
Research Article
J Bacteriol Mycol. 2022; 9(1): 1193.
Prevalence of Multidrug-Resistant Pseudomonas aeruginosa and Risk Factors for their Infections at Intensive Care Units of a Tertiary Hospital in Southern China
Liu J1,4,5, Guo H-W4, Pan Q3, Fu M-Z4, Qiu Y-K4, Wong N-K2 and Huang Y-C4*
1Department of Clinical Laboratory, The First Affiliated Hospital of Hunan University of Medicine, China
2Department of Infection Diseases, Shenzhen Third People’s Hospital, The Second Hospital Affiliated to Southern University of Science and Technology, China
3Shenzhen Key Laboratory of Microbial Genetic Engineering, College of Life Sciences and Oceanology, Shenzhen University, China
4Microbiology Division, Department of Clinical Laboratory, The First Affiliated Hospital of Shantou University Medical College, China
5Department of Clinical Laboratory, Huaihua First People’s Hospital, Huaihua Hospital Affiliated to Nanhua University, China
*Corresponding author: Yuan-Chun Huang, Department of Clinical Laboratory, The First Affiliated Hospital of Shantou University Medical College, No. 57, Changping Road, Shantou, Guangdong, China
Received: December 20, 2021; Accepted: January 24, 2022; Published: January 31, 2022
Abstract
Pseudomonas aeruginosa (PA) is highly significant opportunistic pathogens causing healthcare associated infections (HAIs) in hospital settings, notably at intensive care units (ICUs). The aim of this study was to retrospectively analyze the infection status, prevalence and antimicrobial resistance (AMR) of PA at ICUs of a tertiary care hospital in southern China during a one-year period (2016) and examine the clinical risk factors for HAIs by PA. Multiple-locus variablenumber tandem-repeat (VNTR) analysis (MLVA) method was employed to analyze clonality of the strains. Our results suggested that the resistance of PA in ICUs were higher than in other wards. In terms of resistance to carbapenems, the resistance gene island (blaOXA-1+blaIMP+ant(2’’)-Ia+aac(6’)-Ib) carried in IntI was a salient feature among AMR genes. While PA infections at local ICUs seemed frequent, there were no obvious trends suggestive of outbreaks. Some epidemic strains have apparently thrived locally for substantial periods, as carriers of major AMR genes and virulence factors. For risk factors for HAIs, inappropriate treatment was found to impact empiric antibiotic therapy of PA infections, especially in the case of carbapenems, where patients often did not get proper treatment during hospitalization of more than 30 days. Multifactor analysis shows that ventilator-associated pneumonia (VAP) was an independent risk factor for increasing the 30-day mortality rate in patients. In addition, the use of antimicrobials, duration of hospitalization and use of mechanical ventilation before isolation were independent risk factors for HAIs.
Keywords: Pseudomunas aeruginosa; Antimicrobial resistance; Virluence; Clinical risk factors; Intensive care units (ICUs)
Abbreviations
PA: Pseudomunas aeruginosa; HAIs: Healthcare-Associated Infection; ICUs: Intensive Care Units; MDR PA: Multidrug- Resistant PA; AMR: Antimicrobial Resistance; TTSS: The Type III Secretion System; MV: Mechanical Ventilation; CLSI: The Clinical and Laboratory Standard Institute; VNTR: Variable-Number Tandem-Repeat; MLVA: Multiple-Locus Tandem-Repeat; UPGMA: Unweighted Pair Group Method with Arithmetic Mean; SD: Standard Deviation; HCAI: Healthcare Associated Infection; VAP: Ventilator-Associated Pneumonia
Introduction
Pseudomunas aeruginosa (PA) represents an important cause of healthcare-associated infection (HAIs) in intensive care units (ICUs) [1,2]. Owing to its extraordinary ability to form biofilm and efficiently develop resistance towards broad-spectrum antibiotics, PA has contributed to significant mortality and morbidity in HAIs and thus a heavy burden to health care systems in developed and developing countries alike, including China [3].
Prevalence of multidrug-resistant PA (MDR PA) is on the rise across the globe, with various mechanisms being attributed to the development of antimicrobial resistance (AMR) in MDR PA. The prevalence rates of MDR PA range between 15% and 30%in some geographical areas [4,5]. Of note, the genes of ant(2”)-Ia and aac(6’)- Ib carried by PA lead to increased aminoglycoside resistance [6], while the class B enzymes MBL (IMP) and class D OXA beta-lactamases were the most common ESBLs reported in PA [4,7]. OprD2 protein forms part of the specific pathway for imipenem to enter into PA, and it has ligand specificity with loci specific binding of the imipenem [8]. Among the multitude of virulence determinants of PA, the type III secretion system (TTSS) has been identified as an important contributor to cytotoxicity and PA invasion during infections [9]. TTSS occurs as four cytotoxin genes (exoS, exoU, exoY and exoT), among which the impact of exoS and exoU on pathogen virulence is deemed crucial, whereas exoY and exoT supposedly have minor effects on virulence. The toxA gene is reputedly a principal virulence factor of this bacterium with ADP-ribosylation activity that could halt host protein synthesis and eventually lead to cell death [10]. The frequency of both toxA & exoS genes has been reported to be significantly higher in MDR PA strains isolates from patients with burnt injuries [11]. Additionally, genes carried by integrons usually encode molecules that mediate a variety of resistance mechanisms. Among integrons found in clinically important Gram negative bacteria such as PA, class 1 integron is most common [12].
In terms of risk factors for HAIs, over-prescription and inappropriate use of antimicrobials in the hospital environment clearly drive the development of antibiotic resistance [13]. Inappropriate empiric antimicrobial therapy adversely affected the outcomes of in patients diagnosed with PA infections [14,15]. In this study, we investigated the prevalence of drug-resistant PA in a coastal region (Chaoshan) of southern China, focusing on carriage status of AMR genes and virulence factors, and related clinical data including risk factors of HAIs with PA.
