
Research Article
Austin Biochem. 2016; 1(1): 1002.
Identification of Bacterial Contamination in Fuel Ethanol Fermentation in Southeastern Mexico
Hernández-Heredia S¹, Carrera-Lechuga J², Montor-Antonio JJ¹, Ramírez-Torres A¹, Delfín- García CO², Meza-Villalvazo V², García-Arellano C³ and Moral SD¹*
¹Universidad del Papaloapan, Instituto de Biotecnología, México
²Complejo Agroindustrial de Santander, Carretera Federal Ciudad Alemán-Cosamaloapan, Mexico
³Universidad del Papaloapan, Colonia Ciudad Universitaria, México
*Corresponding author: Sandra Del Moral, Instituto de Biotecnología, Universidad del Papaloapan, Col. Parque Industrial, CP 68301, Tuxtepec, Oaxaca, México
Received: September 28, 2016; Accepted: November 10, 2016; Published: November 14, 2016
Abstract
Ethanol factories are susceptible to bacterial contamination, which decreases their productivity. The main studies about bacterial contamination are realized on distilleries that use corn or beet as raw material, but not molasses of cane sugar as raw. Acetobacter and lactic acid bacteria are the main bacterial contaminant in distilleries that use corn as raw. In this study, the contaminant bacteria in an ethanol factory which use molasses of cane sugar as raw were identified. Seven strains were isolated and belong to the genera Acetobacter, Enterocccus, Klebsiella and Cronobacter. All strains were able to metabolize glucose, mannitol, rhamnose, sucrose, melibiose and arabinose and were resistant to penicillin, but susceptible to enoxacin and netilmicin. This research identifies microorganisms that could be found in distilleries that use molasses as raw in tropical countries.
Keywords: Ethanol factory; Bacteria; Carbohydrates
Abbreviations
MRS: Man, Rogosa and Sharpe; EMB: Eosin Methylene Blue; WLD: Wallerste in Differential; PBS: Phosphate Buffered Saline; PCR: Polymerase Chain Reaction; ONPG: 2-Nitrophenyl-ß- D-Galactopyranoside, ADH: L-Arginine; LCD: L-lysine; ODC: L-ornithine, CIT: Trisodium Citrate; H2S: Sodium Thiosulphate; URE: Urea; TDA: L-tryptophane; VP: Sodium Pyruvate; GEL: Gelatin; GLU: D-Glucose; MAN:D-Mannitol; INO: Inositol; SOR: D-Sorbitol; RHA: L-Rhamnose; SAC: D-Sucrose; MEL: D-Melibiose; AMY: Amyddalin; ARA: L-arabinose; LAC: D-Lactose; MAL: D-Maltose; SAL: Salicin; XYL: D-Xylose; ESC: Esculin; GLY: Glycerol; CEL: D-Cellobiose; MNE: D-Mannose; MLZ: D-Melezitose; RAF: D-Raffinose; THE: D-Trehalose; AK: Amikacine; AM: Ampicillin; CF: Cephalothin; CRO: Ceftriaxone; CL: Chloramphenicol; DC: Dicloxacillin; ENX: Enoxacin; ER: Erythromycin; GE: Gentamicin; NET: Netilmicine; PE: Penicillin; SXT: Trimethoprim-Sulfamethoxazole; CFU: Colony- Forming Unit; S: Susceptible; R: Resistant; I: Intermediate; A10-CAD; B2-CAD; D-CAD; E2-CAD; 7-CAD; 8-CAD: isolated strains
Introduction
Ethanol can serve as an alternative biofuel. It is produced during the fermentation of easily of low cost substrates, such as: corn, sugar cane juice, molasses cane juice, molasses beet juice, cassava, potato, hemicellulose substrates (paper sheet, sawdust) [1]. USA and Brazil are the main ethanol producers at worldwide, corn and sugar cane molasses are used as raw materials, respectively [2].
Ethanol fermentation is carried out by yeast; cell viability of yeast is adversely affected by the acid organics produced by bacteria contaminant (i.e. acetic and lactic acid). Mexico as Latin America use sugar cane and molasses cane juice as biomass for ethanol production, due to its great abundance, easy culture and fermentation [3]. Distilleries in Mexico are located mainly in the southeast where the weather is tropical (40-45°C).
