk Elution and BMD of 88.89% (n=56) (Figure 7). Five (7.94%) isolates were VME and 2 (3.17%) ME. The Colistin Broth Disk Elution test had a sensitivity of 89.58% and specificity of 90.91%. PPV of isolates was 95.56%, and NPV was 80%. This method had a Kappa value of 0.76 (76%), which shows a moderate level of agreement.
Comparison of Agar Dilution to reference BMD method
Overall, 58.7% (n=37) were colistin-resistant with MICs of >2.0 μg/mL. In comparison, 41.27% (n=26) were susceptible, as presented in (Table 2) and (Supplementary Figure 1S(E)). 9.52% (n= 6) of the isolates showed VME, and no ME was detected (Figure 7). The categorical agreement was 90.48%, and kappa statistics was 0.79 (79%). This test had a sensitivity of 87.76% and a specificity of 100%, with a 100% Positive Predictive Value (PPV) and 76.92% Negative Predictive Value (NPV).
Comparison of chromogenic agar to the reference BMD method
Chromogenic agar enhanced the growth of 41(65.08%) of isolates; E. coli = 26, K. pneumoniae = 13 and P. aeruginosa = 2. While it suppressed the growth of 22 (34.92%), E. coli = 17, and K. pneumoniae (Table 3, Supplementary Figure 1S(F)). The method had a sensitivity of 91.49% and a specificity of 90.91% (Figure 7). The CA between chromo-genic agar and BMD method was 90.48%. There were 4 (6.35%) VME and 2 (3.17%) ME. PPV was 95.56%, and NPV was 83.33%. Kappa value for chromogenic agar method showed a moderate level of agreement (0.77, 77%).
Bacteria
Resistance (Growth)
No. /percentageSusceptible (No growth)
No. /percentageE. coli
26 (41.27%)
17 (26.98%)
K. pneumonia
13 (20.63%)
5 (7.94%)
P. aeruginosa
2 (3.17%)
0 (0%)
Table 3: Chromogenic agar susceptibility results.
Comparison of Phoenix to reference BMD method
A total of 42 (66.67%) isolates were colistin-resistant, while 21 (33.33%) were colistin susceptible by Phoenix (Table 2, Supplementary Figure 1S(G)). There was a CA of 95.24% (n=60) between Phoenix and BMD (Figure 7). The VME was 3.17% (n=2), and the ME was 1.59% (n=1). Phoenix method had 95.56% sensitivity and 95.24% specificity, with PPV of 97.73% and NPV of 90.91%. The Kappa value was 0.89 (89%), indicating a strong level of agreement.
Genetic determinant of colistin resistance
As depicted in Table 4, a total of 43 isolates of E. coli were divided into four types based on genetic determinants of colistin resistance. Of these, 26 were plasmid resistance (harbor mcr1-1); 2 were chromosomal modifications; one isolate harbor mcr1-1 plus chromosomal modification, and 14 were sensitive, not presenting any colistin-resistant genetic determinant. Whereas K. pneumoniae has 11 isolates that contain chromosomal colistin resistance, 2 isolates harbored mcr1-1 plus chromosomal modifications, and 5 were sensitive. The resistance of the two isolates of P. aeruginosa was due to the chromosomal modifications.
Organism
Number of isolates
Colistin resistance type
mcr1-1
GMR
mcr1-1 & GMR
Not present
E. coli
43
26
2
1
14
K. pneumonia
18
0
11
2
5
P. aeruginosa
2
0
2
0
0
Total
63
26
15
3
19
Table 4: Genetic determinant of colistin resistance among 63 Gram-negative Bacilli isolates.
Discussion
The use of colistin to treat infections caused by multidrugresistant Gram-negative Bacilli, including E. coli, K. pneumoniae, and P. aeruginosa, is being increasingly described in many countries [28]. Additionally, antimicrobial susceptibility by phenotypic methods, including the colistin method, is currently the cornerstone of Antimicrobial Resistance (AMR) surveillance [17]. Nevertheless, susceptibility testing for colistin remains challenging mainly due to its intrinsic properties. Importantly, there is no agreement between colistin susceptibility testing methodologies [29,30]. The technique most frequently used in routine laboratories is disc diffusion, but it has been shown to be inefficient [17,29,31]. This study tested colistin resistance using seven methods and compared them to BMD as a gold standard, which is the most reliable method [17,19]. However, this method consumes time, effort, and money in routine medical laboratories [18]. Furthermore, the inclusion of representative strains, both susceptible and resistant, is critical in avoiding bias against a positive or negative outcome. Therefore, the strain panel used in this study was selected to ensure that both resistant and sensitive strains are included.