Materials and Methods
Patients and research settings
Databases at the Microbiology Division, Department of Clinical Laboratory, The First Affiliated Hospital of Shantou University Medical College, Shantou City, Guangdong, China, were reviewed to identify patients with PA infections in three types of intensive care units (namely, comprehensive, cardiovascular, and neurosurgery ICUs) within the period of January to December 2016. For patients with multiple episodes of PA infections, only first episodes were analyzed.
Ethical approval
The ethics committee on medical research of the First Affiliated Hospital of Shantou University had evaluated and approved the experimental design of this study.
Study design and clinical data collection
This study was designed as a retrospective study aiming to determine the prevalence, virulence genes and resistance genes in PA isolates as well as PA infection rates, based on data collected by the Department of Clinical Laboratory. Furthermore, we analyzed the impact of inappropriate therapy on patients with PA infections at the ICUs. The main outcome was patient mortality, measured on the basis of 30-day mortality rates. We also assessed secondary outcomes, including the duration of hospitalization and use of invasive procedures. For each patient studied, the following characteristics were recovered from their clinical records: age, gender, date of hospital admission, treatment outcomes as discharge or death (within 30 days from admission), length of total hospital stay, surgery, invasive procedures such as mechanical ventilation (MV), central venous line, urinary catheter, tracheostomy, haemodialysis, catheter enteral or gastric nutrition, and surgical drain during the current hospitalization, underlying conditions such as diabetes mellitus, chronic renal failure, heart failure and cancer, sources of infection, antibiotic use during the current hospitalization, and cases of inappropriate antimicrobial therapies. Antimicrobial therapy was considered to be “appropriate” if the initial antimicrobials, which were administered within 24 hours of acquisition of a culture sample, included at least one antibiotic that was active in vitro. As a universal consensus considered to be lacking, the definition of antibiotic appropriateness used in this study relies on the authoritative guidelines and previous works elsewhere [16].
Identification of isolates and antimicrobial susceptibility testing
A total of 70 non-duplicated strains of PA were used in this study. All isolates were identified by the VITEK 2 COMPACT system (BioMérieux, France) and antimicrobial susceptibility tests were performed with its assemble kit of AST 09 card with the following antibiotics: aminoglycosides (gentamicin, amikacin, tobramycin), carbapenems (imipenem, meropenem), cephalosporins (ceftazidime, cefepime), fluoroquinolones (ciprofloxacin, levofloxacin), penicillins plus Β-lactamase inhibitors (piperacillin-tazobactam), monobactams (aztreonam). PA strains that showed intermediate susceptibility were considered to be resistant. Quality-control protocols were used according to the 2016 guidelines of the Clinical and Laboratory Standard Institute (CLSI). PA ATCC 27853 was used as a quality control strain.
Characterization of drug resistance phenotypes and genotypes
The presence of virulence genes (including exoS, exoU, toxA), class I integron gene, aminoglycoside resistance genes (including ant(2’’)-Ia, aac(6’)-Ib) and Β-lactamase genes (including IMP, OXA, OprD2) were tested by PCR. All primers were based on previously published works as summarized in Table 1. The amplified gene products were sequenced by Sanger method and compared to the sequences deposited in GenBank (http://www.ncbi.nlm.nih.gov/ gene/).
Target
Nucleotide sequence (5'→3')
Size (bp)
Source
IMP
GGAATAGAGTGGCTTAATTCTC
188
[4]
CCAAACCACTACGTTATCT
OXA
ACACAATACATATCAACTTCGC
813
[7]
AGTGTGTTTAGAATGGTGATC
OprD2
GCGCATCTCCAAGACCATG
193
[8]
GCCACGCGATTTGACGGAG
ant(2”)-Ia
TCCAGAACCTTGACCGAAC
700
[6]
GCAAGACCTCAACCTTTTCC
aac(6’)-Ib
GCTCTTGGAAGCGGGGACGG
300
[6]
TCGCTCGAATGCCTGGCGTG
exoS
TCAGGTACCCGGCATTCACTACGCGG
572
[9]
TCACTGCAGGTTCGTGACGTCTTTCTTTTA
exoU
CCTTAGCCATCTCAACGGTAGTC
911
[9]
GAGGGCGAAGCTGGGGAGGTA
toxA
GGTAACCAGCTCAGCCACAT
352
[9]
TGATGTCCAGGTCATGCTTC
IntI
AGTCAGCGGCTTAGATA
457
[12]
GGTGTGGCGGGCTTCGT
Table 1: Specific primers used for PCR.
Multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA)
MLVA analysis was performed to investigate the clonal relationship between isolated PA strains. Gene amplification in MLVA was based on published primers [17] for the following variablenumber- of-tandem-repeats (VNTRs): ms142, ms211, ms212, ms213, ms215, ms217, ms222 for amplifying random DNA fragments. In the clustering analysis, by applying the categorical coefficient (also called Hamming’s distance), this corresponds to an interval of 85 to 100% similarity and unweighted pair group method with arithmetic mean (UPGMA) clustering approaches within BioNumerics. A cutoff value of 50% similarity was applied to define MLVA clusters. Lineages had been arbitrarily numbered according to the order which they were listed in the clustering analysis.
Statistical analysis
Results pertaining to patients’ clinical characteristics and drug resistance were expressed as a percentage of samples. Statistical differences among groups were using the Χ² test with SPSS v.22.0 (SPSS Inc., Chicago, IL). Duration of hospitalization was analyzed by using the Mann–Whitney U-test. Multivariate analysis with binary logistic regression was conducted to examine the associations of risk factors with PA resistance, with control for potential confounders. In addition to duration of hospitalization, all variables with a P-value of <0.05 in the univariate analysis were included in the logistic regression model. A two-tailed P-value of <0.05 was considered to be statistically significant. After establishing the production of some virulence traits and resistance gene geneotypes in different clinical strains of PA, we analyzed the possible correlation between them.