In distilleries, the media is not sterilized and only diluted molasses are used (near 22°Brix). The main contaminants in the fermentation tanks that use corn as raw material are Lactic Acid Bacteria (LAB) mainly Lactobacillus [4]. Although others LAB have been found such as: Leuconostoc, Bifidobacterium, Lactococcus, Pediococcus [5-10]. Acetobacter and Weisella strains have been found also in distilleries; Acetobacter utilize simple carbohydrates and ethanol as carbon source to produce acetic acid [11,12]. These genera affect the process because consume the carbon source, hence, the yield and productivity decrease [7,10,12,13,14]. Besides, organics acid produced by bacteria contaminant as acetic and lactic acid, adversely affect cell viability of yeast [4,15].
To reduce the microbiota contaminant in fermentation broth antimicrobial agents are used. Penicillin, tetracycline, monensin, virginiamycin, polymyxin B [16], hydrogen peroxide, potassium metabisulfite [17], tartaric acid [4] are the most common. Generally, the factory does not make a study to determine which antimicrobial agent could eliminate the contaminant microbiota. This general approach often results in the generation of antibiotic resistant bacteria, making the antibiotics less efficient in the reduction of contaminants. In this paper a microbial community representative of the fermentation and storage tanks of an ethanol factory was studied biochemically and molecularly in order to identifying which antibiotics could remove it. Therefore, this study is relevant for distilleries in tropical countries which produce ethanol via fermentation of molasses from cane juice.
Materials and Methods
Sample collection and bacteria isolation
Samples were collected from fermentation broth every 4 hrs, since pre-fermentation step until fermentation end in ethanol factory. At the molasses storage tank, the samples (500 g) were collected from different sites and homogenized. After, samples were diluted in decimal dilutions in PBS buffer and 100 μL were plated by duplicate on several selective agar mediums: MRS-Itraconazole (Fluka, Germany), WLD (Fluka, Germany) and EMB (Dibico, Mexico). Petri dishes were transported to Molecular Biology Laboratory of the Universidad del Papaloapan and were incubated at 37°C for 48 hrs in CO2 incubator (anaerobic conditions). Colonies with morphologic differences were selected and isolated.
Molecular and biochemical identification
Colonies were characterized at the morphologic level, with biochemical (API 20A and 20E, Biomerieux, France) and molecular tests, Gram’s Method (Hycel, México). To molecular characterization, genomic DNA was extracted using the Ultra Clean microbial DNA isolation kit (MoBIO, USA). The 16S rDNA was amplified with the primers fD1 (CCGAATTCGTCGACAACAGAGTTTGATCCTGGCTCAG) and rD1 (CCCGGGATCCAAGCTTAAGGAGGTGATCCAGCC) by [18]. PCR products were purified, sequenced (Macrogen, Korea) and analyzed through the maximum likelihood method, with Nei Tamura model, and 1500 bootstrap replicates [19].
Antibiotic sensibility testing
Sensitivity testing was performed to the CAT manual (BIORAD), which contains: AK (30 μg), AM (10 μg), CF (30 μg), CRO (30 μg), CL (30 μg), DC (1 μg), ENX (10 μg), ER (15 μg), GE (10 μg), NET (30 μg), 10 U PE and SXT (25 μg). Strains were grown according to supplier recommendations.
Results and Discussion
Microbiota description
Bacteria, yeast and molds were obtained in all growth media, but only bacteria were characterized. Bacteria from the fermentation tank grew mainly on WLD medium, some strains produced turn on agar color, from blue to yellow, due to the acidification of the medium. Yellow area around colony gives an indication of the amount of lactic or acetic acid produced by the colony. On EMB medium grew native yeasts and LAB from molasses on MRS-Itraconazole medium were isolated.