The most widely used method in routine laboratories, disk diffusion, is inefficient; due to weak colistin diffusion in Mueller Hinton agar [32]. Based on our findings, the disk diffusion method using a 10 μg/mL colistin disk was not a reliable method for colistin susceptibility testing with specificity 0% and Kappa value of 0 (0%) (Figure 7). These findings agree with several studies [31,33,34] reporting an inaccurate disk method for determining colistin susceptibility. In addition, an unacceptably high rate of very major errors ranging from 5% to 11% was recorded [31,33,34]. In our study, we did not report any very major errors (0%); however, we observed a high percentage of major errors (31.75%) with a low categorical agreement of 68.25% to the reference BMD method that supports the inaccuracy described by the previous studies.
Lo-Ten-Foe et al. [33]; Tan and Ng. [35]; Behera et al. [36] reported that E-test is a fast and reliable alternative method for determining colistin susceptibility. Our comparison portrayed a strong correlation (Kappa, 83% and Categorical agreement 92%) between broth microdilution and E-test. No false resistant (ME) results by E-test were detected, whereas a 7.94% false susceptible (VME) was exhibited (Figure 7). The E-test has been shown to have high concordance (>96%) in correlation with broth microdilution in previous studies [33,36]. In convergence with our findings, these studies reported one to four percentages of false susceptibility (VME) but no false resistant results (ME).
ComASPTM Sensitest Colistin is a modified BMD method but more straightforward with a shorter turnaround time, as different colistin concentrations are already prepared and exist in the test panels. The outcomes of this method were the most perfect compared to all other tested methods. Our data demonstrated a categorical agreement of 98.4% compared to BMD. Only one isolate was colistinresistant by BMD and susceptible by ComASPTM Sensitest, resulting in a VME value of 1.59%, and no ME was documented (Figure 7). The method has a high specificity of 100%, whereas sensitivity was 97.73%. A PPV and NPV of 100% and 95.24%% were recorded, respectively. The significantly high Kappa value (95%) of ComASPTM Sensitest proposed that this method can be an alternative to BMD. Thus, ComASPTM Sensitest can be accomplished faster with less effort and required accuracy in large laboratories with a high workload. Our findings were concordant with Osei Sekyere, et al. [37]; Carretto et al. [1].
Colistin broth disk elution endorsed by [19] has low cost and needs less effort. According to Simner et al. [38], colistin broth disk elution is a reproducible and accurate method and can alter BMD. Nevertheless, our test results revealed that the Categorical agreement between colistin broth disk elution and BMD is 88.89%. Five isolates with VME and 2 with ME. The method had a moderate agreement (Kappa value of (76%) compared to BMD. The dissimilarity in results could be ascribed to the use of disks produced by different manufacturers.
Colistin agar test was recently introduced to be superior to broth dilution methods in reproducibility and robustness [39] for determining colistin MICs. CLSI [19] lately recommended CAT, an adaptation of the agar dilution process, as a test for colistin resistance in Enterobacterales and P. aeruginosa. No ME was observed with the agar dilution method; however, 9.52% VME was detected mainly among E. coli isolates. Gonzales Escalante et al. [39] suggested adding an extra plate of colistin supplemented with EDTA, a well-known mcr1-1 inhibitor, to the CAT method; this was recently implemented for the precise detection of mcr1-1 producers. Numerous studies have demonstrated excellent agreement between colistin agar dilution and BMD [40,41]. Our study depicted a solid categorical agreement (90.48%) between the agar dilution method and the gold standard BMD (Figure 7).
In this study, we also used CHROMagarTM COL-APSE chromogenic agar to detect colistin resistance. Our numbers show specificity and sensitivity of 91.49% and 90.91%, respectively, whereas a recent study by Sekyere et al. [37] recorded a sensitivity and specificity of 82.05% and 66.67%. On the other hand, Abdul Momin et al. [4] reported specificity and sensitivity of 100% in their study. Sekyere et al. [37] evaluated the media’s performance using different amounts of cultured bacteria; this might be a constraint in our study that needs further investigation. It is remarkable to note that this media suppresses the growth of susceptible strains, but some grew on it, as demonstrated by our study and the previous studies. The CA was 90.48%, where six isolates had different results by the chromogenic agar and BMD. Four isolates showed VME, and two displayed ME. The Kappa value has a moderate level of agreement with 0.77 (77%). Therefore, it reduces the chance of choosing chromogenic agar as an alternative to BMD.