Results and Discussion
Sites of isolation
These 70 strains of PA were isolated from ICUs in 2016, including sputum (66 isolates; 95.78%), blood (2 isolates; 2.11%) and pus (2 isolates; 2.11%), which showed that respiratory infections were the predominant type in PA infections.
Antimicrobial susceptibility testing
The drug susceptibility rates of the isolates were indicated in Table 2. Among 70 P. aeruginosa isolates, 36 (51.43%) and 32 (45.71%) isolates were resistant to imipenem and meropenem, respectively. Thus, a total of 37 isolates (52.86%) resistant to imipenem and/ or meropenem were determined to be resistant to carbapenems. 31(83.78%) of the 37 carbapenem-resistant isolates showed multidrug resistance and 25 isolates were resistant to all antimicrobials tested except polymyxins B. Among 33 carbapenem-susceptible isolates, only 4 isolates (12.12%) were MDR and just one was the XDR PA, which was significantly lower than that among carbapenem-resistant isolates (P < 0.001).
Antimicrobial
Sensitive
Intermediate
Resistant
Piperacillin
50.00% (35/70)
11.43% (8/70)
38.57% (27/70)
Piperacillin/tazobactam
52.86% (37/70)
10.00% (7/70)
37.14% (26/70)
Aztreonam
44.29% (31/70)
14.29% (10/70)
41.43% (29/70)
Ceftazidime
58.57% (41/70)
1.43% (1/70)
40.00% (28/70)
Cefepime
60.00% (42/70)
2.86% (2/70)
37.14% (26/70)
Imipenem
48.57% (34/70)
22.86% (16/70)
28.57% (20/70)
Meropenem
54.29% (38/70)
4.29% (3/70)
41.43% (29/70)
Ciprofloxacin
52.86% (37/70)
5.71% (4/70)
41.43% (29/70)
Levofloxacin
58.57% (41/70)
5.71% (4/70)
35.71% (25/70)
Amikacin
68.57% (48/70)
2.86% (2/70)
28.57% (20/70)
Gentamicin
65.71% (46/70)
1.43% (1/70)
32.86% (23/70)
Tobramycin
67.14% (47/70)
1.43% (1/70)
31.43% (22/70)
Polymyxin B
100.00% (70/70)
0.00% (0/70)
0.00% (0/70)
Table 2: Antimicrobial resistance rates among P. aeruginosa isolates.
Overall, Susceptibility rates to piperacillin/tazobactam, ceftazidime were 52.86% and 58.57% , respectively, higher than those reported previously [18]. The polymyxins B remained active against all these 70 isolates. Antimicrobial susceptibility test revealed that 35(50.00%) PA isolates were MDR PA, among these, 26 isolates (37.14%) were XDRPA. Overall, the correlation could be established between the resistance profiles and the different MLVA genotypes.
Prevalence of resistance genes and class I integrons
All the resistant genes designed in the study and class I integrons was found in the experimental isolates.
19 isolates (27.14%) were simultaneously positive for both OXA- 1, IMP, ant(2’’)-Ia, aac(6’)-Ib and IntI. The isolates with these genes were all the MDR PA, and among these, 18 isolates were XDR PA. Of these, 15 belonged to the same genotype in MLVA typing. PCR assay with primers revealed that 20 (28.57%) of the 70 P. aeruginosa strains were class I integrase positive. Analysis of the integrase PCR product by the test of gene sequencing confirmed class I integron as described previously [19]. The class I integron positive strains of multidrug resistant rate was 38.00% (19/50) and the negative was 14% (7/50), there were significant difference between the two groups (Χ²=40.146, P=0.000). The detection rate of the resistant gene of OprD2 was 64.29% (45/70). But the detection of OprD2 gene deletion strains in this study did not produce significant relationship with the imipenem resistance.
Among the 70 isolates tested, 37 genotypes were identified by the MLVA-PCR; and they were divided into 6 gene clusters (1-6). Cluster 4 included 22 isolates from different patients, which was the main clone type in this study. Out of them 15 isolates harboured IntI, aac(6’)-Ib, ant(2’’)-Ia and OXA-1, IMP.
Detection of the virluence genes
According to PCR, the T3SS effector genes contained in almost each clinical isolate, except for 4 isolates. The amplification of exoS and exoU were more variable with only 1(1.43%) isolates containing both genes. There were 49(70.00%) isolates carried exoS and 18(25.71%) carried exoU. This enabled the strains to be split into three major groups exoS+/exoU-, exoS-/exoU+ and exoS+/exoU+. The toxA (90.0%) was present in almost all isolates. Carriage both toxA and exoS genes were observed in 27 among 35 MDR strains. Meanwhile, there were only 6 strains harboring toxA and exoU (P<0.001), and 2 strains had none of them.
Analyzing the association of exoU or exoS with antimicrobials resistance, there was no significant difference in sensitivity to antimicrobials between the two groups. However, in the 19 strains of PA with resistance genes of OXA-1, IMP and aac(6’)-Ib, ant(2’’)- Ia, there were 17 isolates in exoS+/exoU- mode, but other 2 strains in exoS-/exoU+ mode were with aac(6’)-Ib gene positive (P<0.001).
