
Short Communication
Austin Biochem. 2018; 3(1): 1016.
New Anti-Microbial Molecules for Use in Protecting Food Sources from E.coli Bacteria
Butler B and Darsey JA*
Department of Chemistry, University of Arkansas at Little Rock, USA
*Corresponding author: Darsey JA, Department of Chemistry, Center for Molecular Design and Development and University of Arkansas at Little Rock, USA
Received: May 25, 2018; Accepted: June 07, 2018; Published: June 14, 2018
Abstract
The purpose of this research project is to discover new, naturally occurring, molecules that can be used to inhibit the growth of different bacterial pathogens on food. Specifically, this project focuses on the inhibition of E. coli (Escherichia coli) growth on food, the computational modeling of the structure of environmentally safe drug molecules that can be used to address bacterial growth, and lead to longer shelf lives of foods. The larger impact of the inhibition of bacterial growth on food would be an estimated ten percent increase in the amount of food globally every day that bacterial growth is affected. In cooperation with the Safe Foods Corporation’s existing compounds, which are used to process one hundred and ten million pounds of food world-wide from day to day, this research project should have a significant impact on both local and global food supplies and feed an additional ten million people.
Keywords: E. Coli; Bacterial growth; Safe foods; LogIC50; Molecules
Introduction
Escherichia coli, or E. coli is the most prevalent and well-known organism in the gram-negative bacteria, proteobacteria, phylum (Figure 1). E. coli is one of the most-studied “free -living” microorganisms, in fact, more than seven hundred serotypes of E. coli have been identified [1]. Some strains of E. coli are not harmful and live within human gastro-intestinal systems promoting good digestive health [1,2]. Some strains are used as markers for water contamination [3]. Some strains pose a large threat due to their side-effects. These dangerous pathogenic strains of E. coli can cause toxin producing infections with many different symptoms like severe stomach cramps, bloody diarrhea, and vomiting [3]. Some strains of E. coli can cause urinary tract infections, pneumonia, and other illnesses [3]. Those infected with E. coli usually start to present symptoms three to four days after they eat contaminated foods [3]. Because E. coli can contaminate a large variety of foods as well as water it is always possible for it to cause outbreaks. To handle an outbreak the Food and Drug Administration (FDA) follows a specific procedure. First, they collect pathogen samples from sick people, food itself, and from locations where food is handled. Next, they identify Pathogens through Whole Genome Sequencing to figure out the strain of the pathogen. Then, they compare the pathogen samples to the sequenced pathogen to see how closely they match, and next they act to prevent more people from getting sick [4]. The best way to deal with E. coli is to prevent it from contaminating foods in the first place, and that is what the Safe Foods Corporation seeks to do. Safe Foods develops antimicrobials and equipment options to allow their clients to reduce the risk of food-borne pathogen growth in their production/processing plants. This project, in cooperation with Safe Foods [5], seeks to use computational modeling of different anti-microbial molecules that are shown to inhibit E. coli activity, as well as quantum-mechanical calculations to expediate the process of effective natural antimicrobial discovery and an overall increase in the shelf-life of food. We will use the results of the calculations such as the total energy, dipole moment, and molecular orbital gaps in relation to one another and to IC50 values (values that represent the concentration of an agent to inhibit fifty-percent of a incoming target pathogen’s activity) to gain insight into the effectiveness of each molecule against E. coli based on the correlation of the results in scatter plots. The correlation will be observed using the R² values, good correlation is anything above 0.8, but ideally, we would like to see correlation of 0.9 or higher.
Figure 1: Digital Recreation of Escherichia coli.
Image Source: All You Need to Know About E. Coli. Food Poison Journal.
2015.
Methods
The project began with doing a bit of research to identify molecules that had already been tested against E. coli, as well the IC50 values that are associated with them. We found different research articles that had what we were looking for, but one article was very promising because it was a study that involved twenty-two different molecules [6]. The study sought to observe the E. coli binding site selectivity of different drug molecules, and it also listed the IC50 values for each of these [6]. Using Gaussian 09, a computational molecular modeling program, 3D models of each molecule were made (Figure 2). A chemical compound database called PubChem provided premade structures which we downloaded, opened in Gaussian, checked for accuracy, and adjusted where necessary [7]. We ran an optimization calculation on each molecule in Gaussian that provided total energy, dipole moment, and a molecular orbital spread. Using the molecular orbital spread different energy band gaps were calculated including the HOMO (Highest Occupied Molecular Orbital) LUMO (Lowest Unoccupied Molecular Orbital) Gap and the HUBMO (Highest Unoccupied Bonding Molecular Orbital) LUABMO (Lowest Unoccupied Anti-Bonding Molecular Orbital) Gap.
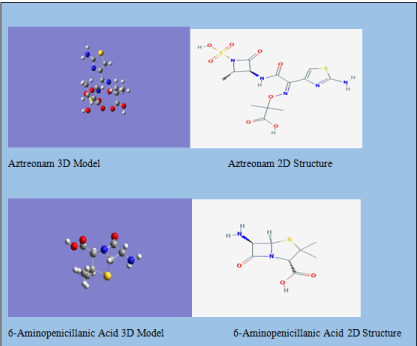
Figure 2: Two of the 3D models of the molecules we ran calculations on and
the 2D structures they are based on.