Figure 1 summarizes the bacterial behavior during pre (0-3 hrs) and fermentation steps (4-28 hrs). Seven strains with different macro and microscopic characteristics were isolated; the largest microbial count was detected at the end of the prefermentation step and in the beginning of fermentation step. At the begin of fermentation step (0 hrs), fermentation broth has around 22° Brix and the fermentation end (24 hrs) has 10° Brix. B2-CAD showed the largest bacterial count for the bioprocess and its concentration was increased in both steps. Unlike A-CAD, B-CAD, C-CAD and E-CAD only were observed during prefermentation step. Instead, D-CAD was observed at the end of the prefermentation and in half of the fermentation step. On the other hand, E2-CAD was only observed at the end of the fermentation. Therefore, B2-CAD can grow as the fermentative yeast of the bioprocess, instead, E2-CAD grow better at fermentation end, when sugar concentration is low. The total microbial concentration was around 3000 CFU/ml, lower than the quantity reported of microbial contaminant from sugar cane juice, where the microbial concentration was approximate 105-108 CFU/ml [4,20]. The low native bacterial concentration observed in this work is an advantage to produce ethanol from molasses and avoid economic lost by contaminant bacteria. B-CAD and C-CAD strains are natives yeast and E-CAD strain is a mold; these isolated were not characterized. A-CAD and E2-CAD were grouped in the clade of Enterococcus, B2-CAD was added in Acetobacter cluster and D-CAD in Cronobacter clade (Figure 2, Table 1). From molasses yeast (9-CAD), bacteria (7-CAD and 8-CAD) and mold (10-CAD) were detected. Bacteria were isolated in MRS-Itraconazole medium and the yeasts and mold of EMB medium. Microbial concentration for each isolated oscillated between 60-560 CFU/ml (Table 2). The bacterial concentration was lowest than fermentation tank, probably by the high concentration of carbohydrates in molasses (around 80° Brix). This shows that the contamination is largest during the fermentation step and the distilleries must focus in this phase. 7-CAD was clustered with the Klebsiella genus and the 8-CAD was not associated with a particular clade (Figure 2, Table 1). Even though Klebsiella has not been found as contaminant typical in distillery factories, it has been found frequently associated with sugar cane as growth factor [21]. Therefore, it could come from there.
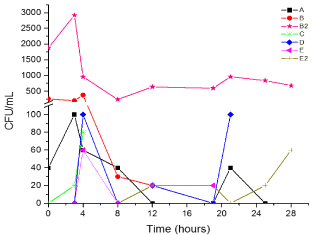
Figure 1: Kinetics of growth of bacteria, yeast and fungi during fermentation
process.
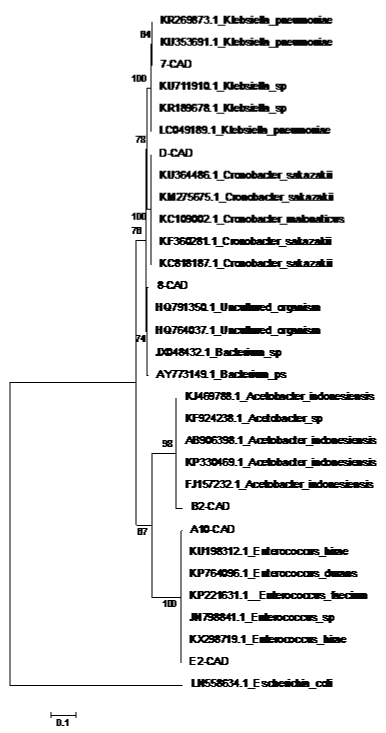
Figure 2: Phylogenetic tree of microbiota isolated from fuel factory.
Isolated strain
Best hit description/ Access number
Cover (%)
Identity (%)
A10-CAD
Enterococcus hirae (KU198312.1)
100
98
B2-CAD
Acetobacter indonesiensis (FJ157232.1)
100
98
D-CAD
Cronobacter (KU364486.1)
100
99
E2-CAD
Enterococcusspp. (KR858847.1)
100
99
7-CAD
Klebsiella (KR269873.1)
100
99
8-CAD
Unculture microorganism
100
96
Table 1: Information general of the local alignment of the sequences obtained of the isolated.