Molecular epidemiology
All isolates were distinguished by 37 different genotypes, 6 clusters by MLVA method (Figure 1). The largest cluster contained 22 isolates but only included genotypes 4, of which 15 isolates carried all the resistance genes (included OXA-1, IMP, and aac(6’)-Ib, ant(2’’)- Ia) and IntI detected, harboring virulence genotype exoS. Most of the strains carrying multidrug resistance genes belonged to the same genotypes. Among the 6 different clusters, the strains of toxA and exoS were detected more in the cluster 4, and were statistically significant compared with other clusters (Χ²=13.555, P=0.019; Χ²=21.222, P=0.001). The strains of exoU were carried more in the cluster 2, which had statistical significance compared with other clusters (Χ²=25.10, P=0.000). In conclusion, there was a significant correlation between the drug-resistant genotypes and the virulence genotypes with the MLVA genotypes.
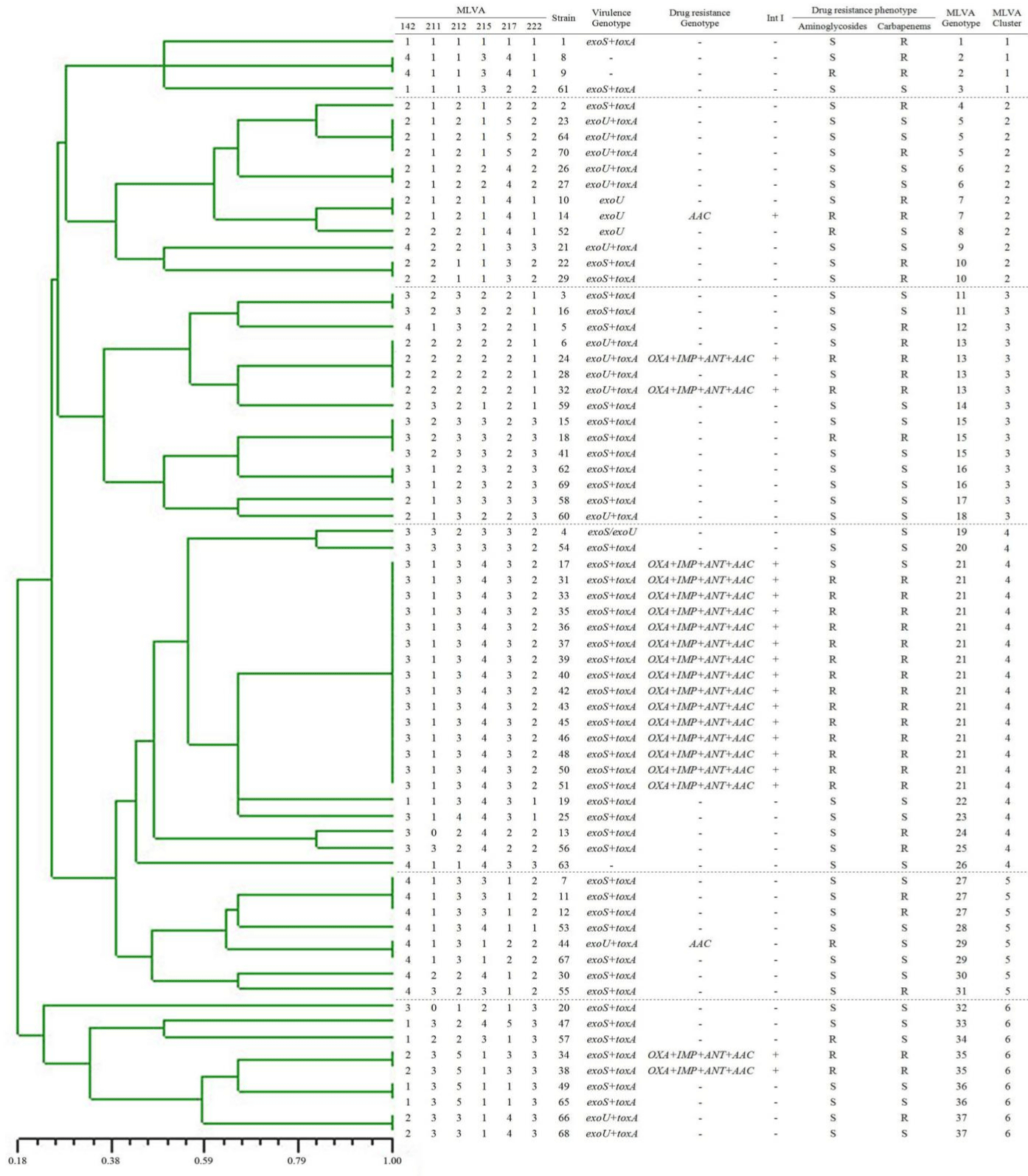
Figure 1: MLVA Genotypic features of 70 Pseudomonas aeruginosa isolates. OXA: blaOXA-1; IMP: blaIMP; ANT: ant(2’’)-Ia; AAC: ant(6’)-Ia.
Clinical data
A total of 70 nonrepetitive patients in ICUs with PA infection were included in the study. The detailed information on factors associated with infection and the relevant demography and clinical characteristics of the study population were summarized in Table 3. Compared with the cohort of patients of the ICU, the mean age (±standard deviation [SD]) of the patients was 55.81±19.97 years, and 55(78.57%) patients were male. Of the 70 patients, 45(64.29%) had healthcare associated infection and the remaining 25 (35.71%) had community infections. The most common underlying diseases were cardiovascular disease (n=35, 50.00%). Comparing with the community infection, patients with healthcare associated infection (HCAI) prone to have a high drug resistance rate, and 40% of the strains were MDR, and even 46.67% of the strains were XDR (Table 4). The healthcare associated infection also had a higher proportion of ventilator-associated pneumonia (VAP). However, patients in ICU with HCAI would often receive empirical therapy before isolation, thus the difference of three kinds of antimicrobials use prior to separation was statistically significant. This also lead to a statistically significant increase in the proportion of patients with healthcare associated infection receiving inappropriate therapy than community infection. The hospitalization time of the patients with healthcare associated infection was significantly longer than that of the patients in the community.