BD Phoenix™ M50. This method showed a CA of 95.24%, VME of 3.17%, and ME of 1.59%. Thus, the sensitivity and the specificity had almost the same values of 95.56% and 95.24%. In addition, the method had a Kappa value of 0.89 (89%), indicating a solid agreement level. In contrast, previous studies showed that Phoenix is a deprived method and has unacceptable and inaccurate results with low specificity and sensitivity [42,43]. Furthermore, a study done by Jayol, et al. [44] mentioned that the Phoenix system could detect plasmid-mediated colistin-resistance bacteria. Conversely, our study results indicated that Phoenix could detect both plasmid and gene-mediated colistin resistance, regardless of the resistance level.
To date, ten mobile mcr genes have been described [9,14,15]. In our study, we did Whole-Genome Sequencing (WGS) for 63 isolates. A high resistance rate to colistin antibiotic was mainly detected among E. coli isolates that harbor mcr1-1 type of genetic determinant of colistin resistance. While most of K. pneumoniae isolates had chromosomal colistin resistance mutation, as demonstrated by Table 4. Genotypic methods, in particular, are unlikely to detect all of the chromosomal mutations responsible for the majority of phenotypic colistin resistance in clinical screening settings [45]. Therefore, a negative PCR molecular test result cannot be used to predict colistin susceptibility because the test cannot rule out the existence of chromosomal mechanisms of resistance or even the presence of a novel mcr gene. To avoid this limitation, mcr genes and identified chromosomal mutations that cause colistin resistance will be possible with WGS. Although strains with novel resistance mechanisms will reduce the sensitivity and negative predictive value, WGS is the most detailed approach for detecting all currently known putative colistin resistance mechanisms. As new resistance mechanisms are described, it will also allow retrospective analysis of sequencing results. The agreement between phenotypic and genotypic test results will increase our understanding of colistin resistance mechanisms.
Conclusion
In conclusion, as we continue to spectate an increasing emergence of colistin resistance in Gram-negative multidrug-resistant bacteria, reliable and rapid colistin susceptibility testing is anticipated. This study found that disk diffusion is still an ineffective method for measuring colistin susceptibility in Gram-negative Bacilli. On the other hand, Phoenix and CompASPT SensiTest colistin methods have remained superior in reproducibility, sturdiness, and simplicity of use with a performance similar to the current recommended BMD procedure. These findings may rationalize and recommend validating these approaches using more samples and including other Gramnegative bacilli to develop a globally accepted consistent method for colistin susceptibility testing. If standardized, these methods can be executed in laboratories with a high workload and colistin resistance surveillance programs. However, our study did not ensure any relation between the type of colistin resistance genetic determinant (chromosomal/plasmid-mediated) and the performance of the specific colistin susceptibility test.
Acknowledgment
This study was supported by (UREP26-024-1-002) grant from the Qatar National Research Fund (QNRF), a member of the Qatar Foundation. The authors would like to thank Hana Abdelrahman and Lubna Abu rub for the technical assistance.
Funding
This research was supported by a grant (UREP26-024-1-002) from the Qatar National Research Fund (QNRF), a member of the Qatar Foundation.
Author Contributions
Conceptualization, N.E. methodology, N.S., H.A., N.M., M.R., K.A., K.A.; software, N.E, N.S and H.A.; validation, N.S. and H.A..; formal analysis, N.E.; investigation, N.E and S.Z .; resources, N.E.; data curation, N.E and S.Z.; writing-original draft preparation, N.S and H. A.; writing-review and editing, N.E. and S. Z; visualization, N.E and S. Z.; supervision, N.E.; project administration, N.E.; funding acquisition, N.E. All authors have read and agreed to the published version of the manuscript”.
References
- Carretto E, Brovarone F, Russello G, Nardini P, El-Bouseary MM, Aboklaish AF, et al. Clinical Validation of SensiTest Colistin, a Broth Microdilution-Based Method to Evaluate Colistin MICs. Journal of Clinical Microbiology. 2018; 56.
- Gwozdzinski K, Azarderakhsh S, Imirzalioglu C, Falgenhauer L, Chakraborty T. An Improved Medium for Colistin Susceptibility Testing. Journal of Clinical Microbiology. 2018; 56.
- Patricia M. Tille In: Bailey & Scott’s diagnostic microbiology. 14th ed. St. Louis, Missouri: Elsevier; 2017: 160-169.