Isolated
Microorganism (Gram)
CFU/ml
7-CAD
Bacterium (-)
520
8-CAD
Bacterium (-)
60
9-CAD
Yeast
560
10-CAD
Fungi
ND
ND: Not Determinated
Table 2: Microbiota isolated from molasses sugar cane.
Biochemical characterization of the bacteria isolated
API 20A and API 20E galleries were used in order to determinate what type of carbohydrate can metabolize the isolated bacteria and if they are presents also in molasses (Table 3). Strains can metabolize several kinds of carbohydrates: monosaccharides (Glu, Lac, Mne), disaccharides (α and β glycosidic linkage) (Sac, Cel), trisaccharides (Raf) and polyols of three and six carbons (Man, Ino, Sor), many of them are present in molasses, as sucrose (30-40%), glucose (4-9%), fructose (5-12%) and dextran (<10%) [22,23]. Thus, contaminant bacteria consume a part of the carbohydrates present in molasses and produce economic losses to the distillery.
Biochemical test
A10-CAD
B2-CAD
D-CAD
E2-CAD
7-CAD
8-CAD
ONPG
-
+
-
+
-
-
ADH
+
+
+
+
+
+
LDC
-
-
+
+
+
+
ODC
+
-
+
-
-
+
CIT
-
-
-
-
-
-
H2S
-
-
-
-
-
-
URE
-
-
+
+
+
-
TDA
-
-
-
-
-
-
IND
-
-
-
-
-
-
VP
-
-
-
-
-
-
GEL
-
-
-
+
-
+
GLU
+
+
+
+
+
+
MAN
+
+
+
+
+
+
INO
+
+
+
+
+
-
SOR
+
+
+
-
+
+
RHA
+
+
+
+
+
+
SAC
+
+
+
+
+
+
MEL
+
+
+
+
+
+
AMY
+
+
+
+
+
+
ARA
+
+
+
+
+
+
LAC
+
+
+
-
+
-
MAL
+
+
+
-
+
-
SAL
-
-
-
-
+
-
XYL
-
-
-
-
-
-
ESC
+
-
+
-
+
+
GLY
-
-
-
-
+
+
CEL
+
-
+
-
+
-
MNE
+
+
-
-
+
+
MLZ
-
-
-
-
-
-
RAF
+
-
+
-
-
-
THE
+
+
-
-
+
+
Table 3: Biochemical behaviour of the bacteria isolated from fermentation broth and molasses.
Acetobacter has been isolated from sugar cane, it metabolizes glucose via the hexose monophosphate pathway and tricarboxylic acid cycle and ethanol to produce organic acids [21,24]. The simple nutritional requirements help to make them almost ubiquitous in some breweries and ethanol, thus becoming one of the most frequent causes of acidity and potential yeast cell death [24]. Acetobacter spp. B2-CADuse the carbohydrates of molasses cane sugar and the ethanol of fermentation tank to produce organic acids.
Lactobacillus is the genus most commonly isolated from ethanol factories which use sugar cane juice or corn [4], however, in this research none strain of this genus was obtained. Enterococcus few times has been found in distilleries, in Poland distilleries(corn as raw) only the 30% of all isolates were Enterococcus and distilleries of cachaça in Brazil only was detected in a fermentation vat (10% of all isolates) [25,26]. In this case, two Enterococcus strains were isolated, A-CAD and E2-CAD (25% of all isolated). Enterococcus fecalis produces lactic acid from molasses [27]. Enterococcus spp. E2-CAD metabolizes various carbohydrates (Table 3) and produces some organic acid, because during its growth on WLD medium turns the color of the medium, from blue to yellow so, acid lactic production could be possible during the fermentation (data not show).
Cronobacter spp. D-CAD metabolizes several carbohydrates (Table 3) and generated a turn into WLD medium, showing its capacity of metabolize molasses to produce organic acids.