Variable
n
Percentage (%)
Gender
Female
15
21.43%
Male
55
78.57%
Age group
<18y
5
7.14%
18-60y
35
50.00%
>60y
30
42.86%
Length of hospital stay (mean days)
=10
8
11.43%
20-Oct
7
10.00%
=20
55
78.57%
Invasive procedures
Mechanical Ventilation
57
81.43%
Tracheostomy
4
5.71%
Central venous catheter
70
100.00%
Surgical drain
17
24.29%
Haemodialysis
2
2.86%
Co-morbidity conditions
41
58.57%
Heart failure
35
50.00%
Cancer
3
4.29%
Diabetes mellitus
10
14.29%
Chronic renal failure
2
2.86%
COPD
8
11.43%
VAP
32
45.71%
Inappropriate therapy
35
50.00%
Antibiotic use prior to separation
42
60.00%
one kind
9
12.86%
two kinds
12
17.14%
three kinds
21
30.00%
Nosocomial infection
45
64.29%
Community infection
25
35.71%
Table 3: Risk factors associated with infection caused by P. aeruginosa in ICUs.
Variable
total, n=70
The HCAI, n=45
The community infection, n=25
P
Drug resistance phenotype
Non-MDR
35(50.00%)
18(40.00%)
17(68.00%)
0.025
MDR
35(50.00%)
27(60.00%)
8(32.00%)
0.873
XDR
26(37.14%)
21(46.67%)
5(20.00%)
0.027
Antibiotic use prior to separation
42(60.00%)
34(75.56%)
8(32.00%)
<0.001
one kind
9(12.86%)
7(15.56%)
2(8.00%)
0.366
two kinds
12(17.14%)
9(20.00%)
3(12.00%)
0.395
three kinds
21(30.00%)
18(40.00%)
3(12.00%)
0.014
Co-morbidity conditions
41(58.57%)
26(57.78%)
15(60.00%)
0.856
Charlson Comorbidity Index (median, IQR)
2.54±2.14
2.64±2.14
2.36±2.16
0.597
Surgery
17(24.29%)
11(24.44%)
6(24.00%)
0.967
Inappropriate therapy
35(50.00%)
27(60.00%)
8(32.00%)
0.025
VAP
32(45.71%)
26(57.78%)
6(24.00%)
0.007
Length of hospital stay (mean days)
56.96±49.03
68.36±50.61
36.44±39.15
0.008
Table 4: Risk factors between HCAI and community infection.
Of the 70 patients with PA infection, 35 (50.00%) patients received inappropriate antimicrobial therapy. To compare the differences between the groups receiving inappropriate and appropriate antimicrobials, their clinical characteristics and the characteristics of the isolated strains’ resistance were shown in Table 5. Regarding to underlying diseases and comorbid conditions, the inappropriate therapy group showed significant associations with the prior hospitalization within 30 days (all P<0.05). Antimicrobials used prior to separation were more frequent in the inappropriate therapy group than they were in the appropriate therapy group (P<0.05). The significant differences were also found with regard to the onset infection between the two groups. When assessing the clinical outcomes of PA infection, the failure rate of inappropriate experience treatment was 37.14% (13/35), and the overall 30-day mortality rate was 34.29% (24/70). The failure rate after empirical antimicrobial treatment was higher in the inappropriate treatment group than the appropriate treatment group (31.43%, 11/35). No significant difference in the 30-day mortality rates between the inappropriate therapy group and the appropriate therapy group (P=0.615). The multinominal logistic regression analysis showed that the use of antimicrobials before separation, prolonged hospitalization days and the use of mechanical ventilation were the independent risk factors of healthcare associated infection. There was a significant correlation between ventilator-associated pneumonia with mortality within 30 days.
Variable
Inappropriate therapy (n=35)
Appropriate therapy (n=35)
P
Age (Year) (mean ± standard deviation)
57.29±19.62
54.34±20.50
0.542
Sex (Male/Female)
27/8
28/7
0.771
Charlson Comorbidity Index (median, IQR)
2.46±2.01
2.63±2.29
0.319
Underlying disease
21(60.00%)
20(57.14%)
0.808
Cardiac disease
19(54.29%)
16(45.71%)
0.473
Liver disease
1
1
-
Renal disease
1
1
-
Respiratory disease
2(5.71%)
6(17.14%)
0.133
Diabetes mellitus
7(20.0%)
3(8.57%)
0.172
Comorbid conditions
Receipt of recent operation
8(22.86%)
9(25.71%)
0.78
Prior hospitalization within 30 days
21(60.00%)
4(11.43%)
0
Antibiotic use prior to separation
26(74.29%)
16(45.71%)
0.015
Polymicrobial infection
22(62.86%)
16(45.72%)
0.15
Indwelling urinary catheters
-
-
-
Central venous catheterization
-
-
-
Invasive procedure
27 (77.14%)
30 (85.71%)
0.356
Onset of infection
Community-acquired
8(22.86%)
17(48.57%)
0.025
HCAI
27/35(77.14%)
18/35(51.43%)
0.025
VAP
18/35(51.43%)
14/35(40.0%)
0.337
Severity of illness
APACHE II Score
21.06±8.61
20.66±11.11
0.387
Table 5: Statistical analysis of clinical factors between inappropriate and appropriate antimicrobials therapy groups.
In our study, carbapenems was the priority in the use of inappropriate treatment accounting for 51.43%, especially for MEM, followed by quinolones (45.71%) Β-lactamase inhibitors (34.29%), amide enzyme inhibitors (17.14%), cephalosporins (5.71%). The average LOS Length of hospital stay (mean days) in the group receiving an inappropriate therapy (68.86 days±54.60 SD) was greater than the one in the group that received an appropriate therapy (45.06 days±40.07 SD), and the difference has statistically significant (p = 0.041).