- Abdul Momin MH, Bean DC, Hendriksen RS, Haenni M, Phee LM, Wareham DW. CHROMagar col-apse: A selective bacterial culture medium for the isolation and differentiation of colistin-resistant gram-negative pathogens. Journal of Medical Microbiology. 2017; 66: 1554-1561.
- McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, et al. Contribution of the PhoP-PhoQ and PmrA-PmrB Two-Component Regulatory Systems to Mg2+-Induced Gene Regulation in Pseudomonas aeruginosa. Journal of Bacteriology. 2006; 188: 3995-4006.
- Fernández L, Jenssen H, Bains M, Wiegand I, Gooderham WJ, Hancock RE. The Two-Component System CprRS Senses Cationic Peptides and Triggers Adaptive Resistance in Pseudomonas aeruginosa Independently of ParRS. Antimicrobial Agents and Chemotherapy. 2012; 56: 6212-6222.
- Ly NS, Yang J, Bulitta JB, Tsuji BT. Impact of Two-Component Regulatory Systems PhoP-PhoQ and PmrA-PmrB on Colistin Pharmacodynamics in Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 2012; 56: 3453-3456.
- Hu Y, Liu F, Lin IY, Gao GF, Zhu B. Dissemination of the mcr-1 colistin resistance gene. The Lancet Infectious Diseases. 2016; 16: 146-147.
- Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious Diseases. 2016; 16: 161-168.
- Gröndahl-Yli-Hannuksela K, Lönnqvist E, Kallonen T, Lindholm L, Jalava J, Rantakokko-Jalava K, et al. The first human report of mobile colistin resistance gene, mcr-1, in Finland. APMIS. 2018; 126: 413-417.
- Shen Z, Wang Y, Shen Y, Shen J, Wu C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. The Lancet Infectious Diseases. 2016; 16: 293.
- Zurfuh K, Poirel L, Nordmann P, Nüesch-Inderbinen M, Hächler H, Stephan R. Occurrence of the Plasmid-Borne mcr-1 Colistin Resistance Gene in Extended-Spectrum-β-Lactamase-Producing Enterobacteriaceae in River Water and Imported Vegetable Samples in Switzerland. Antimicrobial Agents and Chemotherapy. 2016; 60: 2594-2595.
- Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium. Eurosurveillance. 2016; 21.
- Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. MBio. 2019; 10.
- Gelbíčová T, Baráková A, Florianová M, Jamborová I, Zelendová M, Pospíšilová L, et al. Dissemination and Comparison of Genetic Determinants of mcr-Mediated Colistin Resistance in Enterobacteriaceae via Retailed Raw Meat Products. Frontiers in Microbiology. 2019; 10.
- Catchpole CR, Andrews JM, Brenwald N, Wise R. A reassessment of the in-vitro activity of colistin sulphomethate sodium. Journal of Antimicrobial Chemotherapy. 1997; 39: 255-260.
- WHO. The detection and reporting of colistin resistance. 2018.
- Sharma A, Agrawal M. Colistin Susceptibility for Carbapenem-Resistant Gram-Negative Bacilli; Comparative Study of E-test and Vitek 2 Compact™ with Broth Microdilution. Galore International Journal of Health Sciences and Research. 2019; 4: 110-115.
- CLSI. Performance standards for antimicrobial susceptibility testing. 30th. Wayne, PA: Clinical and Laboratory Standards Institute. 2020.
- Hutchens TT. NCCLS: objectives, organization, and activities. Pathologist. 1981; 35: 605-608.
- Gales AC, Reis AO, Jones RN. Contemporary Assessment of Antimicrobial Susceptibility Testing Methods for Polymyxin B and Colistin: Review of Available Interpretative Criteria and Quality Control Guidelines. Journal of Clinical Microbiology. 2001; 39: 183-190.
- CLSI. Performance standards for antimicrobial susceptibility testing. 26th. M100-S23. USA. 2013.
- CLSI. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 9th. M07-A9. Wayne, PA: Clinical and Laboratory Standards Institute, USA. 2012.
- Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. Journal of Antimicrobial Chemotherapy. 2020; 75: 3491-500.
- Alcock BP, Raphenya AR, Lau TT, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Research. 2020; 48: D517-D25.
- Trevethan R. Sensitivity, Specificity, and Predictive Values: Foundations, Pliabilities, and Pitfalls in Research and Practice. Frontiers in Public Health. 2017; 5.