Antibiotics susceptibility
Acetobacter spp. B2-CAD and Enterococcus E2-CAD did not grow on Müller-Hinton broth, thus it was not possible to evaluate the antibiotics susceptibility. The strains tested showed resistance to CF, ER, DC and PE and susceptibility to AK, CRO, CL, ENX, GE and NET (Table 4). It is possible to use some antibiotic to control the contaminant bacteria in minimum inhibitory concentration. Some studies have showed that the most common antibiotics used to control of microbiota on ethanol factories are penicillin, tetracycline, monensin and virginiamycin [28-30] but, all bacteria analyzed in this work were resistant to penicillin, so that this antibiotic should not be used to control the bacterial contamination in this factory. Therefore, it is necessary to do a previous test before use any antibiotic. Another alternative is the synergic combination of diverse chemical substances as benzalkonium chloride and 3, 4, 4-trichlorocarbanilide [15].
Strain
AK
AM
CF
CRO
CL
DC
ENX
E
GE
NET
PE
SXT
Enterococcus sp. A10-CAD
S
R
R
I
I
R
S
R
S
S
R
S
Cronobacter sp. D-CAD
S
I
R
S
S
R
S
R
S
S
R
S
Klebsiella sp. 7-CAD
S
R
I
S
S
R
S
R
S
S
R
S
8-CAD
S
R
R
S
S
R
S
I
I
S
R
R
Table 4: Strains sensibility to different antibiotics.
Conclusion
Acetobacter spp. B2-CAD was the main contaminant strain during the fermentation, although others genera were detected: Enterococcus, Klebsiella and Cronobacter. To control this microbiota through antibiotics is necessary to perform a susceptibility test, to avoid the indiscriminate use of antibiotics. In this study, all strains were resistant to penicillin. To decrease the microbiota isolated in the process AK, CRO, CL, ENX, GE and NET could be used. It is suggested some sanitation method of equipment to diminished contaminant bacterial on fermentation broth. Nowadays, chemical methods of sanitization and lower pH in fermentation broth are being tested to avoid antibiotics use in the ethanol factory evaluated.
Acknowledgement
To ethanol factory for the facilities provided for the realization of this work. Alejandro Alpuche and Elena Vinay for their valuable comments in drafting the manuscript.
References
- Zamora-Hernández T, Prado-Fuentes A, Capataz-Tafur J, Barrera-Figueroa B, Peña-Castro J. Demostraciones prácticas de los retos y oportunidades de la producción de bioetanol de primera y segunda generación a partir de cultivos tropicales. Educación Química. 2014; 25: 122-127.
- Goldemberg J. Ethanol for a Sustainable Energy Future. Science. 2007; 315: 808-810.
- Ortiz-Zamora O, Cortés-García R, Ramírez-Lepe M, Gómez-Rodríguez J, Aguilar-Uscanga M. isolation and selection of ethanol-resistant and osmotolerant yeasts from regional agricultural sources in Mexico. Journal of food process engineering. 2009; 32: 775-786.
- Skinner K, Leathers T. Bacterial contaminants of fuel ethanol production. Journal Industrial Microbiology Biotechnology. 2004; 31: 401-408.
- Rainbow C. Spoilage organisms in breweries. Process Biochemistry. 1971; 6: 15-17.
- JS, Briggs DE, Stevens R, Young TW. Malting and Brewing Science: Hopped ward and beer. 2nd Edition. Chapman and Hall, London. 1982; 741-775.
- Gold R, Meagher M, Hutkins R, Conway T. Ethanol tolerance and carbohydrate metabolism in lactobacilli. Journal of Industrial Microbiology. 1992; 10: 45-54.
- Connolly C. Bacterial contaminants and their effects on alcohol production, in: Jacques K, Lyons T. P, Kelsall D.R, editors, the alcohol texboook, 3ra ed. Nottingham University Press, Nottingham. 1999; 317-334.
- Ingledew WM. Alcohol production by saccharomyces cerevisiae: a yeast primer, in: Jacques K, Lyons TP, Kelsall DR, editors, the alcohol texboook, 3ra ed. Nottingham University Press, Nottingham. 1999; 49-89.
- G-Alegría E, López I, Ruiz J, Saenz J, Fernández E, Zarazaga M, et al. High tolerance of wild Lactobacillus plantarum and Oenococcus oeni strains to lyophilisation and stress environmental conditions of acid pH and ethanol. FEMS Microbiology Letters. 2004; 230: 53-61.