Discussion
PA is an opportunistic human pathogen capable in causing severe infections, especially in ICUs [2,13]. CHINET surveillance of bacterial resistance across China in 2016 showed that the drug resistance rates of PA to imipenem and meropenem were 28.7% and 25.3%, respectively; the drug resistance rates to polymyxin B and amikacin were 0.5% and 8.1%, respectively; the drug resistance rate of the two enzyme inhibitor mixture, gentamicin, ciprofloxacin, ceftazidime, cefepime and piperacillin were less than 20% [18]. Compared with the results in our study, the resistance rates of most drugs were greater than the 2016 China CHINET bacterial resistance monitoring results, except for the slightly lower resistance rate of imipenem and polymyxin B. Of course, this may be related to the fact that the strains we isolated were from the patients in ICUs. It indicated the severity of the antimicrobial resistance rate of PA in ICUs of our study.
The data emphasized the importance of establishing local monitoring for local antimicrobials guide and supported the best treatment. The polymyxin B had the lowest resistance rate in our study, followed by imipenem and aminoglycosides. However, because of the limited clinical application of polymyxin B, imipenem or aminoglycosides were preferred for empirical use, but for patients with severe infections or XDRPA infections, polymyxin B may be considered.
Our results showed considerable genetic variability among the 70 strains, with the detection of 37 distinct genetypes (0.946 of polymorphisms). The genotype cluster 4 contained IntI, OXA-1, IMP and aac(6’)-Ib, ant(2’’)-Ia, which suggested that this specific clone was endemic to the hospital. As a result of persistence over sustained periods, it had become more resistant to antimicrobials. It was speculated that OXA-1+IMP+ant (2 ‘) -Ia+aac (6’) -Ib+IntI was the main resistance gene pattern in 2016. To the best of our knowledge, IntI was closely related to a variety of drug resistance, carrying related drug-resistant gene cassettes that might lead to transmission of resistance as often detected in clinical isolates of PA [12]. Based on previous reports that IntI carried related resistance gene cassettes [20], we speculated that the main resistance gene pattern (OXA- 1+IMP+ant (2 ‘) -Ia+aac (6’) -Ib+IntI) in this study might also be related to the relevant resistance gene cassette carried by the IntI, which had not been reported yet. However, the lack of sequencing of related gene cassettes and IntI was not enough, thus despite this speculation, there remains questions to be clarified in future studies. The IntI might be the main genetic elements of the global resistance transmission of PA. If there is no proper and timely monitoring, the spread of the IntI gene, which may be the main genetic elements of resistance global communication about PA [21], will inevitably complicate treatment of HAIs by MDR PA. Therefore, there is an urgent need not only to rationalize the use of antimicrobials, but also to monitor the dissemination and possible mutation of related resistance genes at the same time. In addition, it is known that the mutation of OprD2 inactivation has become the main mechanism of imipenem resistance [8], but the detection of OprD2 gene deletion strains in this study did not suggest significant relationship with the imipenem resistance.
In the 70 strains of PA clinical isolates, compared with the prevelance of the exoS gene (49/70, 70.00%), the detection rate of exoU gene was lower (18/70, 25.71%), which was similar to the previous report [22]. However, previous studies had shown that not all clinical isolates had the ability to produce ExoS [23], with the overall prevalence of exoS gene in clinical isolates being only approximately 70%. In addition, virulence gene detection rate had obvious difference from the different specimens. The production of ExoS can provide the advantages of PA isolates in respiratory tract colonization or persistent existence [23,24]. In the ICU, there were many cases of colonization of colonized bacteria, which might lead to a significant increase in the detection rate of exoS, which was also important in the mechanism of PA colonization and infection in the respiratory tract.
Interestingly, almost every isolate contained exoS without harboring exoU and vice versa. Besides, only 1 isolate carrying both genes while 4 isolates harboring none. Feltman and coworkers [22] reported these similar findings, indicating it was a nearly universal characteristic of PA isolates, which might owe to the gene altered under the corresponding environmental pressure or related to the enhancement of virulence.
Studies on PA in European and American populations had suggested was the exoU genotype is significantly associated with multidrug resistance and fluoroquinolones resistance compared with the exoS genotype [25,26]. However, there was no significant difference in the resistance between exoU+ genotype with exoS+ genotype in our study, which might be related to the different distribution of population and area. Nevertheless, comparing with the exoU+ genotype, exoS+ genotype strains carried a significantly higher rate of drug resistance, which was different from the previous funding reports. Previous fundings had shown that from a clinical point of view, detection of toxA and eoxS genes in PA clinical isolates might be more significant in drug resistance [11,27]. In our study, 27 strains of MDR (27/35, 77.14%) contained both toxA and eoxS genes at the same time. Compared with non-MDR (22/35, 62.86%), there was not any significant differences could be found in gene prevalence, which was different from other studies [27]. Therefore, we should consider that pathogenicity of PA was multifactorial.
Multi-factor analysis showed that the use of antimicrobials before separation, prolonged hospitalization and mechanical ventilation were independent risk factors for HAIs. At the same time, some studies suggested that the use time of antimicrobials, hospitalization time and tracheal intubation were the main risk factors of healthcare associated infection in PA [26]. These risk factors were interacted to each other. The results of this study showed that the use of antimicrobials before separation, prolonged hospitalization, and mechanical ventilation were risk factors for the occurrence of PA infection in the hospital. The prolonged hospitalization time, the use of antimicrobials, the screening of PA lacking routine ESBL production and the colonization of multidrug-resistant strains in the environment might be the reason of the high resistance rate of PA.