Results
A compilation of all the data, both of which was calculated in Gaussian and the IC50 values, was made into an Excel spreadsheet, and plotted into scatter plots. Each scatter plot compared the different data types, like total energy and dipole moment, to one another, as well as to different functions on the IC50 values, like log and inverse log. Some example plots are HOMO-LUMO Gap vs IC50 MIC, Dipole Moment vs HOMO-LUMO gap, and Inverse IC50 vs Total Energy. In total there were over sixty plots made, and some of these plots can be seen in figures three and four. The plots each included a trend line and a y equals equation that generated an R² value that represented the correlation of the data to each other. There was a high frequency of outliers occurrence in the plots, so outliers were removed from the plots until the R² value improved. Outliers were determined by value within the table, values like “ND” were excluded first, and then values were removed until a better correlation was achieved. The improvement of correlation without outliers can be observed in the plots in (Figure 3). Correlation was determined to be strong if it was above .9, decent if it was above .6, and to be weak if it was below .3. As seen in figure four, the two gaps, HOMO-LUMO and HUBMOLUBMO, have a strong correlation to one another, but the HUBMOLUBMO gap and the LogIC50 value are not that strongly correlated even with the outliers removed. The same can be said about the Total Energy and the Inverse LogIC50 PBP7. Due to the high frequency of outliers a lot of the plots showed very weak correlation and could not be improved without a lack of data point remaining. Only a few plots actually demonstrated good correlation without the removal of outliers, and even fewer were still usable when enough outliers were removed to gain a decent correlation.
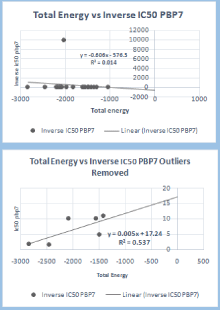
Figure 3: IC50 values were obtained from another study that was already
done [3], and the total energies of the molecules were obtained from
Gaussian 09. Energy Gaps were calculated using the molecular orbital
spread generated by Gaussian and subtraction of the smallest number from
largest number. PBP7 [Penicillin Binding Protein [7].
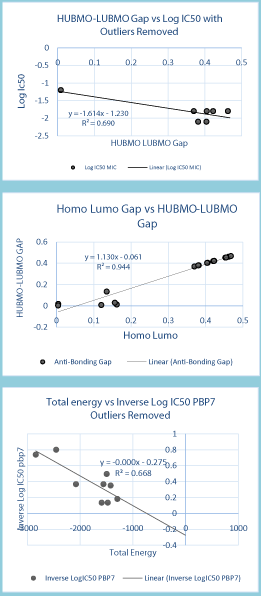
Figure 4: IC50 values were obtained from another study that was already
done [3], and the total energies of the molecules were obtained from
Gaussian 09. Energy Gaps were calculated using the molecular orbital
spread generated by Gaussian and subtraction of the smallest number from
largest number. PBP7 [Penicillin Binding Protein [7].
Discussion
We looked over each of the plots to see what meanings we could garner, and regarding the strongly correlated plots we made a few conclusions. First, from the strong correlation of the orbital gaps we can conclude that gap choice may be insignificant when doing a study like this one. Previously, the HOMO-LUMO gap has been highly regarded as a very important gap, but with the strong correlation between each gap, it may not be the only usable gap. Secondly, with a decent correlation between the HUBMO-LUBMO gap vs the LogIC50 we concluded that the larger this gap is the more effective the molecule might be at inhibiting E. coli activity. For the Total Energy vs the Inverse LogIC50 we concluded that the higher the total energy the lower the effectiveness, or potency, of the molecule. Regarding the other plots, most of the other plots were either too lacking in data points after outlier removal, or they demonstrated a correlation that was too weak to include. A better reference study may have been helpful in finding the IC50 values, and with different values better correlations may have been gained between each of the data types and the functions of those IC50’s. With research of this type (computational molecular modeling) it is important to have a good reference to create new molecules, and this project may prove to be useful in the development of procedures in the future.
Future Work/Studies
Using these results, we will program an Artificial Intelligence program to make predictions based on submitted molecules. Then, using those predictions we can find the most effective molecules to test experimentally. Traditionally anti-microbial compounds must go through lab trials that involve a lengthy process of testing that may leave only a few compounds left. The development of an Artificial Intelligence program that can accurately predict the effectiveness of untested compounds will allow for much faster discovery of antimicrobial compounds that can then be tested in actual lab trials. The AI could act as a screening measure to run the tests on hundreds of compounds at once computationally, and then only those molecules that showed promise would actually be tested in the real-life trials.
Acknowledgment
The authors would like to thank Venkatesh Karrepu, a graduate student, for assisting in running the Gaussian 09 program, as well as the Center for Molecular Design and Development for providing equipment and facilities. Special thanks to Dr. Janet Lanza, Dr. Jim Winter, and Dr. Jerry Darsey for providing research opportunities to students at the University of Arkansas at Little Rock.
References
- Bruce C. “All You Need to Know About E. Coli.” Food Poison Journal. 2015.
- How ‘Good’ Bacteria Make E. Coli Worse. Futurity. 2015.
- E. coli (Escherichia Coli). Centers for Disease Control and Prevention, Centers for Disease Control and Prevention. 2018.
- Center for Food Safety and Applied Nutrition. “Outbreaks: Investigation, Response & Evaluation.” US Food and Drug Administration Home Page, Center for Biologics Evaluation and Research.
- About Us. Safe foods.
- Ozden K, Carlson EE. “Pro?ling of β-Lactam Selectivity for Penicillin-Binding Proteins in Escherichia Coli Strain DC2.” Pro?ling of β-Lactam Selectivity for Penicillin-Binding Proteins in Escherichia Coli Strain DC2. 2015; 59: 2785- 2790.
- The PubChem Project. National Center for Biotechnology Information. PubChem Compound Database, U.S. National Library of Medicine.