- McHugh ML. Interrater reliability: the kappa statistic. Biochemia Medica. 2012; 22: 276-282.
- Biswas S, Brunel J-M, Dubus J-C, Reynaud-Gaubert M, Rolain J-M. Colistin: an update on the antibiotic of the 21st century. Expert Review of Anti-infective Therapy. 2012; 10: 917-934.
- CLSI. Performance standards for antimicrobial susceptibility testing. 27th. M100S. Wayne, PA: Clinical and Laboratory Standards Institute. 2017.
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. 2017.
- Tan TY. Comparison of three standardized disc susceptibility testing methods for colistin. Journal of Antimicrobial Chemotherapy. 2006; 58: 864-867.
- Luis Esaú Ló-J, Christian Rodolfo R-G, Melissa He-D, Claudia Adriana C-C, Rodolfo G-C, Rafael F-C. An alternative disk diffusion test in broth and macrodilution method for colistin susceptibility in Enterobacteriales. Journal of Microbiological Methods. 2019.
- Lo-Ten-Foe JR, de Smet AM, Diederen BM, Kluytmans JA, van Keulen PH. Comparative Evaluation of the VITEK 2, Disk Diffusion, Etest, Broth Microdilution, and Agar Dilution Susceptibility Testing Methods for Colistin in Clinical Isolates, Including Heteroresistant Enterobacter cloacae and Acinetobacter baumannii Strains. Antimicrobial Agents and Chemotherapy. 2007; 51: 3726-3730.
- Galani I, Kontopidou F, Souli M, Rekatsina P-D, Koratzanis E, Deliolanis J, et al. Colistin susceptibility testing by Etest and disk diffusion methods. International Journal of Antimicrobial Agents. 2008; 31: 434-439.
- Tan TY, Ng SY. Comparison of Etest, Vitek and agar dilution for susceptibility testing of colistin. Clinical Microbiology and Infection. 2007; 13: 541-544.
- Behera B, Mathur P, Das A, Kapil A, Gupta B, Bhoi S, et al. Evaluation of susceptibility testing methods for polymyxin. International Journal of Infectious Diseases. 2010; 14.
- Osei Sekyere J, Sephofane AK, Mbelle NM. Comparative Evaluation of CHROMagar COL-APSE, MicroScan Walkaway, ComASP Colistin, and Colistin MAC Test in Detecting Colistin-resistant Gram-Negative Bacteria. Scientific Reports. 2020; 10.
- Simner PJ, Bergman Y, Trejo M, Roberts AA, Marayan R, Tekle T, et al. Two- Site Evaluation of the Colistin Broth Disk Elution Test to Determine Colistin In Vitro Activity against Gram-Negative Bacilli. Journal of Clinical Microbiology. 2019; 57.
- Gonzales Escalante E, Yauri Condor K, Di Conza JA, Gutkind GO. Phenotypic Detection of Plasmid-Mediated Colistin Resistance in Enterobacteriaceae. Journal of Clinical Microbiology. 2019; 58.
- Moskowitz SM, Garber E, Chen Y, Clock SA, Tabibi S, Miller AK, et al. Colistin susceptibility testing: evaluation of reliability for cystic fibrosis isolates of Pseudomonas aeruginosa and Stenotrophomonas maltophilia. Journal of Antimicrobial Chemotherapy. 2010; 65: 1416-1423.
- Turlej-Rogacka A, Xavier BB, Janssens L. et al. Evaluation of colistin stability in agar and comparison of four methods for MIC testing of colistin. European Journal of Clinical Microbiology & Infectious Diseases: Official Publication of The European Society of Clinical Microbiology. 2018; 37.
- Pfennigwerth N, Kaminski A, Korte-Berwanger M, Pfeifer Y, Simon M, Werner G, et al. Evaluation of six commercial products for colistin susceptibility testing in Enterobacterales. Clinical Microbiology and Infection. 2019; 25: 1385-1389.
- Vourli S, Dafopoulou K, Vrioni G, Tsakris A, Pournaras S. Evaluation of two automated systems for colistin susceptibility testing of carbapenemresistant Acinetobacter baumannii clinical isolates. Journal of Antimicrobial Chemotherapy. 2017; 72: 2528-2530.
- Jayol A, Nordmann P, Lehours P, Poirel L, Dubois V. Comparison of methods for detection of plasmid-mediated and chromosomally encoded colistin resistance in Enterobacteriaceae. Clinical Microbiology and Infection. 2018; 24: 175-179.
- Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Frontiers in Microbiology. 2014; 5.