Inappropriate antimicrobials therapy was significantly associated with increased mortality, morbidity, and length of hospital stay [28]. Furthermore, inappropriate empirical antimicrobials therapy was independently associated with higher mortality in patient and inappropriate initial antimicrobials therapy had an adverse effect on survival in patients with gram-negative sepsis [29], however, our study did not find a clear association with mortality. But the results showed that inappropriate treatment significantly increased the mean length of hospital stay compared with those who started receiving appropriate treatment. We observed that 50% of the strains isolated in the ICUs were treated inappropriately. And in contrast to community infections, patients with healthcare associated infections receiving inappropriate treatment was significantly higher with a high increasing incidence of VAP. Multifactor analysis showed a significant correlation between VAP and mortality within 30 days, and VAP was an independent risk factor for increased mortality in 30 of the patients. The use of Mechanical Ventilation as a means of rescue in ICU had also greatly increased the use of ventilator, thus got a high risk of patients with VAP. Current literature suggests that when the infection was effectively controlled along with improved ventilation, the time to use the ventilator should be reduced as much as possible, and the use of non-invasive ventilation can reduce the occurrence of VAP [30].
It was pointed out that the impact of inappropriate empirical antimicrobials therapy depended on the site of the infection. For patients with high risk of infection, inappropriate empiric antimicrobials therapy was identified as an independent risk factor for death [31]. Although most of the specimens were sputum from lower respiratory tract, no mortality was found to be associated with inappropriate treatment. In our study, the most inappropriate empirical antimicrobials was meropenem, which was consistent with a report from Italy showed the highest rate of drug use of meropenem for inappropriate therapy [19]. We suggested that the use of carbapenems and aminoglycoside antimicrobials should be used with caution in the empirical treatment of antimicrobials, especially in the consideration of patients with HCAIs or MDRPA infections. However, no significant statistical significance was found in the use of different kinds of drugs between the inappropriate drug treatment group and the appropriate group which indicated that we still need to guide the application of clinical antimicrobials strictly according to the drug sensitivity report. The appropriateness of empirical treatment cannot be guaranteed. Based on the results of the study, we did not recommend the use of quinolones as a priority in empiric therapy in the study area, which can easily lead to inappropriate therapeutic use. Especially when infected with MDRPA, inappropriate empirical treatment may aggravate the change of drug resistance.
A lack of significant association between inappropriate initial antimicrobial therapy and the outcome of patients with a low-risk source of bacteremia may have been due to a high proportion of catheter removal or early intervention for decompression of biliary or urinary obstruction in the majority of patients. It suggested that nonmedical interventions such as decompression of obstruction or the removal of infection foci were also important aspects of the treatment of infection.
Conclusion
The resistance rates of most drugs in our research were higher than the 2016 China CHINET bacterial resistance monitoring results, except for the slightly lower resistance rate of imipenem and polymyxin B. The drug resistance in this area was severe, and there seems to be a long-term process of colonization or transmission leading to infections at the ICUs with an extensively drug resistant clone type. The resistance genes like OXA-1 IMP, ant(2”)-Ia, aac(6’)- Ib and IntI were correlated with MLVA gene cluster. It is speculated that in 2016, at the ICUs, the related resistance gene island (OXA- 1+IMP+ant(2’’)-Ia+aac(6’)-Ib) carried in IntI was the predominant pattern of drug resistance genes. In the clinical data analysis, multifactor analysis shows that the VAP was an independent risk factor for increasing the mortality rate of 30 days in patients. The use of antimicrobials, length of hospitalization and the use of mechanical ventilation before isolation were independent risk factors for HAIs.
Inappropriate antimicrobials therapy seemed to substantially influence the empiric therapy of PA infections; In particular, use of carbapenems was notably frequent. At the same time, our research showed that patients who usually hospitalized for more than 30 days often did not receive proper treatment. This indicated that clinicians must strictly comply with the drug sensitivity test results in the use of antimicrobials, and use carbapenems with caution in empirical treatment, especially for patients with PA infection who have been hospitalized more than 30 days in the study area.
References
- Keith Poole. Pseudomonas aeruginosa: Resistance to the Max. Frontiers in Microbiology. 2011; 2: 1-13.
- Ribeiro ÁCDS, Crozatti MTL, Silva AAD, Macedo RS, Machado AMDO, Silva ATDA. Pseudomonas aeruginosa in the ICU: prevalence, resistance profile, and antimicrobial consumption. Rev Soc Bras Med Tro. 2020; 53: e20180498.
- Raman G, Avendano EE, Chan J, Merchant S, Puzniak L. Risk factors for hospitalized patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrobial Resistance & Infection Control. 2018; 7: 79-93.
- Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, et al. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clinical Microbiology Reviews. 2019; 32: 19-31.
- Andrew Walkty PLHA, Zhanel GG. Antimicrobial susceptibility of 2906 Pseudomonas aeruginosa clinical isolates obtained from patients in Canadian hospitals over a period of 8 years_ Results of the Canadian Ward surveillance study (CANWARD), 2008-2015. Diagnostic Microbiology and Infectious Disease. 2017; 87: 60-63.
- Holbrook SYL, Garneau-Tsodikova S. Evaluation of Aminoglycoside and Carbapenem Resistance in a Collection of Drug-Resistant Pseudomonas aeruginosa Clinical Isolates. Microbial Drug Resistance. 2018; 24: 1020- 1030.
- Tawfik AF, Shibl AM, Aljohi MA, Altammami MA, Al-Agamy MH. Distribution of Ambler class A, B and D Β-lactamases among Pseudomonas aeruginosa isolates. Burns. 2012; 38: 855-860.
- Cai S, Yiqiang C, Dezhi S, Jinliang K, Yanbin W, Lu AH. Study on the resistance mechanism via outer membrane protein OprD2 and metal Β-lactamase expression in the cell wall of Pseudomonas aeruginosa. Exp Ther Med. 2016; 5: 2869-2872.