- Lushia W, Heist P. Antibiotic resistant bacteria in fuel ethanol fermentations. Ethanol Producer Magazine. May 2005.
- Heist P. Identifying, Controlling the most common Microbial contaminants ethanol producer magazine. May 2009.
- Oliva-Neto P, F Yokoya. Evaluation of bacterial contamination in a fed-batch alcoholic fermentation process. Journal of Microbiology and Biotechnology. 1994; 10: 697-699.
- Thomas K, Hynes S, Ingledew W. Effect of lactobacilli on yeast growth, viability and batch and semi-continuous alcoholic fermentation of corn mash. Journal Applied Microbiology. 2001; 90: 819-828.
- Oliva-Neto P, AL Fabiola, C da Silva Ketrin, F da silva Douglas, FA Ana, Catarina dos Santos. Chemical Inhibition of the Contaminant Lactobacillus fermentum from Distilleries Producing Fuel Bioethanol. 2014; 57: 441-447.
- Red-Bezares B, Sáenz Y, Poeta P, Zarazaga M, Ruiz-Larrea F, Torres C. Assessment of antibiotic susceptibility within lactic acid bacteria strains isolated from wine. International Journal of Food Microbiology. 2006; 111: 234-240.
- Chang IS, Byung HK, Pyong KS. Use of Sul?te and Hydrogen Peroxide to Control Bacterial Contamination in Ethanol Fermentation. Applied and Environmental Microbiology. 1997; 63: 1-6.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S Ribosomal DNA Amplification for Phylogenetic Study. Journal of Bacteriology. 1991; 173: 697-703.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution. 2013; 30: 2725-2729.
- Lucena B, dos Santos B, Moreira J, Moreira A, Nunes A, Azevedo V, et al. Diversity of lactic acid bacteria of the bioethanol process. BMC Microbiology. 2010; 10: 298.
- Tapia-Hernández A, Bustillos-Cristales M, Jiménez-Salgado T, Caballero-Mellado J, Fuentes-Ramírez L. Natural Endophytic Occurrence of Acetobacter diazotrophicus in Pineapple Plants. Microbial Ecology. 2000; 39: 49-55.
- Huber Olbrich. The molasses. Berlin. 1963. p. 130. Publish by Biotechnology-Kempe GmbH. 2006.
- Meade PG. 1986. Manual del Azúcar de Caña. Montaner y Simón S.A Barcelona. p: 305-325.
- Muthaiyan A, Limayem A, Ricke S. Antimicrobial strategies for limiting bacterial contaminants in fuel bioethanol fermentations. Progress in Energy and Combustion Science. 2011; 37: 351-370.
- Boda M, Leja K. The microbiologial situation of distilleries in the Wielkopolsa Rgion in Poland. Polish Journal of Environmental Studies. 2010; 19: 901-906
- Gomes FCO, Silva CLC, Vianna CR, Lacerda ICA, Borelli BM, Nunes AC, et al. Identification of lactic acid bacteria associated with traditional cachaça fermentations. Brazilian Journal of Microbiology. 2010; 41: 486-492
- Wee Y, Kim J, Yun J, Ryu H. Utilization of sugar molasses for economical l (+)-lactic acid production by batch fermentation of Enterococcus faecalis. Enzyme and Microbial Technology. 2004; 35: 568-573.
- Aquarone E. Penicillin and tetracycline as contamination control agents in alcoholic fermentation of sugar cane molasses. Applied Microbiology. 1960; 8: 263-268
- Hynes SH, Kjarsgaard DM, Thomas KC, Ingledew WM. Use of virginiamycin to control the growth of lactic acid bacteria during alcohol fermentation. Journal Industrial Microbiology. 1997; 18: 284-291.
- Stroppa CT, Andrietta MGS, Andrietta SR, Steckelberg C, Serra GE. Use of penicillin and monensin to control bacterial contamination of Brazilian alcohol fermentations. International Sugar Journal. 2000; 102: 78-82.