- Wong-Beringer A, Wiener-Kronish J, Lynch S, Flanagan J. Comparison of type III secretion system virulence among fluoroquinolone-susceptible and -resistant clinical isolates of Pseudomonas aeruginosa. Clin Microbiol Infec. 2008; 14: 330-336.
- Ullah W, Qasim M, Rahman H, Jie Y, Muhammad N. Beta-lactamaseproducing Pseudomonas aeruginosa: Phenotypic characteristics and molecular identification of virulence genes. J Chin Med Assoc. 2017; 80: 173- 177.
- Khosravi AD, Shafie F, Abbasi Montazeri E, Rostami S. The frequency of genes encoding exotoxin A and exoenzyme S in Pseudomonas aeruginosa strains isolated from burn patients. Burns. 2016; 42: 1116-1120.
- Liu M, Ma J, Jia W, Li W. Antimicrobial Resistance and Molecular Characterization of Gene Cassettes from Class 1 Integrons in Pseudomonas aeruginosa Strains. Microb Drug Resist. 2020; 26: 670-676.
- Folic MM, Djordjevic Z, Folic N, Radojevic MZ, Jankovic SM. Epidemiology and risk factors for healthcare-associated infections caused by Pseudomonas aeruginosa. Journal of chemotherapy (Florence). 2020; 2: 1-8.
- Rojas A, Palacios-Baena ZR, López-Cortés LE, Rodríguez-Baño J. Rates, predictors and mortality of community-onset bloodstream infections due to Pseudomonas aeruginosa: systematic review and meta-analysis. Clin Mocrobiol Infec. 2019; 25: 964-970.
- Helio S Sader MDHM. Pseudomonas aeruginosa Antimicrobial Susceptibility Results from Four Years (2012 to 2015) of the International Network for Optimal Resistance Monitoring Program in the United States. Antimicrob Agents Chemother. 2017; 3: 2216-2252.
- Levy Hara G, Kanj SS, Pagani L, Abbo L, Endimiani A, Wertheim HFL, et al. Ten key points for the appropriate use of antibiotics in hospitalised patients: a consensus from the Antimicrobial Stewardship and Resistance Working Groups of the International Society of Chemotherapy. Int J Antimicrobial Ag. 2016; 48: 239-246.
- Vu-Thien H, Corbineau G, Hormigos K, Fauroux B, Corvol H, Clement A, et al. Multiple-Locus Variable-Number Tandem-Repeat Analysis for Longitudinal Survey of Sources of Pseudomonas aeruginosa Infection in Cystic Fibrosis Patients. J Clin Microbiol. 2017; 45: 3175-3183.
- HU Fupin GYZD, Ping XYKM, NI Yuxing SJCY, GUO Sufang WLZF, SU Danhong WRFH, Wenen LYJY, et al. Institute Of Antibiotics, CHINET surveillance of bacterial resistance across China: report of the results in 2016. Chin J Infect Chemother. 2017; 17: 481-491.
- L Ruiz-Martínez. Class 1 integrons in environmental and clinical isolates of Pseudomonas aeruginosa. International Journal of Antimicrobial Agents. 2011; 38: 398-402.
- Ahmadian L, Haghshenas MR, Mirzaei B, Norouzi Bazgir Z, Goli HR. Distribution and Molecular Characterization of Resistance Gene Cassettes Containing Class 1 Integrons in Multi-Drug Resistant (MDR) Clinical Isolates of Pseudomonas aeruginosa. Infect Drug Resist. 2020; 13: 2773-2781.
- Goli HR, Nahaei MR, Rezaee MA, Hasani A, Kafil HS, Aghazadeh M, et al. Role of MexAB-OprM and MexXY-OprM efflux pumps and class 1 integrons in resistance to antibiotics in burn and Intensive Care Unit isolates of Pseudomonas aeruginosa. J Infect Public Heal. 2018; 11: 364-372.
- Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa, Microbiology. 2001; 147: 2659-2669.
- Hauser CMSA. Relative Contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to Virulence in the Lung. Infect Immun. 2004; 12: 6969-6977.
- Suzanne MJ, Fleiszig JPWH, Keith E, Mostov DKTS, Frank ADW. Pseudomonas aeruginosa-Mediated Cytotoxicity and Invasion Correlate with Distinct Genotypes at the Loci Encoding Exoenzyme S. Infect Immun. 1997; 65: 579-586.
- Melissa Agnello AW. Differentiation in Quinolone Resistance by Virulence Genotype in Pseudomonas aeruginosa. Plos One. 2012; 7: 42973-42980.
- Makaoui Maatallah JCAB. Population Structure of Pseudomonas aeruginosa from Five Mediterranean Countries: Evidence for Frequent Recombination and Epidemic Occurrence of CC235. Plos One. 2011; 10: 25617-25628.
- Ivan Mitov TSBM. Prevalence of virulence genes among Bulgarian nosocomial and systic fibrosis isolates of Pseudomonas aeruginosa. Braz J Microbiol. 2010; 41: 588-595.
- Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharm Out. 2014; 10: 441-451.
- Cheol-In Kang SKWB, Eui-Chong Kim MOAK. Bloodstream Infections Caused by Antibiotic-Resistant Gram-Negative Bacilli: Risk Factors for Mortality and Impact of Inappropriate Initial Antimicrobial Therapy on Outcome. Antimicrob Agents Ch. 2004; 2: 760-766.
- Kos VN, Déraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm RA, et al. The Resistome of Pseudomonas aeruginosa in Relationship to Phenotypic Susceptibility. Antimicrob Agents Ch. 2014; 59: 427-436.
- Joo EJ, Kang CI, Ha YE, Park SY, Kang SJ, Wi YM, et al. Impact of inappropriate empiric antimicrobial therapy on outcome in Pseudomonas aeruginosa bacteraemia: a stratified analysis according to sites of infection. Infection. 2011; 39: 309-318.